Abstract
Environmental conditions can shape genetic and morphological divergence. Release of new habitats during historical environmental changes was a major driver of evolutionary diversification. Here, forces shaping population structure and ecotype differentiation (‘pelagic’ and ‘coastal’) of bottlenose dolphins in the North-east Atlantic were investigated using complementary evolutionary and ecological approaches. Inference of population demographic history using approximate Bayesian computation indicated that coastal populations were likely founded by the Atlantic pelagic population after the Last Glacial Maxima probably as a result of newly available coastal ecological niches. Pelagic dolphins from the Atlantic and the Mediterranean Sea likely diverged during a period of high productivity in the Mediterranean Sea. Genetic differentiation between coastal and pelagic ecotypes may be maintained by niche specializations, as indicated by stable isotope and stomach content analyses, and social behaviour. The two ecotypes were only weakly morphologically segregated in contrast to other parts of the World Ocean. This may be linked to weak contrasts between coastal and pelagic habitats and/or a relatively recent divergence. We suggest that ecological opportunity to specialize is a major driver of genetic and morphological divergence. Combining genetic, ecological and morphological approaches is essential to understanding the population structure of mobile and cryptic species.
Keywords: ecological niches, demographic history, population genetics, morphology, bottlenose dolphins
1. Introduction
Environmental variation is a major driver of evolutionary divergence. It can lead to natural selection on environment-associated traits, which can trigger assortative mating, reproductive isolation and ultimately speciation [1]. Adaptive divergence can evolve in allopatry when groups of individuals occur in separated and contrasting environments [2], or in sympatry and parapatry when they have different ecological niches [1,3]. In the absence of geographical barriers to gene flow, habitat and/or prey preferences among groups of individuals can lead to genetic and morphological differentiation. For example, top predators inhabiting neighbouring areas, such as grey wolves in boreal coniferous forest and tundra/taiga, that are known to specialize on different prey, can be genetically and phenotypically differentiated [4]. Other examples include Galapagos sea lions from two distinct rookeries foraging in benthic and pelagic habitats, sympatric populations of killer whales specializing on fish or marine mammals in the North-east Pacific and sympatric generalist and specialist killer whales in the North-east Atlantic (NEA) [5–7]. Similarly, sympatric individuals of several birds and post-glacial temperate lake fish species, showing contrasting morphs adapted to distinct feeding ecology, are at different stages of genetic isolation [8,9].
Current genetic structure and morphological traits may have been driven by both historical and current ecological conditions. Morphological traits can indeed evolve from very short to evolutionary timescales [10,11]. Quaternary glaciation oscillations have had a major role in shaping genetic diversity patterns. Habitat releases during post-glacial periods have created ecological opportunities for evolutionary diversification for many species in the Northern Hemisphere [12]. The magnitude of influence of historical versus current processes on population structure can vary among species [13,14] and both can have an important role. For example, genetic differentiation patterns of arctic canids might be linked to historical climatic conditions, social structure and dispersal behaviours [15]. Preferential dispersal towards a habitat similar to one of juvenile life [16] can generate and maintain evolutionary divergence in highly mobile species. While this process may be ‘imprinted’ in turtles or fishes [17], social learning of foraging techniques for particular prey or habitat may influence dispersal in species having long-term bonds between mothers and calves [4,15]. Individuals may therefore have higher foraging success in familiar habitats where they can use learned hunting techniques, which might enhance their fitness. This process likely limits gene flow and facilitates local adaptation of ecologically distinct groups of individuals [18].
Although highly mobile, cetaceans can show high levels of population structure. This structure is often suggested to be the result of historical processes, social structure or ecological specializations [19,20]. However, genetic studies are rarely correlated with ecology and morphology studies, with the exception of killer whales (reviewed in [21]). To understand the forces shaping the structure of diversity, it is essential to integrate ecological and evolutionary approaches. Bottlenose dolphins in the NEA form two genetically distinct ecotypes: coastal (i.e. generally occurring in waters less than 40 m deep) and pelagic (i.e. mainly sighted in deeper waters). They are hierarchically structured, with two populations within each ecotype [22]. In the coastal ecotype, the Coastal North (CN) population includes individuals sampled around the UK and Ireland, while the Coastal South (CS) population includes individuals of the French and Spanish coasts. The pelagic ecotype is divided into the Pelagic Atlantic (PA) and Pelagic Mediterranean (PM) populations. The forces having shaped this population structure and the divergence of the two ecotypes are not yet understood. The main objective of this study is to address this question using a combination of population genetics and ecological approaches. First, we investigated the most probable population history using approximate Bayesian computation (ABC) [23,24] and correlated the inferences to historical environmental conditions. We tested whether the time frames of ecotype and population formations were compatible with the formation of new ecological niches. Then, we characterized the morphology and ecology (through the analyses of stable isotope (SI) ratios and stomach contents) of the two ecotypes in order to understand how ecotype differentiation is maintained. The SI niche was used as a proxy of the ecological niche [25] with both δ13C and δ34S reflecting the foraging habitat, and δ15N the trophic position during the few weeks preceding sampling [26]. By using complementary approaches, we shed light on how environmental fluctuations and ecological specializations may have shaped genetic and morphological divergence of a highly mobile marine top predator.
2. Material and methods
(a). Genetic inference of population demographic history
(i). Genetic dataset
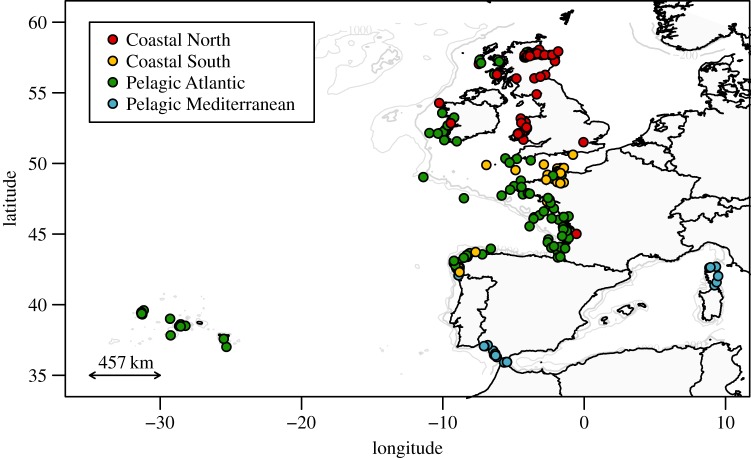
Population history analyses were based on 355 biopsy-sampled or stranded bottlenose dolphins, which were analysed for 25 microsatellites and a 681 base-pair portion of the mitochondrial DNA control region (N = 343) in [22]. In this previous study, each individual was genetically assigned to one of four populations using Bayesian clustering analyses (figure 1).
Figure 1.
Sample locations and genetic populations of bottlenose dolphins included in demographic history analyses.
(ii). Approximate Bayesian computation analysis
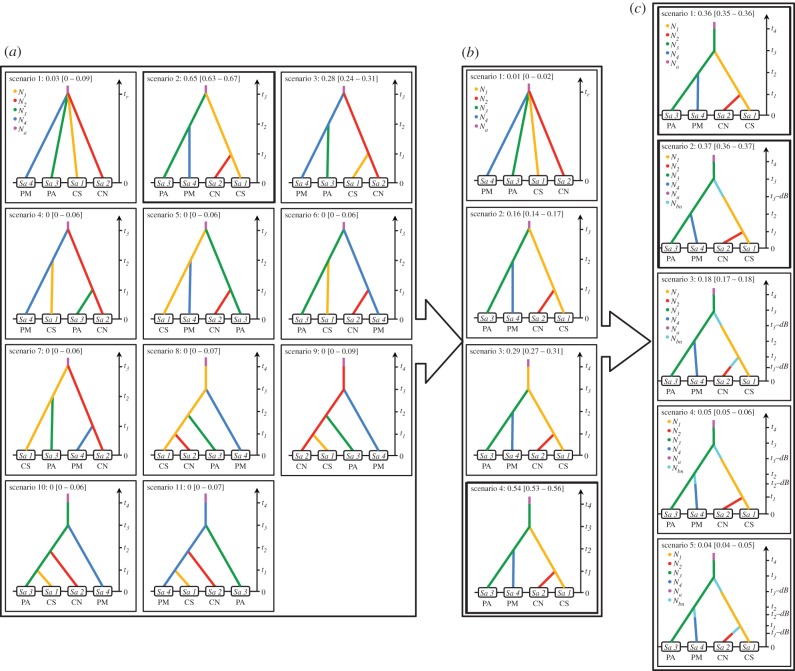
We investigated the demographic history best describing the genetic dataset of the combined microsatellite and mtDNA markers using a coalescent-based ABC approach [23,24]. We stratified the procedure in three steps (figure 2): (i) identify the most likely population tree topologies for our dataset among 11 scenarios describing different potential population topologies (figure 2a); (ii) refine the topology of the best tree (figure 2b) and (iii) test the occurrence of bottlenecks when each population split from its ancestor (figure 2c).
Figure 2.
Schematic diagram of hierarchical ABC analysis to compare various evolutionary histories and divergence scenarios generated and tested using the program DIYABC. PM, Pelagic Mediterranean; PA, Pelagic Atlantic; CS, Coastal South; CN, Coastal North.
For each step, an ABC analysis was conducted using the program DIYABC v. 2.0.4 [27] and included several steps described in the electronic supplementary material, figure S1: (i) coalescent simulations of 106 pseudo-observed datasets (PODs) under each competing scenario and the calculation of summary statistics (SS) describing microsatellites and mtDNA sequences for each POD; (ii) select the best model by estimating the posterior probability (Ppr) of each scenario using a logistic regression on 1% PODs producing SS values closest to the observed ones; (iii) evaluate the confidence in scenario choice by estimating the type I and type II error rates based on simulated datasets; (iv) estimate the marginal posterior distribution of each parameter based on the best model(s); and finally, (v) evaluate the goodness-of-fit of the model–posterior parameter distribution combination with the data.
The parameters defining each scenario (i.e. population sizes, times of population size changes and splits, and mutation rates) were considered as random variables drawn from prior distributions (figure 2; electronic supplementary material, table S1 and appendix S1). For each simulation, DIYABC drew a value for each parameter from its prior distribution and performed coalescent simulations to generate a simulated POD with the same number of gene copies and loci per population as observed. A set of SS were then calculated for each POD and the observed data. A Euclidean distance δ was calculated between the statistics obtained for each normalized simulated dataset and those for the observed dataset [24]. Mutation models for mtDNA and microsatellite loci and the SS used to describe within and among population genetic diversity are detailed in the electronic supplementary material.
(iii). Model selection procedure and confidence in scenario choice
The Ppr of each competing scenario was estimated using a polychotomous logistic regression [28,29] on the 1% of simulated datasets closest to the observed dataset (lowest Euclidean distance δ, see above), subject to a linear discriminant analysis as a pre-processing step (to reduce the dimensionality of the data, [30]). The selected scenario had the highest Ppr value with a non-overlapping 95% confidence interval (95% CI). We evaluated the ability of the ABC analysis to discriminate between tested scenarios by analysing 500 simulated datasets with the same number of loci and individuals as our real dataset. We estimated type I and type II error probabilities following [29].
(iv). Parameter estimation and model checking
We estimated the posterior distributions of each demographic parameter under the best demographic model, by carrying out local linear regressions on the 1% closest of 106 simulated datasets, after a logit transformation to parameter values [24,28]. Following [31], we evaluated whether the best model–posterior distribution combination was more able to reproduce the observed data compared with the alternative scenarios, using the model checking procedure in DIYABC. Model checking was carried out by simulating 1000 PODs under each studied model–posterior distribution combination, with sets of parameter values drawn with replacement from the 1000 sets of the posterior sample. This generated a posterior cumulative distribution function for each simulated SS, from which we were able to estimate the p-value of the deviation of the observed value of each statistic from its simulated distribution under the best demographic model.
(b). Ecological and morphological characterization of ecotypes
Only ecotypes and not all populations were characterized in terms of ecology and morphometry, due to the constraints of tissue and data availability. Sampling was composed of 63 individuals that stranded in the English Channel and the Bay of Biscay from 1991 to 2012, including 21 coastal individuals (18 were genetically assigned to the CS population and three to the CN population) and 42 pelagic individuals from the Atlantic population (representing 32 females, 30 males and one individual of unknown gender, see sampling locations in the electronic supplementary material, figure S2). Morphometric, SI (δ13C, δ34S and δ15N) and stomach content analyses were performed on various animals from this dataset, dependant on the availability of morphometric data, stomach contents and non-decomposed skin samples. All selected individuals had a length greater than 200 cm, to exclude suckling individuals, as neonate δ15N values are up to one trophic level higher than their mothers [32]. All statistics were performed in R 3.0.0 [33].
(i). Morphometric analyses
Ten external morphometric measurements including lengths of appendices and lengths from rostrum to various body parts (L1 to L10; electronic supplementary material, figure S3) were taken by trained observers of the French stranding network. Morphometric analyses were only performed on individuals with no missing measurements and that were not decomposed (Ncoastal = 12 and Npelagic = 27 and Nfemales = 20 and Nmales = 18, Nundetermined = 1). As body length was not significantly different between the two ecotypes (Student's t-test p = 0.28), all measurements were standardized over the total body length (L1) to control for different sizes and ages. As there were no differences between juveniles and adults, all individuals were included in the analyses. First, each ratio was compared between ecotypes using, when appropriate, a Student t-test or a Mann–Whitney–Wilcoxon test. Then, a principal component analysis was performed using the ade4 package [34] to test for morphometric segregation between ecotypes. In addition, to test for a division in the dataset with no a priori assumptions, we performed a maximum-likelihood clustering analysis based on Gaussian mixture models using the mclust package [35]. We used the default settings and the best model was selected by Bayesian information criterion. A discriminant function analysis (DFA) was carried out to find the best combination of standardized variables that separated the two ecotypes, using the ade4 and MASS packages [34,36]. Then, we reassigned individuals to each ecotype using the discriminant function and estimated the rate of correct assignment. All analyses were also performed separately on males and females, due to slight sexual dimorphism.
(ii). Stable isotope analyses
Skin δ13C, δ34S and δ15N values were measured in 40 samples (Ncoastal = 14 and Npelagic = 26, Nfemales = 24, Nmales = 15, Nundetermined = 1). Sample preparation and analyses are detailed in the electronic supplementary material, appendix S2. Sulfur, carbon and nitrogen isotope ratios were determined by a continuous flow mass spectrometer coupled to an elemental analyser. SI values are presented in the conventional δ notation relative to Vienna Pee Dee Belemnite, IAEA-1 and IAEA-2, and atmospheric N2 for δ13C, δ34S and δ15N values, respectively.
Mean differences between coastal and pelagic dolphins and between males' and females' δ13C, δ34S and δ15N were compared using a Student t-test or a Mann–Whitney–Wilcoxon test. SI niches of the two ecotypes were estimated using multivariate, ellipse-based metrics: stable isotope Bayesian ellipses in R [SIBER, 37] implemented in the SIAR package [38]. The standard ellipse area (SEA) is the equivalent of standard deviation for bivariate data. SEA was corrected for sample size (SEAc), which is a robust approach when comparing small and unbalanced sample sizes. SEAB (Bayesian SEA) was calculated using 106 posterior draws to statistically compare niche width between ecotypes [37]. The degree of SEAc overlap between ecotypes was also estimated. Convex hull areas (polygons encompassing all the data points) were also displayed. As described for morphometric analyses, the mixture model-based clustering analysis in the mclust package was used to estimate the most likely number of clusters and assign individuals to each cluster. Individual assignment probabilities were compared to genetic ecotypes.
(iii). Stomach content analysis
Stomach content analysis (Ncoastal = 6, Npelagic = 24 for non-empty stomachs) was conducted to provide a quantitative description of diet and followed a standard procedure for marine top predators [39]. Following [40], analytical methods were based on the identification, quantification and measurements of prey remains, including fish otoliths and bones, cephalopod beaks and crustacean exoskeletons. The dietary importance of each prey species was described by its relative abundance (%N) and by ingested biomass (%M). Relative abundance was defined as the number of individuals of that species found throughout the sample. Biomass was calculated as the product of the average body mass and the number of individuals of the same species in each stomach, summed throughout the entire stomach set. Ninety-five per cent CI around the percentages by number and mass were generated for each prey taxon by bootstrap simulations of sampling errors [41]. The dietary overlap in mass (O) was obtained using the Pianka index [42], which varies from 0 (no overlap) to 1 (complete overlap); values greater than 0.5 are considered to reveal a high overlap.
3. Results
(a). Genetic inference of the population demographic history
We used a three-step procedure to identify the demographic scenario best describing the genetic diversity in the four dolphin populations (figure 2). Among the 11 scenarios tested in the first step (figure 2a), the model SC2 gave the highest fit with the observed data, with a Ppr of 64.7%, (95% CI: [62.6–66.7]). This scenario assumes that CS and PA populations diverged first from an ancestral population, followed by the split of the PM population from the PA population and the CN population from the CS population. The only other scenario receiving significant support, though much lower than SC2, was SC3 with a Ppr of 28%. This scenario assumes a symmetric hypothesis to SC2. All the other scenarios had a Ppr of less than 3%. Therefore, confidence in the SC2 scenario choice was strong. The evaluation of type I error rate (electronic supplementary material, table S2) showed that 68.6% of the datasets simulated with SC2 were correctly identified as being produced by SC2. The false-negative error rate was only observed with SC3 (16.8%) and with SC1 (9.4%). Estimation of the type II error (i.e. false positive) was also very low, especially when considering most of the alternative scenarios, with individual error rates lower than 5% (electronic supplementary material, table S2). The only scenario producing significant type II error rate was SC3, with 22.4% of PODs wrongly selected as being generated by SC2. Overall, excluding SC3, our analyses displayed a strong power (88%) to discriminate among the tested scenarios. A model checking of the goodness-of-fit of the scenario–posterior parameter distributions with the real dataset further indicated that SC2 was the best at reproducing observed SS values (electronic supplementary material, table S2).
Step b (figure 2) of the ABC analysis further refined the population tree (SC2) identified in step a. The scenario where PA is considered as the ancestral population from which CS split fitted the data much better (SC4, Ppr = 54%, 95% CI: [53.0–55.7]) than the scenario where both PA and CS split from the same common ancestral population (SC2, Ppr = 15.7%, 95% CI: [14.2–17.3]). This scenario (SC4, figure 2b) combined with its posterior parameter distributions also provides a better fit with the observed data (see model checking in the electronic supplementary material, table S3).
Step c in the ABC analysis (figure 2) aimed to assess the plausibility of a population bottleneck when each population split from its ancestral population. Of the five hypotheses tested (figure 2c), the scenarios assuming a bottleneck in the CS population (SC2, Ppr = 36.7%, 95% CI: [36.0–37.4]) or no bottleneck (SC1, Ppr = 35.7%, 95% CI: [35.0–36.5]) received the highest support, followed by the scenario assuming a bottleneck in the two coastal populations (SC3, Ppr = 17.7%, 95% CI: [17.0–18.3]). The scenarios assuming a bottleneck in the PM group (SC4) and in all groups (SC5) received significantly lower support (Ppr ≤ 5%; electronic supplementary material, table S4). However, the ABC analysis showed weak power to discriminate between the five scenarios and especially between the first three (electronic supplementary material, table S4). Interestingly, the scenario best able to reproduce the observed data was SC3, assuming a bottleneck in the two coastal populations (electronic supplementary material, tables S4 and S5).
Considering the two most likely scenarios (SC1 and SC2 in figure 2c) and assuming a generation time of 20 years [43], the splitting time between the CS and PA groups (t3, figure 2c) would be approximately 10 320 years BP (yrBP) (95% CI: [4300–47 800]), between PM and PA (t2) approximately 7580 yrBP (95% CI: [2340–22 600]) and between CS and CN (t1) approximately 2560 yrBP (95% CI: [830–6820]). Estimations of the effective population size were the highest in PA (12 200, 95% CI: [6360–14 700], followed by PM (4810, 95% CI: [1500–9200]), CS (2160, 95% CI: [864–3560]) and CN (1990, 95% CI: [678–3660]; electronic supplementary material, table S6).
(b). Morphometric analyses
The most likely number of clusters using morphometric data was one. Univariate and multivariate analyses, except the DFA, failed to discriminate ecotypes both when considering the whole dataset and also sexes separately. The DFA partially discriminated ecotypes, with 74% of dolphins correctly reassigned to their ecotypes (0.89 and 0.88 for males and females, respectively). When variables having the least weights were removed from the analysis, correct assignment rates decreased, highlighting the need for the complete set of variables to partially discriminate ecotypes.
(c). Stable isotope analyses
Pelagic dolphins had higher δ34S (mean ± s.d., 17.9 ± 0.7‰) and lower δ15N (14.2 ± 0.8‰) values than coastal dolphins (δ34S = 14.0 ± 1.0‰, δ15N = 15.7 ± 0.9‰, p < 0.01). There were no significant differences in δ13C values between the two ecotypes (δ13C = −16.2 ± 1.1‰ and −16.7 ± 0.6‰ for coastal and pelagic dolphins, respectively, p = 0.06). No statistical differences were detected between males and females. Isotopic niche spaces of the two ecotypes were distinct. There was no SEAc overlap when considering δ34S and δ13C, and δ34S and δ15N values (figure 3 and electronic supplementary material, figure S4a). Little overlap was found with δ13C and δ15N values (2.1% of the SEAc of the coastal ecotype overlapped with the SEAc of the pelagic ecotype and 5.9% of the SEAc of the pelagic ecotype overlapped with the SEAc of the coastal ecotype; electronic supplementary material, figure S4b). SEAB calculated using Bayesian inference indicated that pelagic dolphins had a narrower niche width than coastal dolphins for δ34S and δ13C, and δ13C and δ15N values (electronic supplementary material, figures S4a, S4b, S5a and S5c; p < 0.01). Niche widths of the two ecotypes were not significantly different when considering δ34S and δ15N values (figure 3 and electronic supplementary material, figure S5b; p = 0.07).
Figure 3.
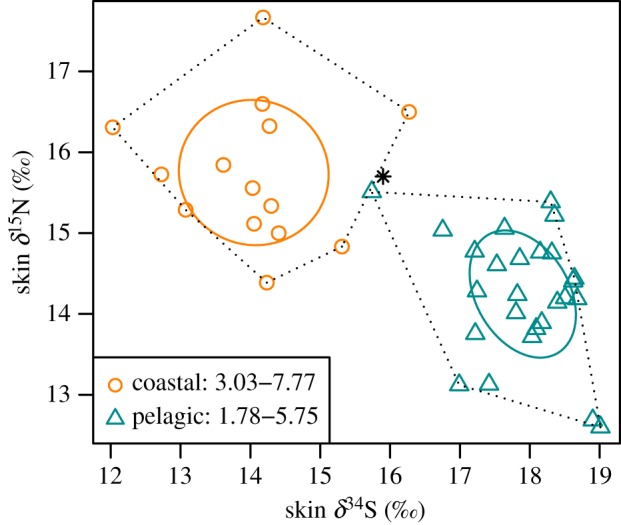
Skin δ34S and δ15N values for genetically determined coastal and pelagic bottlenose dolphins. Solid lines indicate SEAC and dotted lines convex hull areas. Their respective areas values (‰²) are given in the legend. Asterisk indicates the possible migrant (see text). (Online version in colour.)
The most likely number of clusters was two, with individuals assigned with high probability to each cluster (figure 4). The isotopic clustering exactly matched the genetic groups apart from one individual which was classified as coastal with SI analyses but was part of the pelagic genetic group. This individual was photo-identified with coastal resident dolphins in the English Channel over a 2-year period before its death.
Figure 4.
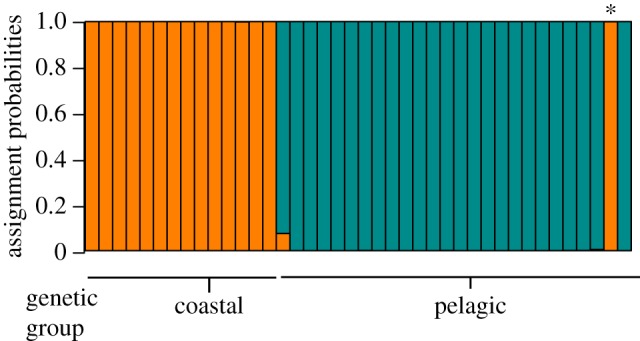
Bar graph of individual assignment probabilities to each of the two isotopic clusters and comparison with genetic groups. Each vertical bar represents one individual. Asterisk indicates the possible migrant (see text). (Online version in colour.)
(d). Stomach content analyses
Despite a large prey diversity (30 species including fish, cephalopods and shrimps), one fish species, hake (Merluccius merluccius), largely dominated the diet of pelagic dolphins with 54.6% of ingested biomass and 24.6% of the relative abundance (electronic supplementary material, table S7). Mackerel (Scomber scombrus) ranked second in terms of ingested biomass with 11.6%M. Then, four other species made up a significant proportion of the diet with a relative abundance of 18%N for blue whiting (Micromesistius poutassou), 10.7%N for pout (Trisopterus spp.), 10.7%N for sprat (Sprattus sprattus) and 10.5%N for scads (Trachurus spp.).
The diet of coastal dolphins appeared less diversified (14 species including fish, cephalopods and shrimps) although this could be linked to a lower sample size. Mullets and pout were the dominant prey items with, respectively, 29.8% and 31.1% of ingested biomass. Sandeels (Ammodytidae) ranked second in terms of relative abundance (33.7%N) but reached 5.2% of the ingested biomass. Thus, the diet of both pelagic and coastal bottlenose dolphins were largely dominated by fish species, however the prey-specific composition varied between the two ecotypes. The niche overlap calculated with the Pianka index is particularly low (0.11 by relative abundance and 0.16 by ingested biomass) strengthening the existence of dietary segregation between coastal and pelagic dolphins.
4. Discussion
(a). Ecologically driven demographic history of bottlenose dolphins in the North-east Atlantic
Ecological conditions likely played a major role in driving genetic divergence of bottlenose dolphins in the NEA. ABC demographic analyses showed that divergence times between coastal and pelagic, and between pelagic Atlantic and Mediterranean bottlenose dolphins were correlated with important historical environmental fluctuations. First, we confirmed the often suggested but never explicitly tested hypothesis of the founding of the coastal populations by the pelagic population [20,22]. The divergence between the two ecotypes likely occurred between the Last Glacial Maxima and the post-glacial period (10 320 yrBP, 95% CI: 4300–47 800). Therefore, the release of the continental shelf when sea ice retreated after 18 000 yrBP may have led to the colonization of coastal habitats by pelagic dolphins. In addition, although the analysis had relatively low power, this colonization was possibly achieved by a small number of individuals (i.e. a founder effect), which was a common pattern during post-glacial periods. More generally, the end of the glaciations in the Northern Hemisphere had a major impact on genetic diversity [12,44].
The divergence between pelagic Atlantic and West Mediterranean populations occurred later (7580 yrBP, 95% CI: 2340–22 600) likely during the Mediterranean ‘Sapropel period’, which was a nutrient-rich period characterized by the deposition of organic-rich sediments on the sea floor. These sediments were formed as a result of increased primary productivity and re-arrangements of water masses, linked to increased freshwater inputs generated by high precipitation rates [45,46]. While this phenomena was particularly intense in the Eastern Mediterranean Sea, other major oceanographic and biological changes occurred simultaneously in the Western part around 8000 yrBP, as a result of increased inflows of Atlantic waters [47]. These new environmental conditions might have created a productive trophic chain favourable for bottlenose dolphins. Interestingly, these conditions were also probably suitable for harbour porpoises (Phocoena phocoena), a small cetacean with high energetic needs. The end of the Sapropel period likely led to the fragmentation of harbour porpoise populations as waters became too oligotrophic and warm for this cold-water affiliated species [48,49]. By contrast, bottlenose dolphins, having a wider range and lower energetic costs [50] are still currently observed in the Mediterranean Sea.
These striking links between changes in environmental conditions and genetic divergences indicate that niche opportunities by the release of new habitats or changes in environmental conditions may be a major driver of genetic divergence, even in highly mobile animals.
By contrast, the separation between the two coastal populations was not linked to a particular climatic event (2560 yrBP, 95% CI: 830–6820). Although it will require further investigations, philopatry or natal-biased dispersal as a result of habitat-specific learned foraging techniques, together with social behaviour, might trigger genetic differentiation as suggested for bottlenose dolphins in the Gulf of Mexico and other mobile social mammals such as killer whales and wolves [4,19,21]. Another hypothesis could be the fragmentation of a coastal meta-population as suggested in [51], which showed that a genetically discrete population in the Humber estuary (east England) disappeared at least 100 years ago. This might be supported by the fact that effective population sizes for coastal populations estimated in DIYABC, which are averaged since their divergence, are 30–40 times larger than the ones obtained using LDNe and ONeSAMP which are based on the last few generations [22]. However, as these results might also be linked to methodological differences [23,24,52,53], these comparisons should be considered with caution and additional evidence is required.
(b). Niche specializations likely maintain genetic divergence between coastal and pelagic ecotypes
As bottlenose dolphins are a highly mobile species and the marine environment has no obvious barriers to gene flow, the opening of new coastal niches after the end of the last glacial period is not sufficient to explain the maintenance of genetic divergence between coastal and pelagic ecotypes. Using two complementary approaches, we showed that current ecological niches of pelagic and coastal bottlenose dolphins were highly segregated. SI signatures and prey species in stomach contents are consistent with a coastal versus pelagic habitat and diet segregation. δ13C and δ15N values are lower in offshore waters than in coastal waters, while δ34S values are higher [54,55]. In addition, prey species occurring in coastal waters are found exclusively in the diet of coastal dolphins, whereas species from the shelf-edge are only found in pelagic individuals. Moreover, δ13C and δ15N signatures of coastal and pelagic prey species were concordant with SI values found for the two ecotypes [54]. Prey species have not been analysed for sulfur isotopes.
The smaller isotopic niche width of pelagic dolphins is consistent with an offshore environment, typically more homogeneous than the mosaic of habitats in coastal areas. In addition, although prey species in pelagic dolphin stomach contents are diverse, they are dominated by large specimens of hake, which are mainly found along the shelf-edge. The main prey of both ecotypes are demersal, thus the main differences is the depth where they are found. Hence, we could hypothesize that different foraging strategies might be used and learned to feed in waters of different depth. Bottlenose dolphins might be philopatric or disperse in habitats similar to their natal ones, as they may be able to use vertically learned or culturally transmitted foraging strategies [56] and target familiar prey, which could enhance their foraging success. This hypothesis has been suggested for other social mammals [15], but rarely with direct evidence of diet/foraging segregation such as in our study (but see [57]). In addition, preferential associations with particular individuals that might be influenced by associations during juvenile life [58] may also reduce dispersal. Hence, ecological specializations at the population level strengthened by social context may maintain genetic divergence in this highly mobile mammal. However, further work is required to investigate niche specializations among populations within ecotypes using a larger sample size. In addition, stability in individual foraging specializations should be investigated using SI analyses in different dentin layers. Nevertheless, our study reinforces the potential of SIs to be a powerful tool in understanding ecologically driven cryptic genetic differentiation in a wide range of taxa [7,57].
Clustering analyses on SI data perfectly matched the genetic structure, except for one individual. This dolphin, which was photo-identified in a coastal area over a 2-year period, had coastal-like isotopic signatures but had been genetically identified as belonging to the pelagic group. Current migration rates are very low between ecotypes [22]. However, as haplotypes are shared between coastal and pelagic dolphins, this individual could possibly be a migrant. Despite niche segregation, some degree of behavioural plasticity might contribute to low levels of gene flow between ecotypes. Ecologically driven complete genetic isolation could be a long process that might never reach completion [10,59].
(c). Absence of strong influence of ecology on external morphological traits
In contrast to our results, pelagic and coastal bottlenose dolphins in other areas of the world showed strong morphological differences. In the North-east Pacific, skulls of coastal bottlenose dolphins had larger rostrum and teeth than pelagic individuals, which might be linked to contrasting diets [60]. In the North-west Atlantic (NWA), coastal individuals were smaller and had proportionally larger flippers than pelagic individuals inhabiting cold open-waters, possibly to provide more manoeuvrability in shallow estuaries or dissipate heat in warmer waters [61]. In addition, while coastal dolphins fed mainly on sciaenid fish, pelagic individuals fed on both fish and squid [62]. In the NEA, several hypotheses might explain the weak morphological differences. First, haplotype network and coalescent-based estimations of divergence times suggested that the differentiation between the two ecotypes occurred more recently in the NEA than in the NWA [22,63], giving less time for morphological divergence. Moreover, coastal and pelagic habitats might be less contrasted in the NEA than in the NWA. In the NWA, environmental conditions might be very different between shallow, enclosed and warm estuaries and cold pelagic waters. By contrast, in the NEA, coastal waters, at the northern range of the species, might be quite similar to pelagic waters in terms of temperature and currents, with the main difference being depth. In addition, both ecotypes fed close to the bottom. Thus, lower differences in ecological selective pressures might contribute to the lack of morphological differentiation. We could not rule out subtler differences that might not be detectable in our relatively small dataset. In addition, differences in skull morphological features should be investigated in the future (as in [60]).
(d). Possible differential stage of speciation in the North Atlantic
We showed that niche creations followed by niche specializations may be major drivers of ecotype differentiation in bottlenose dolphins. Our study emphasizes that understanding the forces shaping genetic and morphological divergences in highly mobile and cryptic animals is only possible thanks to a combination of evolutionary and ecological approaches. They provide complementary information on current and historical timescales. Similar multi-approach studies could help to shed light on divergence patterns in many other species.
At a large scale, bottlenose dolphins might show different stages of speciation throughout their North Atlantic distribution. The speciation process might be ongoing and at an early stage in the NEA and well advanced in the NWA regarding the complete mitochondrial lineage sorting and strong morphological differentiation [61,64]. Variations in the degree of habitat differentiation, contrasting divergence times or behavioural plasticity, may lead to different stages of ecologically driven genetic and morphological divergences for the same species across its range (e.g. for post-glacial fish and killer whales, [8,10,59]). We suggest that environmental opportunity to specialize may be the major factor driving ecological, genetic and morphological divergence.
Supplementary Material
Acknowledgements
We thank Gaël Guillou and Pierre Richard for SI analyses, Tamara Lucas for help with genetic laboratory work, Hélène Peltier for drift-modelling and Amélia Viricel for helpful discussions. We thank everyone that provided samples: Simon Berrow, Joanne O'Brien, Conor Ryan (IWDG, GMIT), Nigel Monaghan (National Museum of Ireland), Andrew Brownlow and Barry McGovern (SAC Inverness), Julie Béesau, Gill Murray-Dickson and Paul Thompson (University of Aberdeen), Rod Penrose (Marine Environmental Monitoring), François Gally (GECC), Réseau National Echouages, Fabien Demaret, Ghislain Doremus, Vincent Ridoux and Olivier Van Canneyt (Pelagis), Eric Alfonsi and Sami Hassani (Océanopolis), Pablo Covelo, Ruth Fernandez, Angela Llavona and Paula Mendez-Fernandez (CEMMA), Ruth Esteban, Pauline Gauffier and Philippe Verborgh (CIRCE), Renaud de Stephanis and Joan Giménez (EBD-CSIC) and Monica A. Silva (IMAR/DOP, WHOI).
Data accessibility
Microsatellite genotypes, mtDNA sequence alignment: Dryad doi:10.5061/dryad.57rr4. Haplotype GENBANK accession numbers: KF650783–KF650837. SI and morphometric data: Dryad doi:10.5061/dryad.v84n1. Stomach content data: electronic supplementary material, table S7.
Funding statement
Funding for sample collection was provided for: the UK by the Cetacean Strandings Investigation Programme funded by DEFRA and the devolved governments in Scotland and Wales; Ireland by National Marine Research Vessels Ship-Time Grant Aid Programme 2010 funded under the Science Technology and Innovation Programme of National Development Plan 2007–2013; Galicia by Direccion Xeral de Conservacion da Natureza-Xunta de Galicia, co-financed with European Regional Development Funds; Andalusia by LIFE ‘Conservación de Cetáceos y tortugas de Murcia y Andalucía’ (LIFE 02 NAT/E/8610); the Azores by TRACE (PTDC/MAR/74071/2006) and MAPCET (M2.1.2/F/012/2011); France by Ministry in charge of environment, Communauté d'Agglomération de la Ville de La Rochelle, Région Poitou-Charentes, Fondation Total and Agence de l'Eau Seine-Normandie.
References
- 1.Schluter D. 2001. Ecology and the origin of species. Trends Ecol. Evol. 16, 372–380. ( 10.1016/s0169-5347(01)02198-x) [DOI] [PubMed] [Google Scholar]
- 2.Mayr E. 1942. Systematics and the origin of species. New York, NY: Columbia University Press. [Google Scholar]
- 3.Dieckmann U, Doebeli M. 1999. On the origin of species by sympatric speciation. Nature 400, 354–357. ( 10.1038/22521) [DOI] [PubMed] [Google Scholar]
- 4.Musiani M, Leonard JA, Cluff HD, Gates C, Mariani S, Paquet PC, Vila C, Wayne RK. 2007. Differentiation of tundra/taiga and boreal coniferous forest wolves: genetics, coat colour and association with migratory caribou. Mol. Ecol. 16, 4149–4170. ( 10.1111/j.1365-294X.2007.03458.x) [DOI] [PubMed] [Google Scholar]
- 5.Hoelzel AR, Dahlheim M, Stern SJ. 1998. Low genetic variation among killer whales (Orcinus orca) in the eastern North Pacific and genetic differentiation between foraging specialists. J. Hered. 89, 121–128. ( 10.1093/jhered/89.2.121) [DOI] [PubMed] [Google Scholar]
- 6.Foote AD, Newton J, Piertney SB, Willerslev E, Gilbert MTP. 2009. Ecological, morphological and genetic divergence of sympatric North Atlantic killer whale populations. Mol. Ecol. 18, 5207–5217. ( 10.1111/j.1365-294X.2009.04407.x) [DOI] [PubMed] [Google Scholar]
- 7.Wolf JBW, Harrod C, Brunner S, Salazar S, Trillmich F, Tautz D. 2008. Tracing early stages of species differentiation: ecological, morphological and genetic divergence of Galapagos sea lion populations. BMC Evol. Biol. 8, 150 ( 10.1186/1471-2148-8-150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudsen R, Primicerio R, Amundsen PA, Klemetsen A. 2010. Temporal stability of individual feeding specialization may promote speciation. J. Anim. Ecol. 79, 161–168. ( 10.1111/j.1365-2656.2009.01625.x) [DOI] [PubMed] [Google Scholar]
- 9.Huber SK, De León LF, Hendry AP, Bermingham E, Podos J. 2007. Reproductive isolation of sympatric morphs in a population of Darwin's finches. Proc. R. Soc. B 274, 1709–1714. ( 10.1098/rspb.2007.0224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berner D, Roesti M, Hendry AP, Salzburger W. 2010. Constraints on speciation suggested by comparing lake-stream stickleback divergence across two continents. Mol. Ecol. 19, 4963–4978. ( 10.1111/j.1365-294X.2010.04858.x) [DOI] [PubMed] [Google Scholar]
- 11.Authier M, Cam E, Guinet C. 2011. Selection for increased body length in Subantarctic fur seals on Amsterdam Island. J. Evol. Biol. 24, 607–616. ( 10.1111/j.1420-9101.2010.02193.x) [DOI] [PubMed] [Google Scholar]
- 12.Hewitt GM. 2000. The genetic legacy of the Quaternary ice ages. Nature 405, 907–913. ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 13.Shikano T, Shimada Y, Herczeg G, Merila J. 2010. History vs. habitat type: explaining the genetic structure of European nine-spined stickleback (Pungitius pungitius) populations. Mol. Ecol. 19, 1147–1161. ( 10.1111/j.1365-294X.2010.04553.x) [DOI] [PubMed] [Google Scholar]
- 14.Johansson M, Primmer CR, Merila J. 2006. History vs. current demography: explaining the genetic population structure of the common frog (Rana temporaria). Mol. Ecol. 15, 975–983. ( 10.1111/j.1365-294X.2006.02866.x) [DOI] [PubMed] [Google Scholar]
- 15.Carmichael LE, Krizan J, Nagy JA, Fuglei E, Dumond M, Johnson D, Veitch A, Berteaux D, Strobeck C. 2007. Historical and ecological determinants of genetic structure in arctic canids. Mol. Ecol. 16, 3466–3483. ( 10.1111/j.1365-294X.2007.03381.x) [DOI] [PubMed] [Google Scholar]
- 16.Davis JM, Stamps JA. 2004. The effect of natal experience on habitat preferences. Trends Ecol. Evol. 19, 411–416. ( 10.1016/j.tree.2004.04.006) [DOI] [PubMed] [Google Scholar]
- 17.Lohmann KJ, Putman NF, Lohmann CMF. 2008. Geomagnetic imprinting: a unifying hypothesis of long-distance natal homing in salmon and sea turtles. Proc. Natl Acad. Sci. USA 105, 19 096–19 101. ( 10.1073/pnas.0801859105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawecki TJ, Ebert D. 2004. Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241. ( 10.1111/j.1461-0248.2004.00684.x) [DOI] [Google Scholar]
- 19.Sellas AB, Wells RS, Rosel PE. 2005. Mitochondrial and nuclear DNA analyses reveal fine scale geographic structure in bottlenose dolphins (Tursiops truncatus) in the Gulf of Mexico. Conserv. Genet. 6, 715–728. ( 10.1007/s10592-005-9031-7) [DOI] [Google Scholar]
- 20.Natoli A, Peddemors VM, Hoelzel AR. 2004. Population structure and speciation in the genus Tursiops based on microsatellite and mitochondrial DNA analyses. J. Evol. Biol. 17, 363–375. ( 10.1046/j.1420-9101.2003.00672.x) [DOI] [PubMed] [Google Scholar]
- 21.de Bruyn PJN, Tosh CA, Terauds A. 2013. Killer whale ecotypes: is there a global model? Biol. Rev. 88, 62–80. ( 10.1111/j.1469-185X.2012.00239.x) [DOI] [PubMed] [Google Scholar]
- 22.Louis M, et al. 2014. Habitat-driven population structure of bottlenose dolphins, Tursiops truncatus, in the North-east Atlantic. Mol. Ecol. 23, 857–874. ( 10.1111/mec.12653) [DOI] [PubMed] [Google Scholar]
- 23.Csilléry K, Blum MGB, Gaggiotti OE, Francois O. 2010. Approximate Bayesian computation (ABC) in practice. Trends Ecol. Evol. 25, 410–418. ( 10.1016/j.tree.2010.04.001) [DOI] [PubMed] [Google Scholar]
- 24.Beaumont MA, Zhang WY, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newsome SD, del Rio CM, Bearhop S, Phillips DL. 2007. A niche for isotopic ecology. Front. Ecol. Environ. 5, 429–436. ( 10.1890/060150.01) [DOI] [Google Scholar]
- 26.Browning NE, Dold C, I-Fan J, Worthy GAJ. 2014. Isotope turnover rates and diet-tissue discrimination in skin of ex situ bottlenose dolphins (Tursiops truncatus). J. Exp. Biol. 217, 214–221. ( 10.1242/jeb.093963) [DOI] [PubMed] [Google Scholar]
- 27.Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30, 1187–1189. ( 10.1093/bioinformatics/btt763) [DOI] [PubMed] [Google Scholar]
- 28.Cornuet JM, Santos F, Beaumont MA, Robert CP, Marin JM, Balding DJ, Guillemaud T, Estoup A. 2008. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24, 2713–2719. ( 10.1093/bioinformatics/btn514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornuet JM, Ravigne V, Estoup A. 2010. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinform. 11, 401 ( 10.1186/1471-2105-11-401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estoup A, Lombaert E, Marin JM, Guillemaud T, Pudlo P, Robert CP, Cornuet JM. 2012. Estimation of demo-genetic model probabilities with approximate Bayesian computation using linear discriminant analysis on summary statistics. Mol. Ecol. Resour. 12, 846–855. ( 10.1111/j.1755-0998.2012.03153.x) [DOI] [PubMed] [Google Scholar]
- 31.Gelman A, Carlin J, Stern H, Dunson D, Vehtari A, Rubin D. 2003. Bayesian data analysis. London, UK: CRC Press. [Google Scholar]
- 32.Fernandez R, Garcia-Tiscar S, Santos MB, Lopez A, Martinez-Cedeira JA, Newton J, Pierce GJ. 2011. Stable isotope analysis in two sympatric populations of bottlenose dolphins Tursiops truncatus: evidence of resource partitioning? Mar. Biol. 158, 1043–1055. ( 10.1007/s00227-011-1629-3) [DOI] [Google Scholar]
- 33.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 34.Dray S, Dufour AB. 2007. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20. [Google Scholar]
- 35.Fraley C, Raftery AE, Murphy TB, Scrucca L. 2012. mclust version 4 for R: normal mixture modeling for model-based clustering, classification, and density estimation. Technical Report No. 597. Department of Statistics, University of Washington.
- 36.Venables WN, Ripley BD. 2002. Modern applied statistics with S, 4th ed New York, NY: Springer. [Google Scholar]
- 37.Jackson AL, Inger R, Parnell AC, Bearhop S. 2011. Comparing isotopic niche widths among and within communities: SIBER—stable isotope Bayesian ellipses in R. J. Anim. Ecol. 80, 595–602. ( 10.1111/j.1365-2656.2011.01806.x) [DOI] [PubMed] [Google Scholar]
- 38.Parnell AC, Jackson AL. 2011. siar: stable isotope analysis in R. R package version 4.1.3 See http://www.CRAN.R-project.org/package=siar.
- 39.Pierce GJ, Boyle PR. 1991. A review of methods for diet analysis in piscivorous marine mammals. Oceanogr. Mar. Biol. 29, 409–486. [Google Scholar]
- 40.Spitz J, Rousseau Y, Ridoux V. 2006. Diet overlap between harbour porpoise and bottlenose dolphin: an argument in favour of interference competition for food? Estuar. Coast Shelf Sci. 70, 259–270. ( 10.1016/j.ecss.2006.04.020) [DOI] [Google Scholar]
- 41.Santos MB, Clarke MR, Pierce GJ. 2001. Assessing the importance of cephalopods in the diets of marine mammals and other top predators: problems and solutions. Fish. Res. 52, 121–139. ( 10.1016/s0165-7836(01)00236-3) [DOI] [Google Scholar]
- 42.Pianka ER. 1974. Niche overlap and diffuse competition. Proc. Natl Acad. Sci. USA 71, 2141–2145. ( 10.1073/pnas.71.5.2141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor BL, Chivers SJ, Larese J, Perrin WF. 2007. Generation length and percent mature estimates for IUCN assessments of cetaceans. Administrative Report LJ-07–01, National Marine Fisheries Service, Southwest Fisheries Science Center.
- 44.Bernatchez L, Wilson CC. 1998. Comparative phylogeography of nearctic and palearctic fishes. Mol. Ecol. 7, 431–452. ( 10.1046/j.1365-294x.1998.00319.x) [DOI] [Google Scholar]
- 45.Rohling EJ, Abu-Zied R, Casford CSL, Hayes A, Hoogakker BAA. 2009. The Mediterranean Sea: present and past. In Physical geography of the Mediterranean Basin (ed. Woodward JC.), pp. 33–67. Oxford, UK: Oxford University Press. [Google Scholar]
- 46.Calvert SE, Nielsen B, Fontugne MR. 1992. Evidence from nitrogen isotope ratios for enhanced productivity during formation of eastern Mediterranean sapropels. Nature 359, 223–225. ( 10.1038/359223a0) [DOI] [Google Scholar]
- 47.Rohling EJ, Dendulk M, Pujol C, Vergnaudgrazzini C. 1995. Abrupt hydrographic change in the Alboran Sea (Western Mediterranean) around 8000 yrs BP. Deep Sea Res. I Oceanogr. Res. Pap. 42, 1609–1619. ( 10.1016/0967-0637(95)00069-i) [DOI] [Google Scholar]
- 48.Fontaine MC, et al. 2014. Postglacial climate changes and rise of three ecotypes of harbour porpoises in western Palearctic waters. Mol. Ecol. 23, 3306–3321. ( 10.1111/mec.12817) [DOI] [PubMed] [Google Scholar]
- 49.Fontaine MC, et al. 2010. Genetic and historic evidence for climate-driven population fragmentation in a top cetacean predator: the harbour porpoises in European water. Proc. R. Soc. B 277, 2829–2837. ( 10.1098/rspb.2010.0412). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spitz J, Trites AW, Becquet V, Brind'Amour A, Cherel Y, Galois R, Ridoux V. 2012. Cost of living dictates what whales, dolphins and porpoises eat: the importance of prey quality on predator foraging strategies. PLoS ONE 7, e50096 ( 10.1371/journal.pone.0050096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nichols C, Herman J, Gaggiotti OE, Dobney KM, Parsons K, Hoelzel AR. 2007. Genetic isolation of a now extinct population of bottlenose dolphins (Tursiops truncatus). Proc. R. Soc. B 274, 1611–1616. ( 10.1098/rspb.2007.0176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tallmon DA, Koyuk A, Luikart G, Beaumont MA. 2008. ONeSAMP: a program to estimate effective population size using approximate Bayesian computation. Mol. Ecol. Resour. 8, 299–301. ( 10.1111/j.1471-8286.2007.01997.x) [DOI] [PubMed] [Google Scholar]
- 53.Waples RS, Do C. 2008. LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 8, 753–756. ( 10.1111/j.1755-0998.2007.02061.x) [DOI] [PubMed] [Google Scholar]
- 54.Chouvelon T, Spitz J, Caurant F, Mendez-Fernandez P, Chappuis A, Laugier F, Le Goff E, Bustamante P. 2012. Revisiting the use of δ15N in meso-scale studies of marine food webs by considering spatio-temporal variations in stable isotopic signatures: the case of an open ecosystem: the Bay of Biscay (North-east Atlantic). Prog. Oceanogr. 101, 92–105. ( 10.1016/j.pocean.2012.01.004) [DOI] [Google Scholar]
- 55.Peterson BJ, Fry B. 1987. Stable isotopes in ecosystem studies. Annu. Rev. Ecol. Syst. 18, 293–320. ( 10.1146/annurev.ecolsys.18.1.293) [DOI] [Google Scholar]
- 56.Cantor M, Whitehead H. 2013. The interplay between social networks and culture: theoretically and among whales and dolphins. Phil. Trans. R. Soc. B 368, 20120340 ( 10.1098/rstb.2012.0340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pilot M, Jedrzejewski W, Sidorovich VE, Meier-Augenstein W, Hoelzel AR. 2012. Dietary differentiation and the evolution of population genetic structure in a highly mobile carnivore. PLoS ONE 7, e39341 ( 10.1371/journal.pone.0039341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stanton MA, Gibson QA, Mann J. 2011. When mum's away: a study of mother and calf ego networks during separations in wild bottlenose dolphins (Tursiops sp.). Anim. Behav. 82, 405–412. ( 10.1016/j.anbehav.2011.05.026) [DOI] [Google Scholar]
- 59.Foote AD, et al. 2013. Tracking niche variation over millennial timescales in sympatric killer whale lineages. Proc. R. Soc. B 280, 20131481 ( 10.1098/rspb.2013.1481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perrin WF, Thieleking JL, Walker WA, Archer FI, Robertson KM. 2011. Common bottlenose dolphins (Tursiops truncatus) in California waters: cranial differentiation of coastal and offshore ecotypes. Mar. Mamm. Sci. 27, 769–792. ( 10.1111/j.1748-7692.2010.00442.x) [DOI] [Google Scholar]
- 61.Hersh SL, Duffield DA. 1990. Distinction of Northwestern Atlantic offshore and coastal bottlenose dolphins based on hemoglobin profile and morphometry. In The bottlenose dolphin (eds Leatherwood S, Reeves RR.), pp. 129–142. San Diego, CA: Academic Press. [Google Scholar]
- 62.Mead JC, Potter CW. 1995. Recognizing two populations of the bottlenose dolphin (Tursiops truncatus) of the Atlantic coast of North America - morphologic and ecologic considerations. IBI Reports, International Marine Biological Research Institute, Kamogawa, Japan 5, 31–44.
- 63.Moura AE, Nielsen SCA, Vilstrup JT, Moreno-Mayar JV, Gilbert MTP, Gray HWI, Natoli A, Möller L, Hoelzel AR. 2013. Recent diversification of a marine genus (Tursiops spp.) tracks habitat preference and environmental change. Syst. Biol. 62, 865–877. ( 10.1093/sysbio/syt051) [DOI] [PubMed] [Google Scholar]
- 64.Hoelzel AR, Potter CW, Best PB. 1998. Genetic differentiation between parapatric ‘nearshore’ and ‘offshore’ populations of the bottlenose dolphin. Proc. R. Soc. Lond. B 265, 1177–1183. ( 10.1098/rspb.1998.0416) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microsatellite genotypes, mtDNA sequence alignment: Dryad doi:10.5061/dryad.57rr4. Haplotype GENBANK accession numbers: KF650783–KF650837. SI and morphometric data: Dryad doi:10.5061/dryad.v84n1. Stomach content data: electronic supplementary material, table S7.