Abstract
Why generalist and specialist species coexist in nature is a question that has interested evolutionary biologists for a long time. While the coexistence of specialists and generalists exploiting resources on a single ecological dimension has been theoretically and empirically explored, biological systems with multiple resource dimensions (e.g. trophic, ecological) are less well understood. Yet, such systems may provide an alternative to the classical theory of stable evolutionary coexistence of generalist and specialist species on a single resource dimension. We explore such systems and the potential trade-offs between different resource dimensions in clownfishes. All species of this iconic clade are obligate mutualists with sea anemones yet show interspecific variation in anemone host specificity. Moreover, clownfishes developed variable environmental specialization across their distribution. In this study, we test for the existence of a relationship between host-specificity (number of anemones associated with a clownfish species) and environmental-specificity (expressed as the size of the ecological niche breadth across climatic gradients). We find a negative correlation between host range and environmental specificities in temperature, salinity and pH, probably indicating a trade-off between both types of specialization forcing species to specialize only in a single direction. Trade-offs in a multi-dimensional resource space could be a novel way of explaining the coexistence of generalist and specialists.
Keywords: mutualism, anemonefish, anemone, trade-off, intraspecific variation
1. Introduction
In nature, species vary in their ability to exploit resources. By definition, on any given resource axis, the distribution of the absolute fitness of specialist species is narrow, while generalists have an equal fitness across the axis [1]. It has classically been proposed that ‘the jack of all trades is the master of none’ [2], which suggests that generalists bear a cost that will reduce their fitness relative to a specialist using the same resource. The availability of two distinct resources in a single ecological dimension, associated with a trade-off in the species ability to exploit these resources, can lead to three evolutionary stable outcomes. The initial species might evolve into: (i) two specialist species, (ii) a single generalist species, and (iii) into a generalist on two resources and a specialist on a single resource [3–6]. Ecological models of food webs (two food sources) predict that the last outcome alone can allow for the coexistence of specialist and generalist species, and this system will reach a stable equilibrium state when both food resources are equally abundant through time [7].
More complex scenarios of generalist–specialist coexistence have been proposed through recent theoretical work [7–9]. For example, stable coexistence of two specialists and a generalist in a two resources system is possible over evolutionary timescale in the presence of environmental variability and different adaptive foraging behaviour [8,9]. In line with the theory, empirical results show that spatial and temporal heterogeneity of the food source enhances generalist and specialist coexistence in two species of Hawaiian stream gobies [10]. Furthermore, formation of cryptic nutritional niches in seven grasshopper species (Melanoplus spp.) with broadly overlapping diets explains the coexistence of generalist herbivores in nature [11]. While this suggests that multiple generalist–specialist coexistence patterns can emerge in heterogeneous systems [9], the evolutionary processes involved in specialization are inevitably more complex when two or more resource dimensions influence the coexistence of species [12].
In multi-dimensional systems, trade-offs occur between resource axes. This can lead to differences in the relative rate of evolutionary response of generalists and specialists [13]. For example, trophic specialist damselfishes (Pomacentridae) maintain in their territories turfs of mutualistic algae on which they feed. Because the fish can only live where the algae can grow, this specialized behaviour is a constraint on the rate of environmental niche evolution that is thus slower in specialized herbivorous damselfishes when compared with more generalized planktivorous relatives [14]. Beside its effects on the rate of evolution, specialization on a specific resource of the system can also influence a species’ ability to specialize in another ecological dimension. For example, aphid parasitoids that are habitat specialists react similarly to the different host species present in the same habitat, whereas habitat generalist species exhibit clear preferences during host selection [15]. Yet, this rare example of specialization dynamics across multiple ecological axes involves only three closely related species without taking into account the evolutionary history of the complete clades. A question that is still standing is whether such dynamics can occur and be persistent over longer evolutionary timescales. This has not, to the best of our knowledge, been investigated.
The clownfishes are an excellent system to examine in detail the evolution of such multi-dimensional specialization over long evolutionary timescale. The 28 currently recognized species [16] are obligate mutualists with sea anemones (Anthozoa; Actinaria) and occur throughout the tropical Indian and Pacific Oceans. The ability of clownfishes to be unharmed by the otherwise stinging tentacles of the sea anemone probably triggered the evolutionary radiation of clownfishes [17]. Host specificity varies in clownfishes from interactions with a single host to potentially all the 10 sea anemone species that host clownfishes [16]. The distributions of clownfish and anemone species span various environmental conditions, allowing the evolution of differential level of environmental specialization associated with these environments. Moreover, the geographical range of anemone species is not a constraint on mutualistic interactions, as eight out of the 10 anemone species are ubiquitous regarding clownfish distributions [18]. However, each anemone species differs in its ecological preferences, such as substrate type, depth or exposure to oceanic currents [16,19], which allows clownfish species to coexist locally as they segregate across hosts and habitats [17,20].
In this study, we test whether a relationship between host-specificity (the number of anemones associated with a clownfish species) and environmental-specificity (expressed as the size of the environmental niche breadth) exists. A positive correlation between environmental and host specificities would suggest little to no trade-off between host and environmental specialization. Increasing environmental niche variation would thus allow species to encounter more mutualistic partners. By contrast, a negative correlation would probably indicate a trade-off between both types of specialization forcing clownfish species to specialize along one of the two resource axes. Host specialist clownfishes would therefore have a wide environmental niche breadth, whereas host generalists would have more narrow suitable environmental conditions. We use a near-complete species-level phylogeny of clownfishes based on seven nuclear markers to address this hypothesis. We estimate the environmental niche of clownfishes with detailed occurrence records and apply recent comparative modelling techniques to test whether the evolution of environmental niche breadth differs between clownfishes that are host specialist and host generalist.
2. Material and methods
(a). Phylogenetic reconstruction
We take advantage of an existing phylogeny of the clownfishes [21]. The dataset includes samples of 41 individuals representing 27 species of clownfish. Five species of Pomacentridae are used as outgroups. Seven nuclear markers (BMP4, Gylt, Hox6a, RAG1, Svep, S7 and Zic1) were sequenced using standard protocols ([21]; for accession numbers, see the electronic supplementary material). We built a consensus for each forward and reverse sequence and trimmed bases with 5% or more error probability. We aligned each marker independently with MAFFT [22] using default settings and concatenated all final alignments in a super-matrix of 6679 nucleotides in length. The phylogeny was reconstructed with MrBayes v. 3.2.2 [23]. The parameters of the nucleotide substitution process were sampled across the nucleotide model space during the Bayesian Markov chain Monte Carlo (MCMC) [24], and, after assessing the fit of all the molecular clock models available in MrBayes with Bayes Factors (BF) [25], a strict clock was applied during the phylogenetic inference. To facilitate the computations of further analyses and because absolute dates are not important in this study, the consensus topology of the inferred phylogenies was scaled to a height of one. We pruned the final tree to keep only one individual for each species. The phylogenies and alignment are deposited in TreeBase (www.treebase.org).
(b). Environmental niche estimation
We estimated the environmental niche of clownfishes with a database of global marine environmental layer at a resolution of 5 arcmin (approx. 9.2 km). This dataset includes 23 geophysical, biotic and climatic variables [26] and has been used in several studies to describe the environmental niche of marine fishes [27,28]. We retrieved clownfishes occurrence records from the Ocean Biogeographic Information System (www.iobis.org, accessed 26 June 2013). From the initial 1609 records, we removed duplicates (multiple occurrences of one species located in a single pixel of the environmental layers) and the species Amphiprion pacificus because only one occurrence was available, resulting in a total of 969 unique records (per species: mean = 35.9, min = 2, max = 166). After extracting the environmental variable values for each occurrence, we ran a principal component analysis (PCA) in R [29] and kept the most meaningful axes of the environmental space for further analyses.
(c). Host-specificity reconstruction
The host specificity of each clownfish species is well documented [16]. We took advantage of an updated version of the interaction table that included the host specificity of recently described species [17]. We used stochastic mapping [30] to map probable realizations of the evolution of host specificity on the clownfishes majority rule consensus tree. We carried on this analysis using the ‘make.simmap’ function available in the R package ‘Phytools’ v. 0.2.9 [31]. It was not possible to directly reconstruct the evolution of host specificity on the tree, because current ancestral state reconstruction methods can handle discrete and continuous traits but not counts. Moreover, a clownfish species interacting with, for example, two species of anemone may not be directly comparable to another clownfish species which interact with two different anemone species. To overcome those difficulties, we first inferred the evolution of interaction of each anemone species separately. Interaction with an anemone was modelled as a binary trait (0 = no interaction, 1 = interaction). We used the ‘all rate different’ model to evolve the interaction along the clownfishes phylogenetic tree and allowed different forward and backward rates between the two states, estimated the prior distribution of the states at the root of the tree and used the MCMC option to set the parameters of the Q matrix. To account for the inherent stochasticity of the process, we performed 10 stochastic mappings replicates. This relatively small number of replications is explained by the large computational requirement of the evolutionary modelling described below but still allows the estimation of the variation in the parameters inference. We stacked the stochastic maps obtained for each anemone species and summed them to obtain the reconstruction of host specificity on the phylogeny ranging from 0 to 10. There are no a priori indications for determining the threshold of potential hosts number that separates host specialists and generalists clownfishes. We therefore tested three different groupings where we defined specialists as interacting with either up to one (group C1), two (C2) and three (C3) anemone species. We defined generalists in each groups as clownfish species that interact with more anemone species than the specialists. Sections of branches with 0 interactions were extremely rare, but methodologically unavoidable. Because the apparition of the mutualism with sea anemones proably preceded the radiation of the clownfishes [17], we grouped those small segments with those having a host specificity of one. We used those rules and simplified the stochastic maps accordingly (figure 1 shows the aggregation of the stochastic mappings for visualization purposes [32]).
Figure 1.
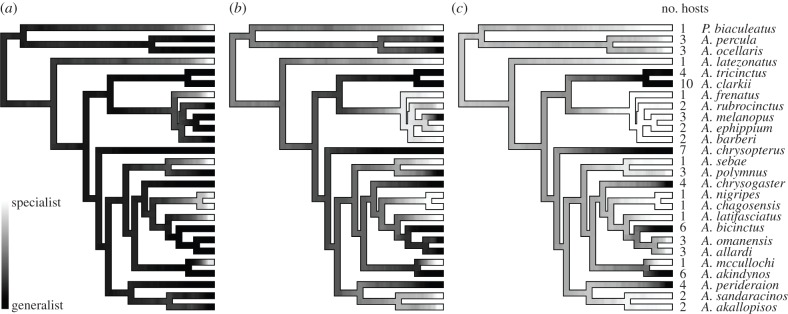
Result of aggregating 100 stochastic character maps (the 10 used in the analyses and 90 others). The degree of shading gives the posterior probability (relative frequency across stochastic maps) of host specialization. Darker branches indicates more generalist mutualistic interactions with sea anemones. While the figure shows a specialist–generalist continuum, the original character maps are binary. Panel (a) shows results of the grouping C1 where specialists are interacting with a single host. Panels (b) and (c) show the results of the groupings C2 and C3, respectively. The number of potential hosts is shown left of the species names. ‘P.’ stands for Premnas and ‘A.’, Amphiprion.
(d). Description and implementation of evolutionary modelling
We used the ‘Ouwie’ R package [33] and a novel approach Kostikova et al. [34] to model the evolution of intraspecific variance in the environmental niche of clownfishes (i.e. niche breadth) and test for an association with host specificity. For both methods, we tested three alternative models: (i) a neutral Brownian model (BM) of evolution of the niche breadth, in which there is no association between host specificity and niche breadth; (ii) a Ornstein–Uhlenbeck model, OU1 with a single niche breadth optimum, which assumes that niche breadth evolves non-neutrally but all species are pulled towards a single optimal value (θ) of niche breadth; and (iii) a model (OUM) with two niche breadth optima (θgeneralists and θspecialists), which assumes that generalist and specialist species have different niche breadths.
Prior to the ‘OUwie’ analysis, we estimated intraspecific variance (i.e. niche breadth) using the maximum-likelihood estimator of a normal distribution on each PC axis. We used the resulting vector of intraspecific variance and the previously described stochastic mappings replicates to estimate the fit of the three evolutionary models in the ‘OUwie’ R package [33] using default settings. Using replicates of the stochastic mappings allowed us to take into account the uncertainty of the ancestral host-specificity reconstruction. We recorded the likelihood of each model and used the Akaike Information Criterion (AIC) to identify the model that best described our data. A major disadvantage of the ‘OUwie’ method is that it does not simultaneously consider both inter- and intraspecific trait evolution but uses niche breadth as a continuous variable with one value per species.
We used a novel approach ([34]; R script available on www.unil.ch/phylo/bioinformatics/jive) to overcome this issue. Unlike conventional comparative methods which use the mean trait value of each species to evaluate evolutionary hypotheses, this method estimates the evolution of a trait by considering all available trait measurements for a given species simultaneously. It, therefore, allows us to explicitly test how intraspecific variance of each species evolves and whether its evolution is associated with a discrete trait. In our case, we used the environmental niche of the species as a continuous variable and the host specificity (specialist versus generalist based on the groupings defined above) as a binary trait. Associated environmental data from all occurrence records obtained from the IOBIS database were used as an input for evolutionary models.
To identify the best-fitting model for the evolution of the niche breadth, we analysed our dataset using an MCMC with thermodynamic integration (TI) under all three models [35]. MCMC with TI allows the estimation of an effect of addition or removal of extra parameters into the model. Under TI, an MCMC sample is drawn from a series of distributions ranging from the posterior to the prior at the two extremes of a path. The path is obtained by altering the acceptance probability by rising the likelihood ratio to the power of a range of values (beta parameter) between 1 and 0. We ran four MCMC chains for 20 000 000 generations, with a burn-in phase of 5 000 000 generations and sampled model parameters each 1000th generation. We used 10 temperature classes given by a beta distribution with β = 0.3 and repeated the TI runs on each of the 10 different stochastic mappings of the three host-specificity groupings. Finally, we averaged the resulting BF across mappings. For each PC axis, we selected a best-fitting model based on the values of BF and ran a simple MCMC chain analysis in order to estimate the parameters of the model. For this conventional MCMC, we allowed two chains to run for 10 000 000 generations with a burn-in phase of 5 000 000 generations and recorded parameters value each 1000th generation. We used the remaining part of the MCMC to calculate the parameters values of a given model. We investigated the adequate convergence of the MCMC chains using Tracer v. 1.4 [36].
3. Results
(a). Environmental niche estimation
The first and second axes of the PCA explain, respectively, 31.3 and 27.2% of the total environmental variation. We do not consider the other axes because they explain only small fractions of variation. The first PC (PC1) shows a gradient of nutrient availability with higher concentrations of nitrates, phosphates and chlorophyll on the negative side of the axis. The second axis shows a gradient of temperature, salinity and pH. Negative values on the second PC axis (PC2) indicate warm waters with relatively low salinity and pH.
(b). Evolutionary modelling
The relative model fits and parameter estimates are comparable between the two modelling techniques. In both analyses, the neutral model for the evolution of niche breadth is consistently rejected for the PC2 axis, but not for the PC1 (tables 1 and 2). For PC2, the performance of OU1 and OUM models differs depending on how specialist clownfishes are defined with regard to the number of host anemones (C1, C2 and C3 groupings, see above). In line with the AIC difference among models recovered in ‘Ouwie’ (table 1), the TI and resulting BF of the novel approach suggests that for C1 grouping the best-fitting model is a single-optimum model, while for C2 and C3 groupings the OUM model has a significantly better fit than the OU1 model (BF > 2; table 2). The estimated parameters suggest that the mode of the niche breadth is twice as large in specialist clownfish species when compared with generalist species (C2: θgeneralists = 3.08 and θspecialists = 5.91; C3: θgeneralists = 2.83 and θspecialists = 4.77). The 95% HPDs provide credibility intervals on estimated parameters (figure 2) and show that, for the PC2, the niche breadth differs significantly between specialist and generalist species.
Table 1.
Comparison of the fit of a Brownian motion (BM) and Ornstein–Uhlenbeck with a single (OU1) or two (OUM) environmental niche breadth optima in OUwie. (The table shows AIC values for each combination of host specialization, environmental PC axis and evolutionary model. Models with the highest AIC support are shown in bold characters.)
| model |
||||
|---|---|---|---|---|
| grouping | PC axis | BM | OU1 | OUM |
| C1 | 1 | 215.35 | 217.92 | 220.71 |
| 2 | 132.61 | 126.94 | 129.35 | |
| C2 | 1 | 215.35 | 217.92 | 220.66 |
| 2 | 132.61 | 126.94 | 124.99 | |
| C3 | 1 | 215.35 | 217.92 | 220.28 |
| 2 | 132.61 | 126.94 | 124.86 | |
Table 2.
Comparison of the fit of a BM and OU1 or OUM environmental niche breadth optima while simultaneously assessing the parameters of intraspecific and interspecific trait evolution. (The table shows BF for each combination of host specialization, environmental PC axis and evolutionary model. Models with the highest BF support are shown in bold characters.)
| model |
||||
|---|---|---|---|---|
| grouping | PC axis | BM | OU1 | OUM |
| C1 | 1 | –230.53 | –235.02 | –231.55 |
| 2 | –152.08 | –149.86 | –150.72 | |
| C2 | 1 | –231.55 | –231.84 | −232.618 |
| 2 | −153.44 | −151.83 | −149.72 | |
| C3 | 1 | −229.99 | −232.54 | −232.94 |
| 2 | −152.68 | −155.50 | −150.95 | |
Figure 2.
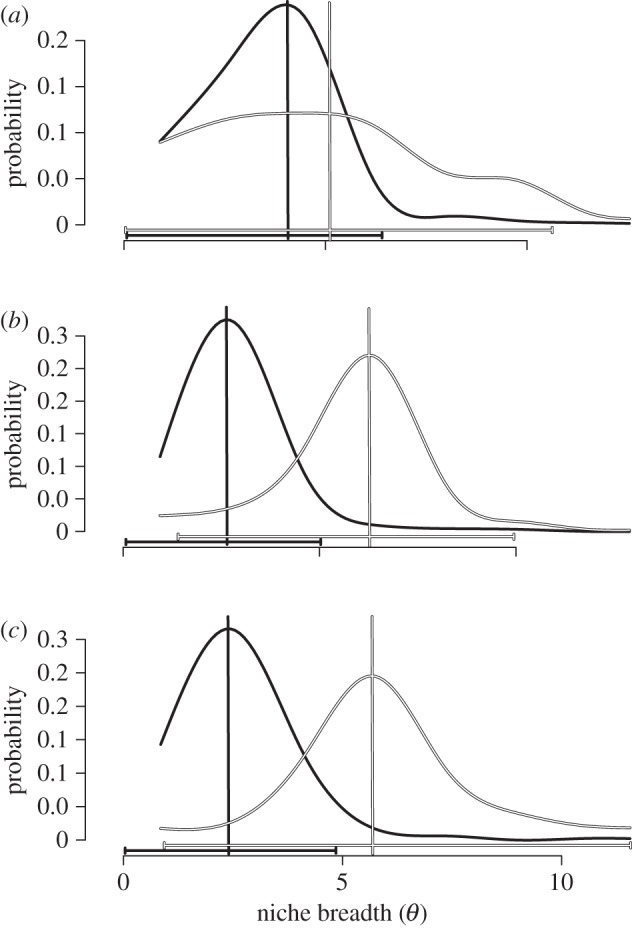
Posterior distributions of niche breadth optima (θ) for the second PC axis. The environmental niche breadth of host generalists species is shown in black and host specialists are in white. Vertical lines show the modes of the distributions and are coloured accordingly. The 95% confidence intervals (CI) are shown below the curves. Results of the grouping C1 are shown in panel (a), C2 in (b) and C3 in (c).
4. Discussion
The evolution of trade-offs between specialist and generalist strategies is a central concern in evolutionary biology. The question is particularly difficult to answer with species involved in host interactions, because species host preferences can overlap to some extent. In this study, we demonstrate that the environmental tolerance regarding temperature, salinity and pH of host-specific clownfish species are broad when compared with the host generalist species. Such specialization trade-offs across multiple resource axes (i.e. niche trade-off) offers a potential explanation to the coexistence of generalists and specialists.
Environmental niche specificity is a well-known correlate of latitudinal and elevational gradients. For example, host specialization in Papilionoidea butterflies decreases at higher elevations suggesting that either oscillating abiotic conditions, lower herbivore pressure or both act to enforce and reduce host-specificity [37]. Similarly, loss of mutualism with ants is observed in Lycaenidae species at high elevations where costs of mutualism are also high [38]. Similar patterns of change from specialists to generalists when climbing the latitudinal gradient are observed in various taxonomic groups [39], yet, in clownfishes, we do not find that climate is significantly correlated with patterns of host specialization (see the electronic supplementary material). While the tropical distribution of clownfishes spans only a restricted part of the latitudinal gradient in the tropics, we still observe substantial differences in environmental specialization among species. The lack of relationship between climate and host specialization may suggest that the selective pressure of abiotic conditions is overridden by specialization trade-offs across multiple resource axes.
Our modelling results suggest that specialization trade-offs across multiple resource axes plays an important role in the evolutionary trajectories of species. Indeed, we find that increased host specialization in clownfishes is associated with the evolution of decreased environmental specialization, expressed here as the breadth of environmental tolerances. By contrast, host generalist clownfish species are in general very specific in their habitat preferences. One likely interpretation is that generalist–specialist trade-offs across multiple resource axes act as a compensatory (balancing) mechanism that allows long-term coexistence of species in the presence of congeneric competition. Similarly, a trade-off between habitat and host suitability for larval development in an aphid parasitoid is constrained by the level of host specificity [15], resulting in lower affinity of host-specific parasites with particular habitats. We consider the relatively weak support for the alternative model (OUM; table 1) of the OUwie analysis sufficient evidence to reject the simpler models, because the results of both OUwie and our method are completely consistent. A key factor to consider is the relatively small sample size, which leads to reduced AIC and BF differences between models.
The effect of host specificity on niche breadth is, however, only recovered on PC2. This axis shows a gradient between cold, saline and basic to warmer, less saline and less basic water, while the PC1 axis is related to nutrient availability. Anthropogenic disturbances such as sediment run-off from land increase the concentration of nutrients in coral reefs [40,41]. This could explain why we recover an evolutionary trade-off between host and environmental niche specificity only on PC2. Indeed, this human-induced change is extremely recent on an evolutionary timescale. While an increased concentration of nutrients probably poses new selective pressures on clownfish species, not enough time has passed for any adaptive response to be picked by our comparative methods. On the contrary, temperature in coral reefs has been historically more stable [42], which probably explains why we recover an evolutionary pattern for PC2. Furthermore, more environmentally focused studies would be necessary to fully resolve this issue.
In clownfishes, part of the variation in host variation can be explained by ecological character displacement, which is regarded as a key driver of evolutionary diversification in many groups [43]. Indeed, in natural communities, syntopic clownfish species that have completely overlapping preferences with respect to their anemone host(s) usually occur in different habitats [20]. While the general outcome of ecological character displacement is phenotypes correlated to resource usage [43], we cannot rule out solely on the base of such observation, that this pattern is caused by other processes [44]. Clownfishes do show a relationship between phenotype and host use [17], yet most clownfish species still have partial to completely overlapping anemone host preferences. Competition for preferred hosts probably plays a role in clownfishes diversification, but our results still suggests that other resource axes are also important in shaping clownfish species assemblages. Our results echo a simulation model in which spatial structure and correlated evolution of ecological preference traits create complex fitness landscapes that allow the coexistence of multiple specialist and generalists on four resources [45].
In this study, we treat all anemone species as equally advantageous to the clownfishes. While the general reciprocal benefits of the clownfish–anemone mutualism are well known [46,47], it is unknown whether some anemone species are more beneficial to the resident clownfishes than other. Anemone variation in size or morphology could change the efficiency of the protection provided by the stinging tentacles, making particular anemone species a safer haven than others. Indeed, juveniles of a host generalist clownfish species can occupy different anemone species than adults. For example, in the case of Amphiprion bicinctus, which occurs in the Red Sea, adults prefer a host in which they can fully conceal their body, while juveniles, which are smaller in size, accommodate well in smaller anemones [48]. It is unclear whether this situation occurs in other regions [49] where the species density of clownfish species is higher (A. bicinctus is the only clownfish species occurring in the Red Sea and juveniles thus compete only with adults). Yet, such variation would be very similar to resource abundance varying in time or space [10]. Further theoretical research is necessary to understand how the variation in resource abundance in a system with two or more resource axes could impact on specialist–generalist coexistence.
5. Conclusion
A plethora of empirical and theoretical studies explains generalists and specialists coexistence [6,9,10,45], yet, most studies focus on systems with a single resource. Coexistence is, for example, made possible by variations in the spatial [10] and temporal [50] availability of the common resource. Our results show that when niche trade-offs are spread across multiple resource axes, the coexistence of generalist and specialist clownfish species is possible and likely to be sustainable over evolutionary times. Examples of species sorting over several resources axes are intriguingly scarce but exist [15,45]. It is unclear whether the paucity of study cases is owing to the rarity of the process or if it truly represents an area of research requiring more attention.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Jana Vamosi, Susanne Renner and Scott Armbruster for the invitation to participate to this special issue. We are grateful to two anonymous reviewers, whose constructive comments helped to improve the manuscript.
Data accessibility
DNA sequences: Genbank accessions KF774317–774435; KF819364–819385. Phylogenetic data: TreeBase accession no. S15377.
Funding statement
This work was funded by the University of Lausanne research fund and a grant 31003A-138282 from the Swiss National Science Foundation to N.S. It further received support for computational work from the Vital-IT facilities from the Swiss Institute of Bioinformatics.
References
- 1.Remold S. 2012. Understanding specialism when the jack of all trades can be the master of all. Proc. R. Soc. B 279, 4861–4869. ( 10.1098/rspb.2012.1990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacArthur RH. 1972. Geographical ecology: patterns in the distribution of species. New York, NY: Harper & Row. [Google Scholar]
- 3.Levins R. 1963. Theory of fitness in a heterogeneous environment. II. Developmental flexibility and niche selection. Am. Nat. 97, 75–90. ( 10.1086/282258) [DOI] [Google Scholar]
- 4.Lawlor LR, Maynard Smith J. 1976. The coevolution and stability of competing species. Am. Nat. 110, 79–99. ( 10.1086/283049) [DOI] [Google Scholar]
- 5.Kisdi E, É Kisdi. 2002. Dispersal: risk spreading versus local adaptation. Am. Nat. 159, 579–596. ( 10.1086/339989) [DOI] [PubMed] [Google Scholar]
- 6.Abrams PA. 2006. Adaptive change in the resource-exploitation traits of a generalist consumer: the evolution and coexistence of generalists and specialists. Evolution 60, 427–439. ( 10.1111/j.0014-3820.2006.tb01124.x) [DOI] [PubMed] [Google Scholar]
- 7.Wilson DS, Yoshimura J. 1994. On the coexistence of specialists and generalists. Am. Nat. 144, 692–707. ( 10.1086/285702) [DOI] [Google Scholar]
- 8.Egas M, Dieckmann U, Sabelis MW. 2004. Evolution restricts the coexistence of specialists and generalists: the role of trade-off structure. Am. Nat. 163, 518–531. ( 10.1086/382599) [DOI] [PubMed] [Google Scholar]
- 9.Abrams PA. 2006. The prerequisites for and likelihood of generalist–specialist coexistence. Am. Nat. 167, 329–342. ( 10.1086/499382) [DOI] [PubMed] [Google Scholar]
- 10.Kido MH. 1997. Food relations between coexisting native Hawaiian stream fishes. Environ. Biol. Fish. 49, 481–494. ( 10.1023/A1007323103260) [DOI] [Google Scholar]
- 11.Behmer ST, Joern A. 2008. Coexisting generalist herbivores occupy unique nutritional feeding niches. Proc. Natl Acad. Sci. USA 105, 1977–1982. ( 10.1073/pnas.0711870105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stireman JO, Singer MS. 2003. What determines host range in parasitoids? An analysis of a tachinid parasitoid community. Oecologia 135, 629–638. ( 10.1007/s00442-003-1235-2) [DOI] [PubMed] [Google Scholar]
- 13.Whitlock MC. 1996. the red queen beats the jack-of-all-trades: the limitations on the evolution of phenotypic plasticity and niche breadth. Am. Nat. 148, S65–S77. ( 10.1086/285902) [DOI] [Google Scholar]
- 14.Litsios G, Pellissier L, Forest F, Lexer C, Pearman PB, Zimmermann NE, Salamin N. 2012. Trophic specialization influences the rate of environmental niche evolution in damselfishes (Pomacentridae). Proc. R. Soc. B 279, 3662–3669. ( 10.1098/rspb.2012.1140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stilmant D, Bellinghen C, Van Hance T, Boivin G. 2008. Host specialization in habitat specialists and generalists. Oecologia 156, 905–912. ( 10.2307/40309579) [DOI] [PubMed] [Google Scholar]
- 16.Fautin DG, Allen GR. 1997. Anemonefishes and their host sea anemones. Perth, Australia: Western Australian Museum. [Google Scholar]
- 17.Litsios G, Sims CA, Wüest RO, Pearman PB, Zimmermann NE, Salamin N. 2012. Mutualism with sea anemones triggered the adaptive radiation of clownfishes. BMC Evol. Biol. 12, 212 ( 10.1186/1471-2148-12-212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ollerton J, McCollin D, Fautin DG, Allen GR. 2007. Finding NEMO: nestedness engendered by mutualistic organization in anemonefish and their hosts. Proc. R. Soc. B 274, 591–598. ( 10.1098/rspb.2006.3758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fautin DG. 1981. The clownfish sea anemones: Stichodactylidae (Coelenterata: Actinaria) and other sea anemones symbiotic with Pomacentrid fishes. Trans. Am. Philos. Soc. 71, 3–115. ( 10.2307/1006382) [DOI] [Google Scholar]
- 20.Elliott JK, Mariscal RN. 2001. Coexistence of nine anemonefish species: differential host and habitat utilization, size and recruitment. Mar. Biol. 138, 23–36. ( 10.1007/s002270000441) [DOI] [Google Scholar]
- 21.Litsios G, Pearman PB, Lanterbecq D, Tolou N, Salamin N. In press The radiation of the clownfishes has two geographical replicates. J. Biogeogr. [Google Scholar]
- 22.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. ( 10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huelsenbeck JP, Larget B, Alfaro ME. 2004. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 21, 1123–1133. ( 10.1093/molbev/msh123) [DOI] [PubMed] [Google Scholar]
- 25.Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP. 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the hymenoptera. Syst. Biol. 61, 973–999. ( 10.1093/sysbio/sys058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyberghein L, Verbruggen H, Pauly K, Troupin C, Mineur F, De Clerck O. 2012. Bio-ORACLE: a global environmental dataset for marine species distribution modelling. Glob. Ecol. Biogeogr. 21, 272–281. ( 10.1111/j.1466-8238.2011.00656.x) [DOI] [Google Scholar]
- 27.Parravicini V, et al. 2013. Global patterns and predictors of tropical reef fish species richness. Ecography 36, 1254–1262. ( 10.1111/j.1600-0587.2013.00291.x) [DOI] [Google Scholar]
- 28.Stuart-Smith RD, et al. 2013. Integrating abundance and functional traits reveals new global hotspots of fish diversity. Nature 501, 539–542. ( 10.1038/nature12529) [DOI] [PubMed] [Google Scholar]
- 29.R Development Core Team 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.Rproject.org) [Google Scholar]
- 30.Huelsenbeck JP, Nielsen R, Bollback JP. 2003. Stochastic mapping of morphological characters. Systematic Biology 52, 131–158. ( 10.1080/10635150390192780) [DOI] [PubMed] [Google Scholar]
- 31.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 32.Revell LJ. 2013. Two new graphical methods for mapping trait evolution on phylogenies. Methods Ecol. Evol. 4, 754–759. ( 10.1111/2041-210X.12066) [DOI] [Google Scholar]
- 33.Beaulieu JM, Jhwueng D-C, Boettiger C, O'Meara BC. 2012. Modeling stabilizing selection: expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evolution 66, 2369–2383. ( 10.1111/j.1558-5646.2012.01619.x) [DOI] [PubMed] [Google Scholar]
- 34.Kostikova A, et al. In press Bridging inter- and intraspecific trait evolution with hierarchical Bayesian approach. Syst. Biol. [DOI] [PubMed]
- 35.Lartillot N, Philippe H. 2006. Computing Bayes factors using thermodynamic integration. Syst. Biol. 55, 195–207. ( 10.1080/10635150500433722) [DOI] [PubMed] [Google Scholar]
- 36.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellissier L, Fiedler K, Ndribe C, Dubuis A, Pradervand J-N, Guisan A, Rasmann S. 2012. Shifts in species richness, herbivore specialization, and plant resistance along elevation gradients. Ecol. Evol. 2, 1818–1825. ( 10.1002/ece3.296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellissier L, Litsios G, Fiedler K, Pottier J, Dubuis A, Pradervand J-N, Salamin N, Guisan A. 2012. Loss of interactions with ants under cold climate in a regional myrmecophilous butterfly fauna. J. Biogeogr. 39, 1782–1790. ( 10.1111/j.1365-2699.2012.02743.x) [DOI] [Google Scholar]
- 39.Schemske DW, Mittelbach GG, Cornell HV, Sobel JM, Roy K. 2009. Is there a latitudinal gradient in the importance of biotic interactions? Annu. Rev. Ecol. Evol. Syst. 40, 245–269. ( 10.1146/annurev.ecolsys.39.110707.173430) [DOI] [Google Scholar]
- 40.Szmant AM. 2002. Nutrient enrichment on coral reefs: Is it a major cause of coral reef decline? Estuaries 25, 743–766. ( 10.1007/BF02804903) [DOI] [Google Scholar]
- 41.Bellwood DR, Hughes TP, Folke C, Nyström M. 2004. Confronting the coral reef crisis. Nature 429, 827–33. ( 10.1038/nature02691) [DOI] [PubMed] [Google Scholar]
- 42.Randall JE. 1998. Zoogeography of shore fishes of the Indo-Pacific Region. Zool. Stud. 37, 227–268. [Google Scholar]
- 43.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 44.Stuart YE, Losos JB. 2013. Ecological character displacement: glass half full or half empty? Trends Ecol. Evol. 28, 402–408. ( 10.1016/j.tree.2013.02.014) [DOI] [PubMed] [Google Scholar]
- 45.Birand A, Vose A, Gavrilets S. 2012. Patterns of species ranges, speciation, and extinction. Am. Nat. 179, 1–21. ( 10.1086/663202) [DOI] [PubMed] [Google Scholar]
- 46.Mebs D. 2009. Chemical biology of the mutualistic relationships of sea anemones with fish and crustaceans. Toxicon 54, 1071–1074. ( 10.1016/j.toxicon.2009.02.027) [DOI] [PubMed] [Google Scholar]
- 47.Szczebak JT, Henry RP, Al-Horani FA, Chadwick NE. 2013. Anemonefish oxygenate their anemone hosts at night. J. Exp. Biol. 216, 970–976. ( 10.1242/jeb.075648) [DOI] [PubMed] [Google Scholar]
- 48.Huebner L, Dailey B, Titus B, Khalaf M, Chadwick N. 2012. Host preference and habitat segregation among Red Sea anemonefish: effects of sea anemone traits and fish life stages. Mar. Ecol. Progr. Series 464, 1–15. ( 10.3354/meps09964) [DOI] [Google Scholar]
- 49.Fautin DG. 1991. The anemonefish symbiosis: what is known and what is not. Symbiosis 10, 23–46. [Google Scholar]
- 50.Östergård H, Hambäck PA, Ehrlén J. 2009. Responses of a specialist and a generalist seed predator to variation in their common resource. Oikos 118, 1471–1476. ( 10.1111/j.1600-0706.2009.17540.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: Genbank accessions KF774317–774435; KF819364–819385. Phylogenetic data: TreeBase accession no. S15377.


