Abstract
Adaptive radiation (AR) is a key process in the origin of organismal diversity. However, the evolution of trait disparity in connection with ecological specialization is still poorly understood. Available models for vertebrate ARs predict that diversification occurs in the form of temporal stages driven by different selective forces. Here, we investigate the AR of cichlid fishes in East African Lake Tanganyika and use macroevolutionary model fitting to evaluate whether diversification happened in temporal stages. Six trait complexes, for which we also provide evidence of their adaptiveness, are analysed with comparative methods: body shape, pharyngeal jaw shape, gill raker traits, gut length, brain weight and body coloration. Overall, we do not find strong evidence for the ‘stages model’ of AR. However, our results suggest that trophic traits diversify earlier than traits implicated in macrohabitat adaptation and that sexual communication traits (i.e. coloration) diversify late in the radiation.
Keywords: trait evolution, convergence, niche overlap, ecological specialization, gill raker
1. Introduction
Adaptive radiation (AR) is the rapid diversification of an evolutionary lineage into an array of species as a consequence of their adaptation to various ecological niches and is thought to be responsible for a great deal of the taxonomic, morphological and ecological diversity on Earth [1–3]. ARs are triggered by ecological opportunity through the colonization of novel environments or the evolution of key innovations, opening up new adaptive zones for organisms to specialize and diversify into [1]. Darwin's finches on the Galapagos archipelago [4], anole lizards on the islands of the Caribbean [5] and the species-flock of cichlid fishes in the East African Great Lakes [6] are famous examples of extant ARs. Studying such outbursts of organismal diversification has revealed a number of putatively general features of AR (reviewed in [2]). Among those is the observation that diversification sometimes proceeds more rapidly in the initial phases of an AR (‘early burst’, EB scenario), but slows down with the filling up of ecological niche space as more species form. However, the generality, or even frequency, of this pattern was recently called into question [7]. Another observation is that the invasion of niche space by diversifying organisms is not random. Different aspects of the environment have been proposed to influence diversification in different phases throughout the course of an AR. Accordingly, there should be temporal stages of AR, in which specialization to available niches, and hence diversification, is predominantly based on different adaptive traits or trait complexes [8].
In vertebrates, for example, it is regularly observed that clades forming early in an AR are subdivided ecologically with respect to macrohabitat specializations [2,8]. Here, we define macrohabitat as a geographically extensive part of the environment encompassing considerable ecological variation, e.g. the benthic or limnetic zones of a lake. This first ‘stage’ is evident in established ARs, such as Lake Malawi cichlids [9], and also in very recent, incipient ARs, like crater-lake cichlids [10] or three-spine stickleback [11]. Subsequently, in a second stage, specialization occurs primarily with respect to either more spatially restricted microhabitats or resources therein, as found in e.g. some Anolis lizards [5] and Darwin's finches [12]. According to Streelman & Danley's [8] general vertebrate model, the most closely related species within an AR often differ in little else than signalling characters, like nuptial coloration. This third and final stage of an AR is observed, for example, in parrotfish [13], Lake Malawi cichlids [9] and elephantfish [14]. It is important to note that these stages are not necessarily discrete and that selection pressures which dominate in one period of a radiation are probably also acting at other times, albeit to a lesser degree [9]. While these hypothesized stages are apparent in some groups, the existence of stages is less clear in other ARs such as Hawaiian drosophilids [15], potentially emphasizing this feature to be unique to vertebrate radiations and definitively calling for further in-depth quantitative evaluations of the ‘stages model’.
In vertebrate ARs, it also appears that the sequence of stages relative to another might differ. In Phylloscopus Old World leaf warblers [16] or extinct actinopterygians [17], for example, habitat divergence followed trophic divergence. Overall, it is unclear how pervasive the phenomenon of ‘AR in stages’ actually is in nature. Although theoretical work [18–20] points to the model having merit, empirical testing is hampered by the need to obtain data for a number of traits for many member species of an AR. Synthesizing different studies of trait evolution into a test of the ‘stages model’ is further complicated by the lack of taxonomic overlap between studies, unclear phylogenetic relationships, the study of different traits, and/or the application of different analytical approaches.
Nonetheless, several predictions can be derived from the hypothesis of ‘AR in stages’ and tested given the appropriate data: (i) adaptive traits or trait complexes will differ in their amount of phylogenetic signal and time-dependence of their diversification; (ii) the ordering of traits or trait complexes by their time-dependence of diversification should mirror the hypothesized order of stage-wise dominating selection pressures (first: macrohabitat, second: microhabitat and resources, third: sexually selected characters; [8]); and (iii) traits or trait complexes that are involved in specialization to available ecological niches early in an AR should have attained a larger between-species difference, if standardized by the variation within species.
Here, we test these predictions in the AR of cichlid fishes of Lake Tanganyika (LT), East Africa. We investigate the evolution of ecologically and reproductively relevant traits in 51 representative species using phylogenetic comparative methods. Our dataset comprises the majority of cichlid tribes present in LT and a reasonable fraction of the species, including the most abundant ones coexisting in the lake's southern basin. It also covers most of the ecological specializations found in LT cichlids, e.g. epilithic algae grazing, scale eating, fish hunting, invertebrate picking, as well as sand, rock or open water dwelling species. Trait data for body shape, size and weight, lower pharyngeal jaw (LPJ) bone shape and weight, as well as stable isotope data, and a robust multi-marker phylogeny are available from the study of Muschick et al. [21]. For this study, we combined these previous data with new data on gut length, brain weight, gill raker structure and coloration. Trait evolution in LT cichlids has been the focus of previous studies [21–28]. However, most studies considered only one or few traits in isolation and did not comprehensively compare multiple traits in the context of ecology and phylogeny (but see [25]), and the ‘stages model’ has not yet been tested explicitly in LT cichlids.
In this study, we first test for a phenotype–environment correlation in the traits and in the trait complexes under examination by using stable isotope ratios as a proxy for macrohabitat and trophic niche. Such a phenotype–environment correlation is an inert feature of an AR and informs about the adaptive nature of the traits in question [1]. We then quantify the overlap between species in morphological trait space as proxy for their degree of specialization in the respective trait or trait complex. Correlations among trait complexes that take into account phylogenetic relationships are also examined. Finally, by explicitly fitting models of trait evolution to the trait data and molecular phylogeny, we evaluate the merit of the ‘stages model’ of AR for LT cichlids.
2. Material and methods
(a). Sampling
We collected trait data for 51 LT cichlid species, which is approximately one-quarter of the endemic species of this lake. The dataset comprises 36 genera (of 53) and 10 of the 16 tribes [29] (see the electronic supplementary material) described for LT cichlids and is thus representative of the phylogenetic, morphological, ecological and behavioural diversity. In subsets of specimens (electronic supplementary material, table S1), we measured six trait ‘complexes’: body shape, the LPJ apparatus, the gill raker apparatus, brain weight, intestine tract length and colour. The data for body and LPJ shape, stable isotopes and phylogenetic relationships were taken from [21]. Measurements of gill raker traits, gut length, brain weight and scoring for coloration were newly generated for this study.
(b). Choice of traits and their ecological relevance
Body shape is important in swimming performance and manoeuvrability and has been shown to correlate with macrohabitat (i.e. on a benthic–limnetic axis) in cichlids [21]. The LPJ is part of the cichlids' pharyngeal jaw apparatus, i.e. a second set of jaws in the cichlids throat used to manipulate the food items taken up by the oral jaws [30,31]. Another important component of the feeding apparatus are gill rakers, which are used to filter and sort food items in the buccal cavity in many groups of fishes [32,33–35] (see electronic supplementary material, figure S1). These bony protrusions on the gill arches have been little studied in cichlids, as opposed to other evolutionary model systems such as stickleback [32]. After uptake, mastication and filtering, food items reach the intestinal tract where enzymatic digestion takes place and nutrients are absorbed. Herbivorous species specialized on resources of low digestibility, e.g. algae and plants, usually have longer intestines resulting in a longer retention time for improved digestion [27]. Brain size is known to show a strong allometric relationship with body size over a large range of organisms [36], but residual variation and shifts in relative sizes of brain parts have been hypothesized to have adaptive value [37,38]. Body coloration in cichlids can differ greatly between species, even between closely related ones or populations of the same species [39], and is important not only in mate recognition, reproductive behaviour, intraspecific aggression, but also camouflage and mimicry [40].
(c). Trait data
The gill raker trait assessments essentially followed previous investigations in three-spine stickleback [32,34]. Brain tissue was removed from the neurocranium in the field and stored in ethanol or RNAlater; in the laboratory, preserved fish brains were drained and dried at 60°C overnight and subsequently weighed to the nearest milligram. To investigate gut length, we removed the entire alimentary canal (‘gut’) from the anus to the posterior end of the stomach and measured its length to the nearest millimetre. In order to evaluate body coloration, we adopted and modified an existing colour-scoring scheme for cichlids [41].
In the following, we describe data re-used from Muschick et al. [21]: phylogenetic relationships were derived from the enforced molecular-clock phylogeny by pruning it to the 51 species included here. Body shape information was assessed on the basis of landmarks derived from photographs using tpsDig [42], procrustes aligned in MorphoJ [43] and analysed in R [44]; LPJ shape information was obtained in a similar way, but including a sliding process of semi-landmarks in tpsRelw [45]. Stable isotope data were used as proxies for macrohabitat-related specialization on a benthic–limnetic axis (δ13C) and for specialization to the trophic niche (‘microhaitat’, δ15N; e.g. [46]).
Prior to statistical analyses, we log transformed all trait values, apart from landmark procrustes coordinates, gill raker counts and coloration scores, using the phyl.resid function of the R package Phytools [47]. Further details to trait data and their statistical treatment are provided in the electronic supplementary material.
(d). Correlations between traits and the ecological niche
As a test for ecological specialization, we evaluated the correlation of trait values with ecological niches, where stable isotope ratios of carbon and nitrogen were used as niche proxies. The relationship of body and LPJ shape and gut length with stable isotope ratios has been investigated before [21,27], but for gill raker morphology in cichlids this is the first demonstration to our knowledge. We correlated the first principal component (PC) of a PC analysis (PCA) comprising both stable isotope ratios, as well as each element separately, with the first PC for each trait (respective scaled trait values for univariate traits, neither corrected for phylogenetic relationships) and accounted for phylogenetic relationships between species using phylogenetic generalized linear models as implemented in the R package Caper [48].
(e). Ecological specialization and overlap between species
We used plots of linear discriminants (LDs) to illustrate each species' position in morphospace for the multivariate data, and boxplots to illustrate the univariate data. Next, we calculated the between-species distances for each trait (see the electronic supplementary material for details). To compare the relative overall separation of species for each trait, we used ‘species’ as the independent variable in multivariate analyses of variance (MANOVA) for multivariate traits and in analyses of variance (ANOVA) for univariate traits, respectively. Here, we could not include colour as a trait, as no within-species measurements were available. We used Wilks' λ to assess species overlap in the MANOVA, and F-values in the ANOVA. As Mahalanobis distances are scaled by the within-group variance, we used them as a generalized measure of trait divergence, which can be compared among traits. To reveal the ordering of traits by their attained trait distance, we implemented breakpoint regression models following [14]. We estimated breakpoints and respective linear relationships for segments using the function segmented in the Segmented R package [49]. For multivariate traits, we calculated the Mahalanobis distances as described above. Univariate trait values were scaled with the averaged within-species variance.
(f). Phylogenetic tests for stages of adaptive radiation
To test for apparent stages in diversification, we fitted models of trait evolution to the trait axes derived from our transformation of raw trait values (see above) using the fitContinuous function in the R package Geiger [50] (number of random starting points = 1000, simultaneous estimation of standard error). To describe the more general process of trait evolution, we fitted three macroevolutionary models to our data: (i) the Brownian motion model assumes trait values to evolve according to a diffusion process, resulting in trait similarity between species being mainly dependent on the amount of shared ancestry; (ii) the Ornstein–Uhlenbeck model of trait evolution simulates attraction to a single optimum of trait values, with the alpha parameter indicating the strength of this attraction; and (iii) the white noise model does not assume a covariance structure in the data owing to phylogeny, it is equivalent to drawing trait values from a single normal distribution. We also assessed the time-dependence of trait evolution using two models. The δ model by Pagel [51] was used to assess the relative contribution of trait evolution early in the radiation versus late in the radiation. Here, values below 1 indicate that trait evolution occurred primarily along the more basal branches in the phylogeny, whereas values greater than 1 indicate trait evolution predominantly in younger subclades. Second, the a parameter of the EB model (also known as ACDC, for ‘accelerating–decelerating’ [52]) implemented in Geiger was used to test for accelerating or decelerating rates of trait evolution across the phylogeny. Negative values indicate a slowdown in trait evolution, while positive values identify acceleration. For an estimate of phylogenetic signal in the data, we calculated Pagel's λ [51], where λ ranges from 0 to 1 and higher λ values mean stronger phylogenetic signal in the trait data. As a second measure of the pervasiveness of phylogenetic signal in our trait data, we calculated Blomberg's K statistic [52,53] using the phylosignal function in the R package Picante v. 1.6–1 [54]; K > 1 indicates stronger resemblance of species than expected under a Brownian model of trait evolution, while values less than 1 point to a greater evolutionary malleability of the trait.
If the diversification of LT cichlids was indeed driven by different selection pressures in the hypothesized order of stages, we would expect this to be reflected in parameter estimates and the fit of the macroevolutionary models to the data. The time-dependent models should fit reasonably well and differ in their parameter estimates between traits, pointing to different temporal maxima of divergence. To robustly deduct a temporal dependence of diversification, however, the Ornstein–Uhlenbeck model of trait evolution would need to be rejected [7], as selection to a single trait optimum with differing strength could mimic such time-dependence.
3. Results
(a). Correlations between traits and the ecological niche
The ecological relevance of traits can be assessed by their correlation with parameters describing the ecological niche. Here, we used stable isotope ratios of carbon (C) and nitrogen (N) as proxies for macrohabitat preference and trophic position of species, respectively (figure 1 and table 1). Body shape is significantly correlated with δ13C, but not with δ15N. The correlation with δ13C becomes weaker and insignificant when phylogenetic relationships are taken into account (by phylogenetic generalized least squares (PGLS)). LPJ shape and gill raker morphology correlate significantly with both isotopic signatures, but more strongly with δ13C. Phylogenetic correction decreases these correlations in LPJ for both isotopes and in gill rakers for δ15N, while it reinforces the correlation between δ13C and gill rakers. Gut length correlates very strongly with both isotopic signatures. Here, both correlations are even more pronounced in the PGLS analysis. Brain weight and coloration stand in stark contrast to the aforementioned traits in that they either do not correlate with ecological niche proxies, or, in the case of δ13C and colour, only with marginal significance.
Figure 1.
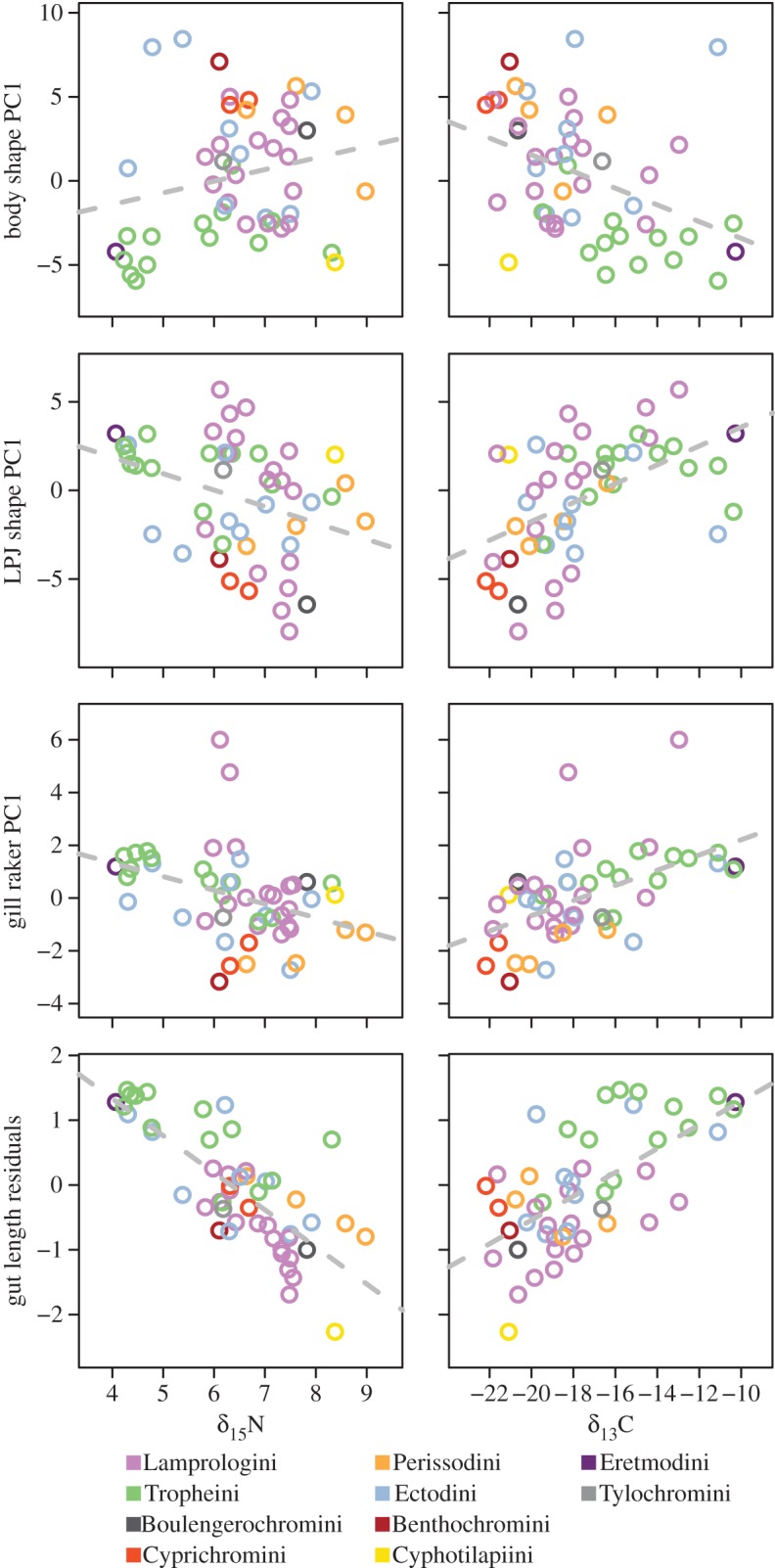
Phenotype–environment correlations. Species means of trait values (see ‘Material and methods’ for details) were plotted against species means of δ13C and δ15N. δ13C is considered to be an indicator of macrohabitat use, with higher or lower values reflecting a benthic or limnetic carbon source, respectively. δ15N is a proxy for trophic level, with larger values reflecting a higher trophic position.
Table 1.
Phenotype–environment correlation. (Correlations of species means for the first principal component of residual trait data (phylogeny not accounted for) and the stable isotope ratio of either carbon or nitrogen (Pearson's correlation coefficient). Phylogenetically generalized least-squares (PGLS) analysis was used to remove the phylogenetic signal potentially present in this correlation. Correlations that remained significant after table-wide adjustment of p-values (after Bonferroni) are given in italics.)
| body shape | LPJ shape | gill raker | gut length | brain weight | colour | ||
|---|---|---|---|---|---|---|---|
| δ13C | Pearson's correlation | −0.41 | 0.51 | 0.53 | 0.63 | 0.12 | 0.28 |
| p-value | 0.0031 | 0.0002 | 0.0001 | 0.0000 | 0.3878 | 0.0497 | |
| PGLS correlation | −0.12 | 0.38 | 0.72 | 2.03 | 0.02 | 0.59 | |
| p-value | 0.3752 | 0.0278 | 0.0203 | 0.0011 | 0.9817 | 0.0591 | |
| δ15N | Pearson's correlation | 0.22 | −0.35 | −0.37 | −0.77 | −0.22 | −0.18 |
| p-value | 0.1197 | 0.0137 | 0.0076 | 0.0000 | 0.1216 | 0.2165 | |
| PGLS correlation | −0.06 | −0.12 | −0.31 | −1.06 | −0.21 | 0.01 | |
| p-value | 0.1940 | 0.0563 | 0.0040 | 0.0000 | 0.3718 | 0.9611 |
Between traits, partial Mantel tests using phylogenetic distance as a covariate revealed weak to moderate and predominantly positive relationships (table 2). However, after correction for multiple comparisons, only a subset of the correlations remained statistically significant. The strongest correlations were found between gut length and brain weight (partial Mantel statistic = 0.5, p < 0.0001), and between gill raker traits and LPJ traits (0.42, p < 0.0001). Gut length and LPJ shape correlated positive with a coefficient of 0.24. No correlation was evident between colour and any other trait.
Table 2.
Trait complex covariation among Tanganyikan cichlids. (Results of partial Mantel tests accounting for phylogenetic distance. Mahalanobis distances were calculated for traits with intraspecific variance, and Manhattan distance was calculated for body coloration. Correlations are given below the diagonal, p-values above. Comparisons that are significant after adjustment for multiple comparisons (after Bonferroni) are given in italics.)
|
p-value |
||||||
|---|---|---|---|---|---|---|
| Mantel statistic | body shape | LPJ shape | gill raker | gut length | brain weight | colour |
| body shape | 0.0004 | 0.0044 | 0.0035 | 0.0286 | 0.595 | |
| LPJ shape | 0.22 | 0.0001 | 0.0002 | 0.0015 | 0.5824 | |
| gill raker | 0.15 | 0.42 | 0.1744 | 0.0027 | 0.7944 | |
| gut length | 0.15 | 0.24 | 0.05 | 0.0001 | 0.2837 | |
| brain weight | 0.11 | 0.19 | 0.18 | 0.5 | 0.7686 | |
| colour | −0.02 | −0.02 | −0.06 | 0.03 | −0.06 | |
(b). Ecological specialization and overlap between species
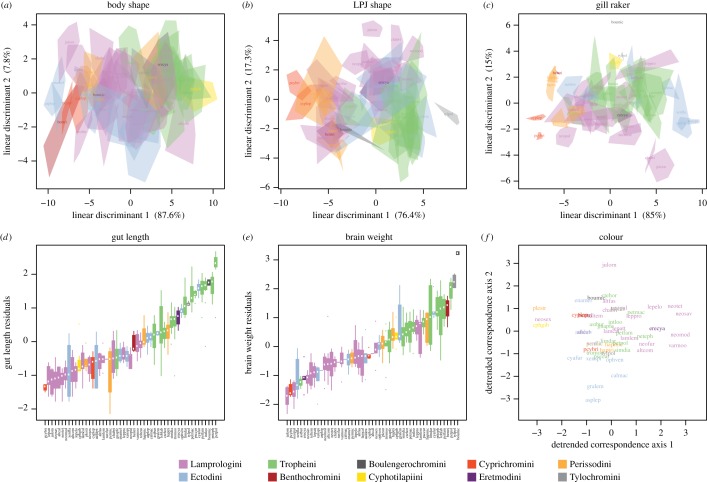
The LT cichlid species examined are at least somewhat, and often strongly, separated in the traits studied here (figure 2). The axes along which species are most separated (scaled by within-species variance) were highlighted by a LD analysis. In body shape space (figure 2a), species show fewer areas of between-species overlap than in the other two multivariate traits, LPJ (figure 2b) and gill rakers (figure 2c). The between-species overlap is most apparent in gill raker space. Gut length (figure 2d) and brain weight (figure 2e) also separate species in morphospace, with overlap between species being less pronounced in gut length. The 14 algae-eating species in the dataset clearly show increased relative gut lengths compared with the non-algae-eating species. Overlap in colour space could not be examined, but some clustering is apparent (figure 2f). On the tribe-level, traits also do show separation, with partitioning of morphospaces being most apparent between the most species-rich LT tribes Lamprologini, Tropheini and Ectodini.
Figure 2.
Niche overlap and divergence between species of cichlids. (a–c) Species overlap on the first two axes from a LD analysis for each multivariate trait. Polygons for species are defined by the most extreme individual and coloured by tribe identity. Amount of between-group variance explained by each axis is given in parentheses. (d,e) Species overlap for the univariate traits gut length and brain weight, sorted by median. (f) Relative position of species in colour space as defined by the first two axes of a detrended correspondence analysis.
MANOVA and ANOVA show that means of trait axes are significantly different between species for all traits (electronic supplementary material, table S2). Wilks' λ indicates that almost all the variance in the multivariate traits is accounted for by species identity. Compared with body and LPJ shape, gill rakers show increased, but still minor, unexplained variance, interpretable as niche overlap (Wilks' λ = 0.0121). The lower F-value in the analysis of relative brain weight compared to gut length corroborates the larger overlap between species in this trait (electronic supplementary material, table S2). In summary, the collective results indicate that the species are well differentiated in all traits, but most strongly in body shape.
(c). Trait evolution and test for the ‘stages model’
Recall that the ‘stages model’ of AR predicts that macrohabitat-related traits, such as body shape, diversify early in the radiation (stage 1). Successively, trophic traits (stage 2) and then traits involved in sexual communication (stage 3) would come to dominate diversification. By fitting models of trait evolution to our data, we evaluated the plausibility of a scenario of diversification in stages in LT cichlids. We assessed which macroevolutionary model fits the data best by comparing the sample-size corrected Akaike information criterion (AICc). The different body shape dimensions derived from the phylogenetic PCA (pPCA) show the best fit to different models of trait evolution: body shape dimension 1 is best approximated by the Pagel's λ model, whereas dimensions 2 and 3 are best approximated by Ornstein–Uhlenbeck and Pagel's δ models, respectively. However, while the model fit difference for dimension 1 is moderately pronounced (min. ΔAICc = 1.47), the model selection is ambivalent in the other body shape dimensions. This is similar for LPJ shape, where the min. ΔAICc is 0.47 for dimension 1 and even lower for dimensions 2 and 3. Gill raker traits, gut length and brain weight all show a phylogenetic signal very similar to that produced by a Brownian motion-like trait evolution (electronic supplementary material, table S3), evidenced by Blomberg's K and Pagel's λ parameter being close to 1, and best fit of the Brownian motion model of trait evolution. Coloration appears to be best fitted by a model of Brownian trait evolution, too. However, in contrast to most other traits, coloration dimensions are reasonably well fitted by the ‘white noise’ model, as well. This might indicate a lack of phylogenetic covariance structure in this trait. We used Pagel's δ model and the EB model to reveal variation in the rate of trait evolution across the timeframe of the radiation. All traits show an acceleration in trait evolution as evidenced by positive a values, as well as a concentration of trait evolution comparatively early in the radiation, shown by δ values more than 1 (electronic supplementary material, table S3). Among the six trait complexes, colour appears to be most rapidly evolving. Of the remaining traits, body shape evolution appears to accelerate in the course of the radiation, followed by LPJ shape. Gill rakers, gut length and brain weight show comparatively little acceleration. However, in no case was the fit of a time-dependent model significantly better than either the Brownian motion or the Ornstein–Uhlenbeck model of trait evolution (ΔAICc > 2). Thus, we have to limit our tentative conclusions about time dependency of trait evolution to the above-stated fits of macroevolutionary scenarios with our data.
In quantifying phylogenetic signal, our results are in agreement with those of Wagner et al. [27], who reported a Pagel's λ of 0.995 for gut length, and of Gonzalez-Voyer et al. [24] who reported a Pagel's λ of 0.71 for brain weight. Both studies have investigated LT cichlids, but included less and different species than this study. Clabaut et al. [22] find that body shape is best predicted by trophic niche and water depth, which is corroborated by our analyses here.
Breakpoint regression models for distances of residual trait values (phylogeny not being accounted for) scaled by within-species variance against phylogenetic distance show the initial increase in trait divergence with evolutionary divergence. After the estimated breakpoint, linear models indicate no further increase in trait divergence, and trait distance appears to be uncorrelated to phylogenetic distance (electronic supplementary material, figure S3).
4. Discussion
Our examination of the stages model of AR in LT cichlids is based on data for six trait complexes analysed within a phylogenetic framework using 51 representative species. We also examined the ecological relevance of these traits using stable isotope data, specialization to ecological niches and niche overlap, as well as trait covariation.
(a). Phenotype–environment correlation
All traits apart from brain size and coloration showed correlation with the ecological niche as approximated by stable isotope data. Overall residual brain size as measured in this study does not appear to correlate with ecological niche. However, it seems likely that cognitive demands regarding specialization to macrohabitats and trophic niches differ such that an effect on brain evolution would be expected [55]. For a smaller dataset of LT cichlids, a significant correlation between diet and brain weight was found [56], but their approximation of the ecological niche differs considerably from ours as it relied on qualitative descriptions from the literature. In general, specialization to available niches might involve changes in the relative sizes of brain substructures [57,58] such that the measure of whole brain weight is too crude to characterize differences.
In contrast to brain weight, gut length is strongly correlated with the trophic niche, and also with our proxy for the macrohabitat niche, δ13C. This is not surprising given the demands of a herbivore's diet on the digestive system. Herbivores are also more likely to acquire their carbon signature from the littoral realm, as this is where epilithic algae occur, explaining the correlation with our macrohabitat proxy. An example of an exception to this is Cyathopharynx furcifer (Cyafur), a plankton feeder with a limnetic carbon signature. Figure 2e illustrates this clearly with herbivorous species being those with the highest relative gut length. The gill raker data also meet the expectation of a significant correlation with ecological niche. Coloration does correlate with marginal significance with the macrohabitat niche. The colour dimorphism found in some species has been correlated with microhabitat specializations [59,60], so this pattern could also be expected on the species level. However, convergence through mimicry might obscure patterns [61,62]. More precise quantification of colour and its relationship to niche specialization is thus required.
(b). Ecological specialization and overlap between species
By scaling trait values by the average within-species variance, we standardized our measurements to allow comparisons across traits and to infer the relative contribution to niche specialization. We find that traits varied in their overlap between species. While body shape shows signs of a more recent divergence compared with trophic traits, it is the trait attaining the largest between-species trait distances if scaled by within-species variance. Also, species overlap less in body shape morphospace compared to trophic traits. This suggests a higher degree of specialization of species in their macrohabitat niche than in trophic characters, such as the pharyngeal jaw apparatus. Trophic traits attain less between-species standardized trait distance, which could be interpreted as trophic traits being less evolutionarily constrained. Another possible explanation is that the adaptive landscape of microhabitats differs between geographically separated sites, which might inflate the morphospace taken up by a given species if individuals from different sites are analysed together.
(c) Trait. covariation
Analysing trait covariation while controlling for phylogenetic relatedness reveals several pairwise comparisons to be significantly correlated. Gut length and brain weight covary—most probably due to both being trophic adaptations, for example to herbivory, which could also impose specific cognitive demands due to habitat complexity. The weaker correlation of body and LPJ shape is somewhat unexpected, as body shape is thought of conferring adaptation to macrohabitat, whereas the pharyngeal jaw apparatus is mainly involved in trophic adaptation. However, food types and abundances differ not only between microhabitats but also between macrohabitats, and, thereby, macrohabitat and trophic adaptations might covary. The correlation between LPJ shape and gill raker traits is not surprising, given that both traits together are responsible for the uptake and the processing of food items, possibly leading to functional constraints. In addition, developmental constraints may further limit independency of trait evolution in the case of LPJ and gill rakers, as both trait complexes derive from gill arch constituents.
(d). Trait evolution and test for the ‘stages model’
The predictions derived from the ‘stages model’ were met to different degrees. In general, we did not find definitive evidence for the existence of discrete stages in the AR of cichlids in LT, because the time-dependent models of phenotypic evolution (EB model and Pagel's δ) were not supported over others. However, some of the patterns observed were nonetheless consistent with the ‘stages model’. For example, traits were found to exhibit different amounts of phylogenetic signal and the timing of diversification varied among traits (supporting our prediction (i)). The ordering of trait diversification (prediction (ii)), with the caveat of not being able to reject the Ornstein–Uhlenbeck model, did not mirror our expectations in the amount of phylogenetic signal. The ordering of the relative amounts of attained relative between-species divergence (scaled by within-species divergence) is in agreement with our prediction from the ‘stages model’ and the findings of the phylogenetic model fitting.
The time-dependence of traits analysed here was not unequivocally evident. Simpler models, not invoking a change in the rate of phenotypic evolution within the timeframe of the AR, received greater support. That said, the time-dependent models (EB model and Pagel's δ) often did fit reasonably well even compared to the respective best-fit model (ΔAICc < 2). Parameter estimates for those two models differed between traits, but did not conform to the expectation of an apparent ordering of trait evolution in the form of macrohabitat-related traits first, then microhabitat-related (in our case: trophic) traits, and, finally, sexual communication traits. Instead, our parameter estimates revealed a different, tentative ordering of trait evolution in LT cichlids that was sometimes but not unambiguously corroborated by the fit of the respective macroevolutionary models: it appears that the evolution of trophic traits is less accelerated across the radiation than either body shape or coloration, suggesting that there are temporal stages of phenotypic evolution in the AR. The ordering, however, is somewhat different to the ‘stages model’, with trophic traits diverging first, followed by macrohabitat-related traits, and, finally communication traits.
The same conclusion can be drawn from the comparison of the amount of phylogenetic signal in each trait complex. Traits characterized by their functionality in feeding (LPJ, gill raker, gut) showed a stronger phylogenetic signal than traits used in macrohabitat adaptation (body shape) or communication (colour). In fact, coloration showed no significant phylogenetic signal at all, which is in agreement with the stages model [8] and is to be expected given current arguments that the evolution of sexual characters is often rapid and unconstrained. On the other hand, the temporal patterns of phenotypic evolution in trophic relative to macrohabitat-related traits do not conform to the ‘stages model’. All results of our model fitting approach point to less phylogenetic signal and, hence, a more recent divergence in body shape traits compared with trophic traits. A similar scenario, with diversification into macrohabitats coming second, has been suggested for Phylloscopus warblers [16], and, more recently, for two extinct fish radiations [17]. The latter authors presented a ‘head-first’ scenario, in which fish head morphology consistently diversified prior to trunk morphology. Assuming head morphology to be a predominantly trophic trait, and trunk morphology responding mainly to selection pressures exerted by macrohabitats, the order of trait evolution in those radiations appears to follow the same trend as we uncover here. Note, however, that such trait complexes are probably not independent. Body shape, for example, also includes information on head shape, so that a signal of adaptation to diet in head shape is probably captured by body shape data too. Thus, the signal in body shape is perhaps biased towards a higher similarity with trophic traits, a possibly general problem in analyses like this. An alternative interpretation of our results is that coloration was a target of selection throughout the radiation, whereas body shape and, especially, the trophic trait complexes LPJ, gill rakers and gut length were involved in certain (initial) stages only [63].
Yet another explanation for our findings is that convergent evolution within the AR of LT cichlids [21] caused an ‘erosion’ of the phylogenetic signal in certain trait complexes, possibly accumulating with time since the climax of divergence. Convergence might also be a potential explanation of why divergence with respect to macrohabitat was suggested as the initial stage of morphological evolution in the first place [8,9]. Low within-species but large between-species variance in body shape make this trait appear well suited for taxonomic inference, but a lack of phylogenetic signal would lead to erroneous taxonomic groupings. In cases where convergent evolution has in this way hampered the traditional reconstruction of phylogenetic relationships that did not include molecular data, stages of AR could have been suggested spuriously. By relying on taxonomic affiliations derived from characters implicated in, for example, habitat or resource specializations, recurrent adaptations would not be recognized as such. In this case, the impression of a temporal order of phenotypic evolution within ARs would be an artefact, as has been shown in bower-building cichlids of Lake Malawi [64]: molecular phylogenetic analyses revealed that some genera were actually not monophyletic to the result that the revised trait distribution suggested trophic morphology to diverge consistently earlier in the AR compared to mate recognition traits (i.e. bower shape). Similarly, in LT cichlids such systematic revisions have been common as reliable molecular phylogenies have become available that uncovered cases of convergent evolution (e.g. [21,65]).
(e). Limitations of our approach
The inability of our study to conclusively reject or support a model of AR in stages in LT cichlids has several plausible explanations, including study design, methodological limitations or biological processes. Concerning study design, the basal tribes Trematocarini and Bathybatini, both predominantly inhabiting the open waters, are under-represented in our dataset. Including more species of those tribes might help generate more definitive results with respect to the ‘stages model’.
Methodologically, our approach analysed body shape as a whole, not discriminating between head- and trunk shapes. Therefore, trophic adaptations in head shape could be represented in our assessment of body shape evolution (see above). However, Muschick et al. [21] have shown that the major axis of body shape evolution in LT cichlids discriminates deep-bodied versus elongated morphs, reflecting macrohabitat adaptation also in other fish groups [10,66]. Additionally, although our dataset is certainly rich relative to today's standards and can readily address questions about ecological specialization and the order of divergence of traits early in the radiation, the number of taxa used might be insufficient to reliably discriminate between scenarios of recent trait evolution (e.g. [67]). Also in terms of methodology, the fitting of evolutionary models implicitly uses reconstructed ancestral phenotypes, which may be inaccurate. Additional information from fossils would be highly useful to verify these estimates [68]. Furthermore, evolutionary change might be underestimated in cases where later changes curb earlier ones, to the end that differences in the rate of evolution between traits might become blurred [69].
5. Conclusion
In this study, we examined the time- dependence of trait evolution and diversification in the species-flock of cichlid fishes in East African LT to test whether this AR proceeded in discrete stages, as has been proposed earlier for vertebrate ARs. Although we do not find strong evidence for the classic stages model of AR in LT cichlids, we find that—contrary to earlier predictions—trophic traits diversified earlier in the radiation than traits related to macrohabitat specializations, whereas sexual communication traits (i.e. coloration) appear to have diversified late. The lack of power in our approach to discriminate between plausible macroevolutionary hypotheses emphasizes the need for even more comprehensive comparative studies, which would benefit from the addition of fossil data.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank the organizers of the special issue, J. Vamosi, S. Renner and S. Armbruster, for inviting us to participate; the associate editor J. Vamosi, R. Gillespie and an additional anonymous referee for valuable comments; our collaborators at the Department of Fisheries, Republic of Zambia, for support in the field and research permits; and H. H. Büscher and various other members of the Salzburger laboratory for help during fieldwork and comments on the manuscript.
Ethics statement
All experiments have been performed under permits issued by the cantonal veterinary office in Basel and the Lake Tanganyika Research Unit, Department of Fisheries, Zambia.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Funding statement
M.M. received funding from the Swiss National Science Foundation (SNSF) and the University of Basel, and a travel stipend from the European Science Foundation's (ESF) ‘Frontiers of Speciation Research’ programme. P.N. and W.S. were both supported by the European Research Council (ERC, grants NatHisGen as well as INTERGENADAPT and CICHLID∼X, respectively). W.S. received further support through the University of Basel and the Swiss National Science Foundation (grant nos. 3100A0_138224 and CRSII3_136293). M.R. was supported by a Swiss National Science Foundation (SNSF) Sinergia grant (CRSII3_136293) awarded to W.S. and a Swiss Academy of Sciences (SCNAT) travel grant. Parts of the samples have been collected on an expedition to LT supported by the National Geographic Society.
References
- 1.Schluter D. 2000. The ecology of adaptive radiation. New York, NY: Oxford University Press. [Google Scholar]
- 2.Gavrilets S, Losos JB. 2009. Adaptive radiation: contrasting theory with data. Science 323, 732–737. ( 10.1126/science.1157966) [DOI] [PubMed] [Google Scholar]
- 3.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press. [Google Scholar]
- 4.Lack D. 1947. Darwin’s finches. Cambridge, UK: Cambridge University Press; (S.l.) [Google Scholar]
- 5.Losos JB. 2009. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Berkeley, CA: University of California Press. [Google Scholar]
- 6.Fryer G, Iles TD. 1972. The cichlid fishes of the great lakes of Africa: their biology and evolution. Edinburgh, UK: Oliver and Boyd. [Google Scholar]
- 7.Harmon LJ, et al. 2010. Early bursts of body size and shape evolution are rare in comparative data. Evolution 64, 2385–2396. ( 10.1111/j.1558-5646.2010.01025.x) [DOI] [PubMed] [Google Scholar]
- 8.Streelman JT, Danley PD. 2003. The stages of vertebrate evolutionary radiation. Trends Ecol. Evol. 18, 126–131. ( 10.1016/S0169-5347(02)00036-8) [DOI] [Google Scholar]
- 9.Danley PD, Kocher TD. 2001. Speciation in rapidly diverging systems: lessons from Lake Malawi. Mol. Ecol. 10, 1075–1086. ( 10.1046/j.1365-294X.2001.01283.x) [DOI] [PubMed] [Google Scholar]
- 10.Barluenga M, Stölting KN, Salzburger W, Muschick M, Meyer A. 2006. Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature 439, 719–723. ( 10.1038/nature04325) [DOI] [PubMed] [Google Scholar]
- 11.Nagel L, Schluter D. 1998. Body size, natural selection, and speciation in sticklebacks. Evolution 52, 209–218. ( 10.2307/2410936) [DOI] [PubMed] [Google Scholar]
- 12.Grant PR, Grant BR. 2007. How and why species multiply: the radiation of Darwin’s finches. Princeton, NJ: Princeton University Press. [Google Scholar]
- 13.Streelman JT, Alfaro M, Westneat MW, Bellwood DR, Karl SA. 2002. Evolutionary history of the parrotfishes: biogeography, ecomorphology, and comparative diversity. Evolution 56, 961–971. ( 10.1111/j.0014-3820.2002.tb01408.x) [DOI] [PubMed] [Google Scholar]
- 14.Arnegard ME, McIntyre PB, Harmon LJ, Zelditch ML, Crampton WG, Davis JK, Sullivan JP, Lavoue S, Hopkins CD. 2010. Sexual signal evolution outpaces ecological divergence during electric fish species radiation. Am. Nat. 176, 335–356. ( 10.1086/655221) [DOI] [PubMed] [Google Scholar]
- 15.Kambysellis MP, Ho K-F, Craddock EM, Piano F, Parisi M, Cohen J. 1995. Pattern of ecological shifts in the diversification of Hawaiian Drosophila inferred from a molecular phylogeny. Curr. Biol. 5, 1129–1139. ( 10.1016/S0960-9822(95)00229-6) [DOI] [PubMed] [Google Scholar]
- 16.Richman AD, Price T. 1992. Evolution of ecological differences in the Old-World leaf warblers. Nature 355, 817–821. ( 10.1038/355817a0) [DOI] [PubMed] [Google Scholar]
- 17.Sallan LC, Friedman M. 2012. Heads or tails: staged diversification in vertebrate evolutionary radiations. Proc. R. Soc. B 279, 2025–2032. ( 10.1098/rspb.2011.2454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavrilets S, Vose A. 2005. Dynamic patterns of adaptive radiation. Proc. Natl Acad. Sci. USA 102, 18 040–18 045. ( 10.1073/pnas.0506330102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavrilets S, Vose A. 2009. Dynamic patterns of adaptive radiation: evolution of mating preferences. In Speciation and patterns of diversity (eds Butlin R, Bridle J, Schluter D.), pp. 102–126. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 20.Gavrilets S. 2004. Fitness landscapes and the origin of species. Princeton, NJ: Princeton University Press. [Google Scholar]
- 21.Muschick M, Indermaur A, Salzburger W. 2012. Convergent evolution within an adaptive radiation of cichlid fishes. Curr. Biol. 22, 2362–2368. ( 10.1016/j.cub.2012.10.048) [DOI] [PubMed] [Google Scholar]
- 22.Clabaut C, Bunje PM, Salzburger W, Meyer A. 2007. Geometric morphometric analyses provide evidence for the adaptive character of the Tanganyikan cichlid fish radiations. Evolution 61, 560–578. ( 10.1111/j.1558-5646.2007.00045.x) [DOI] [PubMed] [Google Scholar]
- 23.Hoerner ME. 2011. Testing for differences in rates of speciation, extinction, and morphological evolution in four tribes of cichlids endemic to Lake Tanganyika, East Africa. Evolution 65, 3398–3412. ( 10.1111/j.1558-5646.2011.01390.x) [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez-Voyer A, Winberg S, Kolm N. 2009. Distinct evolutionary patterns of brain and body size during adaptive radiation. Evolution 63, 2266–2274. ( 10.1111/j.1558-5646.2009.00705.x) [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Voyer A, Kolm N. 2011. Rates of phenotypic evolution of ecological characters and sexual traits during the Tanganyikan cichlid adaptive radiation. J. Evol. Biol. 24, 2378–2388. ( 10.1111/j.1420-9101.2011.02365.x) [DOI] [PubMed] [Google Scholar]
- 26.Duponchelle F, Paradis E, Ribbink AJ, Turner GF. 2008. Parallel life history evolution in mouthbrooding cichlids from the African Great Lakes. Proc. Natl Acad. Sci. USA 105, 15 475–15 480. ( 10.1073/pnas.0802343105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner CE, McIntyre PB, Buels KS, Gilbert DM, Michel E. 2009. Diet predicts intestine length in Lake Tanganyika's cichlid fishes. Funct. Ecol. 23, 1122–1131. ( 10.1111/j.1365-2435.2009.01589.x) [DOI] [Google Scholar]
- 28.Kidd MR, Duftner N, Koblmuller S, Sturmbauer C, Hofmann HA. 2012. Repeated parallel evolution of parental care strategies within Xenotilapia, a genus of cichlid fishes from Lake Tanganyika. PLoS ONE 7, e31236 ( 10.1371/journal.pone.0031236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koblmuller S, Sefc KM, Sturmbauer C. 2008. The Lake Tanganyika cichlid species assemblage: recent advances in molecular phylogenetics. Hydrobiologia 615, 5–20. ( 10.1007/s10750-008-9552-4) [DOI] [Google Scholar]
- 30.Liem KF, Osse JWM. 1975. Biological versatility, evolution, and food resource exploitation in African cichlid fishes. Am. Zool. 15, 427–454. ( 10.1093/icb/15.2.427) [DOI] [Google Scholar]
- 31.Liem KF. 1973. Evolutionary strategies and morphological innovations: cichlid pharyngeal jaws. Syst. Zool. 22, 425–441. ( 10.2307/2412950) [DOI] [Google Scholar]
- 32.Berner D, Adams DC, Grandchamp AC, Hendry AP. 2008. Natural selection drives patterns of lake-stream divergence in stickleback foraging morphology. J. Evol. Biol. 21, 1653–1665. ( 10.1111/j.1420-9101.2008.01583.x) [DOI] [PubMed] [Google Scholar]
- 33.Smith TB, Skulason S. 1996. Evolutionary significance of resource polymorphism in fishes, amphibians and birds. Annu. Rev. Ecol. Syst. 27, 111–133. ( 10.1146/annurev.ecolsys.27.1.111) [DOI] [Google Scholar]
- 34.Gerking SD. 1994. Feeding ecology of fish. San Diego, CA: Academic Press. [Google Scholar]
- 35.Sanderson SL, Cheer AY, Goodrich JS, Graziano JD, Callan WT. 2001. Crossflow filtration in suspension-feeding fishes. Nature 412, 439–441. ( 10.1038/35086574) [DOI] [PubMed] [Google Scholar]
- 36.Striedter GF. 2005. Principles of brain evolution. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 37.Barton RA, Harvey PH. 2000. Mosaic evolution of brain structure in mammals. Nature 405, 1055–1058. ( 10.1038/35016580) [DOI] [PubMed] [Google Scholar]
- 38.Sol D, Bacher S, Reader SM, Lefebvre L. 2008. Brain size predicts the success of mammal species introduced into novel environments. Am. Nat. 172, S63–S71. ( 10.1086/588304) [DOI] [PubMed] [Google Scholar]
- 39.Baric S, Salzburger W, Sturmbauer C. 2003. Phylogeography and evolution of the Tanganyikan cichlid genus Tropheus based upon mitochondrial DNA sequences. J. Mol. Evol. 56, 54–68. ( 10.1007/s00239-002-2380-7) [DOI] [PubMed] [Google Scholar]
- 40.Maan ME, Sefc KM. 2013. Colour variation in cichlid fish: developmental mechanisms, selective pressures and evolutionary consequences. Semin. Cell Dev. Biol. 24, 516–528. ( 10.1016/j.semcdb.2013.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzburger W, Niederstätter H, Brandstätter A, Berger B, Parson W, Snoeks J, Sturmbauer C. 2006. Colour-assortative mating among populations of Tropheus moorii, a cichlid fish from Lake Tanganyika, East Africa. Proc. R. Soc. B 273, 257–266. ( 10.1098/rspb.2005.3321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohlf FJ. 2008. tpsDig, v. 2.11. Stony Brook, NY: Department of Ecology and Evolution, SUNY. [Google Scholar]
- 43.Klingenberg CP. 2011. MorphoJ: an integrated software package for geometric morphometrics. Mol. Ecol. Resour. 11, 353–357. ( 10.1111/j.1755-0998.2010.02924.x) [DOI] [PubMed] [Google Scholar]
- 44.R Core Team 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 45.Rohlf FJ. 2010. tpsRelw, v. 1.49 Stony Brook, NY: Department of Ecology and Evolution, SUNY. [Google Scholar]
- 46.Vander Zanden MJ, Vadeboncoeur Y. 2002. Fishes as integrators of benthic and pelagic food webs in lakes. Ecology 83, 2152–2161. ( 10.1890/0012-9658(2002)083[2152:FAIOBA]2.0.CO;2) [DOI] [Google Scholar]
- 47.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 48.Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Isaac N, Pearse W. 2012. caper: Comparative analyses of phylogenetics and evolution in R, v. 0.5 See http://CRAN.R-project.org/package=caper.
- 49.Vito M, Muggeo R. 2008. segmented: an R package to fit regressions model with broken-line relationships. R News 8, 20–25. [Google Scholar]
- 50.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131. ( 10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 51.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 52.Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 53.Blomberg SP, Garland JT. 2002. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 15, 899–910. ( 10.1046/j.1420-9101.2002.00472.x) [DOI] [Google Scholar]
- 54.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for intergrating phylogenies and ecology. Bioinformatics 26, 1463–1464. ( 10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 55.Kotrschal K, Van Staaden MJ, Huber R. 1998. Fish brains: evolution and environmental relationships. Rev. Fish Biol. Fish. 8, 373–408. ( 10.1023/A:1008839605380) [DOI] [Google Scholar]
- 56.Gonzalez-Voyer A, Winberg S, Kolm N. 2009. Social fishes and single mothers: brain evolution in African cichlids. Proc. R. Soc. B 276, 161–167. ( 10.1098/rspb.2008.0979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez-Voyer A, Winberg S, Kolm N. 2009. Brain structure evolution in a basal vertebrate clade: evidence from phylogenetic comparative analysis of cichlid fishes. BMC Evol. Biol. 9, 238 ( 10.1186/1471-2148-9-238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huber R, van Staaden MJ, Kaufman LS, Liem KF. 1997. Microhabitat use, trophic patterns, and the evolution of brain structure in African cichlids. Brain Behav. Evol. 50, 167–182. ( 10.1159/000113330) [DOI] [PubMed] [Google Scholar]
- 59.Mboko SK, Kohda M. 1995. Pale and dark dichromatism related to microhabitats in a herbivorous Tanganyikan cichlid fish, Telmatochromis temporalis. J. Ethol. 13, 77–83. ( 10.1007/BF02352566) [DOI] [Google Scholar]
- 60.Kohda M, Hori M. 1993. Dichromatism in relation to the trophic biology of predatory cichlid fishes in Lake Tanganyika, East-Africa. J. Zool. 229, 447–455. ( 10.1111/j.1469-7998.1993.tb02647.x) [DOI] [Google Scholar]
- 61.Hori M, Watanabe K. 2000. Aggressive mimicry in the intra-populational color variation of the Tanganyikan scale-eater Perissodus microlepis (Cichlidae). Environ. Biol. Fishes 59, 111–115. ( 10.1023/A:1007657419083) [DOI] [Google Scholar]
- 62.Schelly R, Takahashi T, Bills R, Hori M. 2007. The first case of aggressive mimicry among lamprologines in a new species of Lepidiolamprologus (Perciformes: Cichlidae) from Lake Tanganyika. Zootaxa 1638, 39–49. [Google Scholar]
- 63.Salzburger W. 2009. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol. Ecol. 18, 169–185. ( 10.1111/j.1365-294X.2008.03981.x) [DOI] [PubMed] [Google Scholar]
- 64.Kidd MR, Kidd CE, Kocher TD. 2006. Axes of differentiation in the bower-building cichlids of Lake Malawi. Mol. Ecol. 15, 459–478. ( 10.1111/j.1365-294X.2005.02787.x) [DOI] [PubMed] [Google Scholar]
- 65.Salzburger W, Meyer A, Baric S, Verheyen E, Sturmbauer C. 2002. Phylogeny of the Lake Tanganyika cichlid species flock and its relationship to the Central and East African haplochromine cichlid fish faunas. Syst. Biol. 51, 113–135. ( 10.1080/106351502753475907) [DOI] [PubMed] [Google Scholar]
- 66.Krabbenhoft TJ, Collyer ML, Quattro JM. 2009. Differing evolutionary patterns underlie convergence on elongate morphology in endemic fishes of Lake Waccamaw, North Carolina. Biol. J. Linn. Soc. 98, 636–645. ( 10.1111/j.1095-8312.2009.01305.x) [DOI] [Google Scholar]
- 67.Boettiger C, Coop G, Ralph P. 2012. Is your phylogeny informative? Measuring the power of comparative methods. Evolution 66, 2240–2251. ( 10.1111/j.1558-5646.2011.01574.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slater GJ, Harmon LJ, Alfaro ME. 2012. Integrating fossils with molecular phylogenies improves inference of trait evolution. Evolution 66, 3931–3944. ( 10.1111/j.1558-5646.2012.01723.x) [DOI] [PubMed] [Google Scholar]
- 69.Losos JB. 2011. Seeing the forest for the trees: the limitations of phylogenies in comparative biology. Am. Nat. 177, 709–727. ( 10.1086/660020) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.