Abstract
The ‘dilution effect’ (DE) hypothesis predicts that diverse host communities will show reduced disease. The underlying causes of pathogen dilution are complex, because they involve non-additive (driven by host interactions and differential habitat use) and additive (controlled by host species composition) mechanisms. Here, we used measures of complementarity and selection traditionally employed in the field of biodiversity–ecosystem function (BEF) to quantify the net effect of host diversity on disease dynamics of the amphibian-killing fungus Batrachochytrium dendrobatidis (Bd). Complementarity occurs when average infection load in diverse host assemblages departs from that of each component species in uniform populations. Selection measures the disproportionate impact of a particular species in diverse assemblages compared with its performance in uniform populations, and therefore has strong additive and non-additive properties. We experimentally infected tropical amphibian species of varying life histories, in single- and multi-host treatments, and measured individual Bd infection loads. Host diversity reduced Bd infection in amphibians through a mechanism analogous to complementarity (sensu BEF), potentially by reducing shared habitat use and transmission among hosts. Additionally, the selection component indicated that one particular terrestrial species showed reduced infection loads in diverse assemblages at the expense of neighbouring aquatic hosts becoming heavily infected. By partitioning components of diversity, our findings underscore the importance of additive and non-additive mechanisms underlying the DE.
Keywords: dilution effect, diversity–disease relationship, biodiversity, Batrachochytrium dendrobatidis
1. Introduction
Biodiversity loss is happening at increasingly rapid rates and changing the distribution of organisms around the globe [1]. Biodiversity declines alter several features of communities, including the number of species (species richness), identity of species (species composition), their relative abundances (species evenness) and species interactions [2]. These features, in concert, drive key mechanisms responsible for ecosystem functioning, such as primary production, competition, predation and disease dynamics [3–7].
A number of recent studies have shown that declines in biodiversity can lead to increases in disease risk [7–10], but the underlying mechanisms leading to this pattern, and their generality, are complex and controversial [11–13]. This phenomenon of high host diversity reducing disease, termed the dilution effect (DE), can arise through several potential mechanisms by which diversity affects transmission, including encounter reduction, susceptible host regulation, infected host mortality and recovery augmentation (reviewed in [9]). These mechanisms have in common that both host interactions and the identity of the species influence the likelihood of transmission and disease. Encounter reduction—one clear mechanism that lowers transmission rates and thus leads to DE—operates in communities in a couple of ways. First, the proportion of susceptible and immune hosts may change with shifts in diversity, leading to reduced encounter rates between infected and uninfected hosts [9]. Second, diverse host communities may partition niche space more finely due to local adaptation, specialization and competition [14], and may in turn experience reduced encounter rates and transmission among conspecifics, and reduced pathogen spillover across host species [5]. Identifying the drivers of transmission most affected by biodiversity loss and their relative contribution to wildlife diseases has been challenging, despite their importance for effective wildlife management and disease forecasting. Without a clear understanding of these mechanisms, we will not be able to predict general patterns of disease dynamics in nature.
The mechanisms behind diversity–disease relationships are in many ways parallel to other important processes driving ecosystem functioning or performance [15–17]. Studies in the field of biodiversity–ecosystem function (BEF) demonstrate that a decrease in species diversity can reduce primary productivity [16–18] and increase herbivory [19] through a variety of additive and non-additive mechanisms. Within the BEF literature, additive mechanisms are those entirely driven by host composition, such that the ecological response of any species in a diverse assemblage can be predicted by its response in monoculture and its relative abundance in the mixed community [20]. Additive mechanisms also apply to diversity–disease relationships [5], and as defined, a necessary condition is that host species will respond identically to disease in single-host and mixed assemblages. The sampling effect is a common additive mechanism that applies to both BEF and DE [6,16]. It states that highly diverse assemblages have a higher probability of including at least one species with extreme ecological characteristics that can substantially affect ecological responses such as primary productivity [15,21] and/or pathogen transmission.
While additive mechanisms almost certainly play a role in disease dynamics, many important mechanisms for BEF and diversity–disease relationships are non-additive. Non-additive mechanisms occur when the ecological response of a given species in mixed assemblages cannot be predicted by how it responds in uniform populations. A non-additive mechanism commonly identified in the field of BEF arises due to interspecific differences in resource utilization with downstream effects on primary productivity [16]. Non-additive mechanisms also apply to diversity–disease relationships [5], and can result from host interactions and differential habitat use to alter disease response of host species in diverse assemblages. Two non-additive ecological mechanisms that have been widely studied in BEF, facilitation/inhibition and niche partitioning, are jointly referred to as ‘complementarity’ [17]. In facilitation/inhibition, heterospecific neighbours control damage to a particular plant species by attracting or repelling herbivores (e.g. associational susceptibility/resistance) [22]. In niche partitioning, species have complementary habitat use or resource utilization, and thus diversity often has a positive influence on primary productivity [3]. Although complementarity clearly applies to BEF studies on primary productivity, its potential role in DE is not intuitive, because host species do not complement each other in order to obtain higher or lower infection loads. However, parallel processes do exist. For example, differential habitat use among host species can affect disease risk if species diversity causes a reduction in niche overlap, and thereby decreases host contact rates and transmission.
In addition to complementarity, selection is a second component of diversity that can potentially affect diversity–disease relationships [5]. Selection measures the disproportionate impact of one particular species (or guild) in diverse assemblages compared with its performance in uniform populations [21], and therefore it can be driven by both additive and non-additive mechanisms [15,16]. Selection can occur due to one species's ability to become disproportionately less infected (thus becoming locally dominant over multiple generations) at the expense of neighbouring species becoming heavily infected (and becoming locally extinct) [16]. Therefore, selection does not depend just on species frequencies (the so-called sampling effect), but also on a variety of host species interactions (non-additive mechanisms) [16].
Here, we used an amphibian host–pathogen system to identify mechanisms underlying diversity–disease relationships. We partitioned the net effect of host diversity on pathogen infection loads and measured the relative contribution of ‘complementarity’ and ‘selection’ to disease risk. We experimentally exposed tropical amphibians to a panzootic strain of the chytrid fungus Batrachochytrium dendrobatidis (Bd) in single- and multi-host treatments. This epidermal pathogen has a broad host range among amphibians [23], and is implicated in population declines and species extinctions worldwide [24–27]. We used seven wild-collected tropical amphibian species that fall along a continuum of breeding mode and habitat use, ranging from fully terrestrial to mostly aquatic. Our specific goals were to (i) test whether the mechanisms of complementarity and selection, or a combination of both, drive diversity–disease relationship in our study system, and (ii) identify the contribution of species composition to pathogen dynamics. Our work demonstrates the application of principles of BEF to disease ecology. This perspective offers an accurate and fine-scale measurement of the effects of biodiversity on disease and enhances our mechanistic understanding of diversity–disease outcomes. Both of these goals are increasingly critical with the rapid anthropogenic acceleration of biodiversity loss.
2. Material and methods
(a). Host species
We captured adult anurans of seven locally abundant species in October 2012 from Parque Estadual da Serra do Mar—Núcleo Santa Virginia in the Brazilian Atlantic Forest (−23.35° S, −45.16° W). We assigned a host aquatic index (AI) to each species (adapted from [28]), which quantifies the amount of time spent in aquatic environments summed across different amphibian life stages. Because Bd is a water-borne fungal pathogen, AI also serves as a relative measure of species-specific exposure and transmission probability in natural communities [29,30]. Our seven focal host species ranged from exclusively terrestrial species (i.e. direct developers) occupying forest leaf-litter ((AI = 0) Brachycephalus pitanga (PIT) and Ischnocnema parva (PAR); Brachycephalidae), to species breeding in aquatic habitats but occupying the arboreal stratum ((AI = 1) Dendropsophus minutus (MIN), Scinax hayii (HAY) and Hypsiboas bandeirantes (BAN); Hylidae), to species breeding in aquatic habitats and occupying the margins of streams and other bodies of water ((AI = 2) Physalaemus cuvieri (CUV), Leptodactylidae; Hylodes phyllodes (PHY), Hylodidae). Thus, our focal taxa represent a full gradient of host AI found in the natural environment ranging from terrestrial (AI = 0) to highly aquatic (AI = 2).
(b). Focal pathogen
We investigated the effect of host diversity on dynamics of the amphibian chytrid fungus (Bd), which has caused population declines in hundreds of amphibian species in the Neotropics [24,25], North America [31], Europe [32] and Australia [24,26]. Bd can be transmitted by frog-to-frog contact, but it is also spread through host contact with aquatic environmental reservoirs [33]. Because many amphibian species share the same aquatic breeding sites (e.g. streams and ponds), interspecific transmission is likely, as free-living zoospores can be acquired from the water and initiate the infection of amphibian skin [34]. Therefore, the mode of transmission could influence process in both BEF and disease dynamics. For the present experiment, we used a global panzootic Bd strain (CLFT023) isolated from the Atlantic Coastal Forest, state of Minas Gerais [35].
(c). Experimental design
Each experimental unit consisted of a rectangular terrarium (40 × 29 × 13.5 cm) with terrestrial habitat at one end of the container (autoclaved moist Sphagnum) and aquatic habitat at the other end. To control for host density, each of our experimental units (single-host or multi-host) included four individual amphibians. We replicated single-host treatments four times for each of the seven host species, totalling 28 experimental units. We randomly assigned four unrepeated host species (species richness = 4) to each multi-host treatment, totaling 25 unique host assemblages.
To clear potential Bd infections from the field, we treated experimental animals with Itraconazole (0.01% solution) for 7 days [36] prior to beginning the experiment. A randomly selected subsample of hosts (n = 40) tested negative after the clearing protocol. For the experimental infection, we cultured Bd strain CLFT023 [35] in tryptone agar Petri plates at approximately 19°C for 7 days. We harvested Bd by flooding plates with distilled water and waiting for approximately 3 h for zoospores to release. We then pooled inoculum from plates, quantified zoospores with a haemocytometer and added 106 zoospores in 250 ml of dechlorinated water to the wet end of each experimental unit. This protocol ensured comparable exposure across replicates. We added the amphibians to the terrestrial habitat of each experimental unit and kept temperatures at 19.74°C ± 0.55 s.d. on a 12 L : 12 D cycle.
We monitored amphibians daily and fed them pinhead crickets (Gryllus cf. assimilis) ad libitum. We swabbed all individuals and terminated the experiment on the 18th day post-infection. This period encompasses approximately five replication cycles of Bd [34] and is sufficient for the pathogen to reach peak infections in susceptible amphibians [37,38]. During the course of the experiment, we swabbed dead or dying animals and removed them from the experimental units. We tested swabs for Bd in duplicate using Taqman qPCR [39,40] with standards of 0.1, 1, 10, 100 and 1000 zoospore genomic equivalents (GE) to determine the infection intensity of Bd in each individual host.
(d). Partitioning the effects of host diversity on Batrachochytrium dendrobatidis
A seminal paper by Loreau & Hector [16] described a quantitative framework for calculating the contribution of selection and complementarity to the net diversity effect on primary producers. This framework has been widely used in BEF studies to examine the impact of plant diversity on yields and herbivory [17–19]; to the best of our knowledge, this study is its first application to disease ecology. The net diversity effect (here redefined as net effect of host diversity) is calculated using the following equation:
where 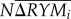 measures complementarity, Ncov(ΔRY, M) measures selection, N = number of host species in the multi-host assemblages, Mi = Bd infection loads of species i in single-host treatments and ΔRY = RYOi − RYEi = the deviation from expected relative Bd infection loads of species i in the multi-host assemblage [16].
measures complementarity, Ncov(ΔRY, M) measures selection, N = number of host species in the multi-host assemblages, Mi = Bd infection loads of species i in single-host treatments and ΔRY = RYOi − RYEi = the deviation from expected relative Bd infection loads of species i in the multi-host assemblage [16].
The complementarity component measures changes in the average infection in diverse host assemblages relative to weighted average loads of host species in uniform populations. Negative values imply that average infection loads are lower in multi-host assemblages than predicted by the infection loads of each component host species in single-host systems. Positive values indicate that average infection loads are higher in multi-host assemblages than predicted by the infection loads of each component species in single-host systems.
Selection, so named because its application is based on Price's general theory of selection, is measured by a covariance function [41]. In disease ecology, selection can occur when positive or negative diversity–disease relationships are leveraged by the disproportionate impact of one particular species (or guild) in diverse assemblages. Positive selection values indicate that species normally carrying high infection loads in uniform populations obtain even higher infection loads in multi-host assemblages at the same time as other species in the community become less infected or show minor changes in loads. By contrast, negative values for selection indicate that host species normally carrying high infection loads in uniform populations obtain disproportionately lower loads while in diverse assemblages. Because selection measures the disproportionate impact of a species in mixture compared with its performance in single-host treatments, it is expected that species composition (sampling effect) as well as host species interactions will impact this component of diversity. Selection and complementarity add up to the net effect of host diversity; in this paper, we maintain this terminology for consistency across fields.
(e). Statistical analyses
We compared average Bd infection intensity (log10 transformed) between diversity treatments (single-host and multi-host) and among the three categories of host AI using a standard least-square general linear model (GLM). We used stratified models with individuals nested within experimental units (nested ANOVA) [42,43]. We tested whether selection, complementarity and the net effect of host diversity were positive, negative or neutral by observing whether 95% confidence intervals overlapped zero. In addition, we used a t-test to compare average host AI between assemblages showing positive and negative complementarity or selection. We did not analyse prevalence data because more than 96% of the hosts became infected with Bd during the experiment. Mortality was low in both single-host (n = 3) and multi-host treatments (n = 6). Nonetheless, because mortality affects total host densities, which was otherwise controlled in our experiment, we repeated the analyses and quantified complementarity, selection and the net host diversity effect while excluding assemblages that experienced mortality.
3. Results
Bd infection loads were reduced by 66.5% in multi-host compared with single-host treatments (multi-host: least-square mean, LSM = 61.892 zoospore GEs; 1.171 logGE; single-host: LSM = 184.612 GE; 1.354 logGE; F = 4.434, p < 0.039; figure 1a). We found that this significant reduction in Bd infection loads in diverse host assemblages was driven by the complementarity component (figure 1b). Our measures of selection, however, showed both positive and negative values (of lower intensity) across our mixed host assemblages (figure 1b). Combined, complementarity and selection resulted in a net effect of host diversity reducing Bd infection loads (mean = −0.695 logGE; −0.040, –1.351 CI). These results remained unaltered after assemblages that experienced mortality were omitted from calculations (electronic supplementary material, table S1).
Figure 1.
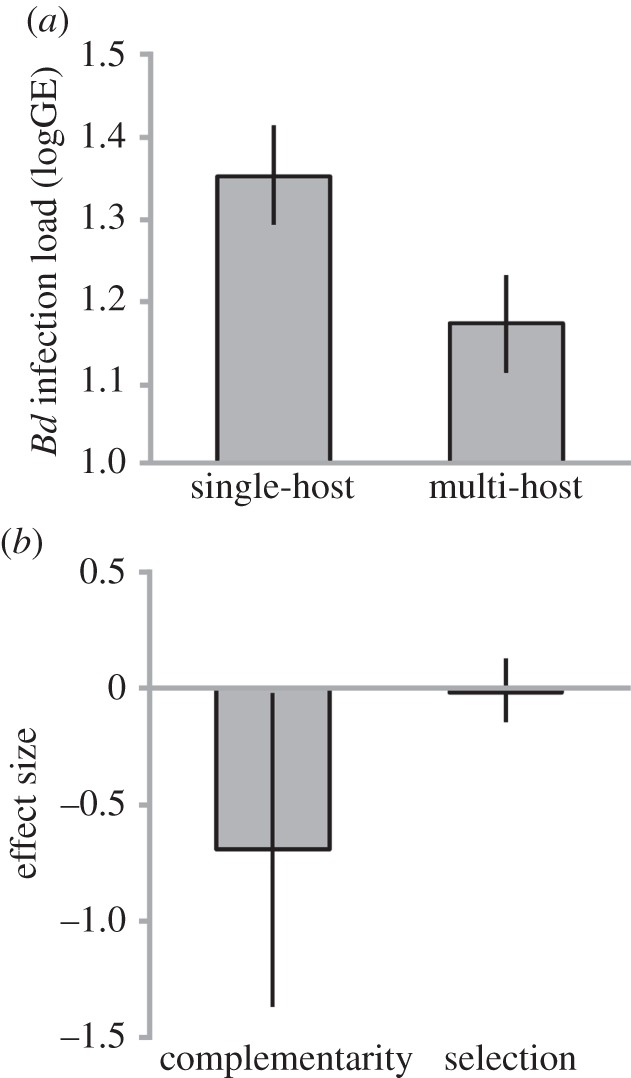
Effects of host diversity on Bd infection loads. (a) Average Bd infection loads in single- and multi-host treatments (mean ± 95% s.e.). (b) Net effect of host diversity partitioned into the two components complementarity and selection.
As expected, host species with a high AI carried higher Bd infection loads than terrestrial hosts independent of diversity treatment (F = 11.288, p = 0.001; figure 2). Furthermore, most host species showed a decrease in Bd infection loads in multi-host assemblages (figure 2). Our test for the effect of host identity on the observed DE showed no association between average host AI and strength of complementarity across diverse host assemblages (F = 0.003, p = 0.954). Conversely, host species composition significantly predicted the direction of selection. Specifically, multi-host assemblages where selection was positive were composed of species with lower host AI when compared with assemblages where selection was negative (F = 8.267, p = 0.008; electronic supplementary material, table S2; figure 3). This pattern was strongly influenced by the terrestrial host Brachycephalus pitanga, which showed a decrease in infection loads as neighbouring aquatic host species experienced pathogen amplification (figure 3).
Figure 2.
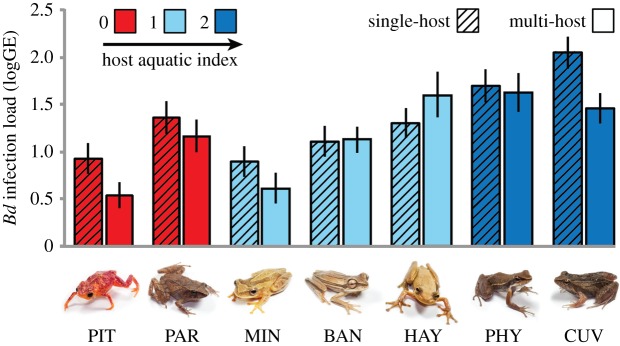
Average Bd infection load across single- and multi-host treatments (least-square mean ± s.e.). Species abbreviations: B. pitanga, PIT; I. parva, PAR; D. minutus, MIN; H. bandeirantes, BAN; S. hayii, HAY; H. phyllodes, PHY; P. cuvieri, CUV. Colours represent host AI ranging from fully terrestrial (AI = 0) to mostly aquatic (AI = 2).
Figure 3.
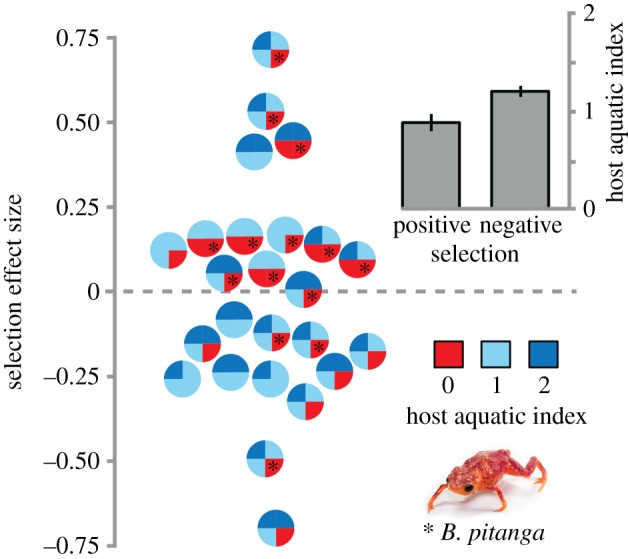
Selection component across 25 unique and equally diverse host assemblages. Bars represent average host AI (±95% s.e.) for assemblages experiencing positive and negative selection (graph jittering added to avoid overlapping data points). Assemblages containing the aposematic pumpkin toadlet (Brachycephalus pitanga) are highlighted with an asterisk. Colours represent host AI ranging from fully terrestrial (AI = 0) to mostly aquatic (AI = 2).
4. Discussion
Our experiment demonstrated that diversity can reduce Bd infection loads in amphibians through a non-additive mechanism that falls under the umbrella of complementarity (sensu BEF). Specifically, lower Bd transmission among host species due to reduced shared habitat use was the likely mechanism leading to the observed DE at the community level. Furthermore, the highly variable selection component in our experiment indicated that the presence of a particular host species in diverse assemblages can disproportionately increase or decrease infection loads in neighbouring hosts through direct association. Therefore, non-additive and additive mechanisms of diversity were tightly linked as drivers of chytridiomycosis in our experimental system.
Previous foundational work attributes the DE to a combination of additive and non-additive mechanisms [10]. Nevertheless, isolating the effect of biodiversity per se from the impact of host species composition is often a challenging task in natural systems due to the correlative nature of field-collected data. In many cases, anthropogenic habitat change is the proximate force selectively removing host species with high degrees of ecological specialization, and thus habitat generalist hosts often become dominant in depauperate communities [44–47]. In the case of Lyme disease, biodiversity loss promotes dominance of the habitat generalist Peromyscus leucopus, a highly competent host of the pathogen Borrelia burgdorferi [44]. For West Nile virus infections, host diversity and community competence are tightly negatively correlated, such that depauperate host communities are dominated by competent reservoirs [46]. Likewise, biodiversity loss increases transmission of the parasitic trematode Ribeiroia ondatrae because highly competent amphibian hosts dominate species-poor communities [47]. These studies found evidence for non-additive mechanisms (shifts in host species interactions) as well as a strong additive mechanism driving DE (numerical dominance of a competent host species). In our study system with randomly assembled host communities, diversity was negatively associated with disease risk when total host density was controlled for. We expect that the DE we observed experimentally would be even stronger in the wild if biodiversity loss in a real system simultaneously leads to changes in density and in community composition (e.g. by favouring superspreaders or species that induce continuous re-infection in neighbouring host species). Even though our experimental study does not perfectly mimic the natural assembly and disassembly of amphibian communities, it provides a quantitative framework for the relative contribution of two important components of diversity to disease dynamics.
By partitioning the net effect of host diversity, we quantified the relative contribution of complementarity and selection. The main mechanism leading the observed community-level DE falls under the umbrella of complementarity (sensu BEF) [16], where diversity per se led to lower infection loads in diverse assemblages. Niche theory predicts that species in diverse assemblages will compete for resources such as space, and thus benefit from reduced overlap in habitat use [14]. Therefore, lower niche overlap can cause both host encounter reduction and decreased exposure to the aquatic pathogen reservoirs such as Bd (reviewed in [9]), thus having a potential impact on both density- and frequency-dependent Bd transmission [48,49]. A second potential mechanism by which complementarity can lead to DE is inhibition through associational resistance among particular host species [22]. However, our community-level measures of Bd infection loads in diverse assemblages were not strongly driven by a particular combination of species, as we did not find an association between host composition and the intensity of complementarity.
Even though host species composition did not explain the intensity and direction of complementarity, we found it to be important in explaining selection. Specifically, we found positive values for selection in assemblages containing both terrestrial and aquatic hosts, and negative values in assemblages dominated by aquatic hosts (figure 3). This seems counterintuitive because terrestrial host species carry lower infection loads while in single-host treatments, and thus cannot be directly responsible for positive selection. However, the presence of the terrestrial aposematic pumpkin toadlet (Brachycephalus pitanga) may indirectly cause a disproportionate increase in infection loads of one or few neighbouring aquatic host species (figure 3). Brachycephalus pitanga secretes tetrodotoxin (a potent neurotoxin) from its skin, and it is possible that this species deterred more susceptible aquatic hosts from the dry terrestrial habitat, thus disproportionately increasing their exposure to Bd in the aquatic environment. Alternatively, pumpkin toadlets may compromise the ability of neighbouring hosts to fight infections through chemical interference [50] in a way similar to allelopathy in plants. According to our results, B. pitanga could potentially increase the likelihood of local extinction in aquatic hosts and, over multiple generations, become the dominant host species. We must highlight, however, that this final outcome of selection was not captured by our short-term experiment, as competitive exclusion could not take place during the length of our study. Nevertheless, the multiple aspects of host species interactions highlight the importance of measuring both complementarity and selection, allowing us to propose further hypotheses about potential mechanisms for species- and community-level processes leading to pathogen dilution or amplification.
Our results, combined with previous empirical laboratory studies [51,52], support the DE in amphibian–Bd systems. By contrast, our previous field-based empirical studies found strong support for Bd amplification [53]. Using field-collected data, we reported a positive relationship between amphibian species richness and Bd infection, after accounting for the effects of land cover and climate [53]. Two habitat generalist amphibians from Costa Rica (the rain frog, Craugastor fitzingeri) and Australia (the stony creek frog, Litoria lesueuri) showed higher Bd occurrence, prevalence and infection loads in diverse communities. We hypothesized that natural species-rich communities are more likely to include competent hosts for Bd than depauperate ones, increasing pathogen transmission. For instance, natural species-rich communities include a higher proportion of stream-dwelling specialists [54,55] that often carry higher Bd infection intensities in the wild [25,29]. Most host species in these diverse natural communities may have had a higher likelihood of suffering Bd spillover from the highly infected stream dwellers, such as species of Atelopus in Central America [25] and Taudactylus in Australia [56]. Combined, these findings illustrate that studies investigating diversity–disease relationships will show contrasting results when non-additive and additive effects of diversity are not quantified independently. Because observational field studies cannot fully disentangle the impact of species interactions from additive effects, laboratory-controlled experiments will continue to be important to understand mechanisms of species interactions driving wildlife diseases.
Global biodiversity is declining sharply, due in large part to anthropogenic habitat change and emerging diseases [4,57,58]. Therefore, understanding the mechanisms by which biodiversity alters disease dynamics can considerably advance the field of disease ecology and has important implications for conservation of natural populations. Our results indicate that shifts in host interactions and habitat use—both mechanisms of complementarity—can drive DE. In our study system, dilution was probably driven by interactions in diverse assemblages that reduced host contact rates and Bd transmission. Partitioning the net effect of host diversity on disease across several unique communities, rather than relying on the correlative effects of host species richness, evenness and composition, will allow us to identify specific mechanisms of diversity–disease relationships and test for their generality across host communities. Finally, our study shows that the application of methods from BEF can facilitate new avenues in experimental design and data analysis, with important theoretical implications for the field of disease ecology, and practical implications for understanding and predicting wildlife epidemics.
Supplementary Material
Acknowledgements
We thank A. Agrawal, S. McArt, S. Cook-Patton and B. Dalziel for feedback on experimental design and analyses; A. Agrawal, M. F. K. Becker, S. Claflin, S. McArt, S. Cook-Patton, two anonymous reviewers, and Zamudio and Leibold laboratory members for feedback on the manuscript; and R. Martins, M. Aguiar Passos, A. B. C. Lima, J. Ruggeri, D. Genari and T. A. Pires for field assistance.
Ethics statement
Research permits were provided by Instituto Chico Mendes da Conservação da Biodiversidade–Brazil (Permits 29964-3; 17242-3), Instituto Florestal do Estado de São Paulo (Permit 260108-010.479/2012), Universidade Estadual Paulista (UNESP) Comissão de Ética no Uso de Animal (Permit 7180), US Fish & Wildlife Services (Permit 2013MI1337329) and the Cornell University Institutional Animal Care and Use Committee (Protocol 2010-0069).
Data accessibility
Data available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.s4h7h.
Funding statement
Our work was funded by grants from the National Science Foundation (DEB-1209382 to C.G.B.; DEB-0542848 to K.R.Z.), Atkinson Center for a Sustainable Future (to C.G.B.), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Fulbright (grant 2157-08 to C.G.B.), Department of Ecology and Evolutionary Biology at Cornell University (to C.G.B.), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2011/51694-7 to L.F.T., 2012/04160-0 to C.L., 2008/50928-1 to C.F.B.H.) and Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (BJT 312895/2014-3 to C.G.B., L.F.T. and C.F.B.H.).
References
- 1.Pimm SL, Russell GJ, Gittleman JL, Brooks TM. 1995. The future of biodiversity. Science 269, 347–350. ( 10.1126/science.269.5222.347) [DOI] [PubMed] [Google Scholar]
- 2.Chapin FS, et al. 2000. Consequences of changing biodiversity. Nature 405, 234–242. ( 10.1038/35012241) [DOI] [PubMed] [Google Scholar]
- 3.Tilman D. 1999. The ecological consequences of changes in biodiversity: a search for general principles. Ecology 80, 1455–1474. [Google Scholar]
- 4.Daszak P, Cunningham AA, Hyatt AD. 2000. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287, 443–449. ( 10.1126/science.287.5452.443) [DOI] [PubMed] [Google Scholar]
- 5.Ostfeld RS, Thomas M, Keesing F. 2009. Biodiversity and ecosystem function: perspectives on disease. In Biodiversity, ecosystem functioning, and human wellbeing: an ecological and economic perspective (eds Naeem S. et al), pp. 209–216. New York, NY: Oxford University Press. [Google Scholar]
- 6.Johnson PTJ, Thieltges DW. 2010. Diversity, decoys and the dilution effect: how ecological communities affect disease risk. J. Exp. Biol. 213, 961–970. ( 10.1242/jeb.037721) [DOI] [PubMed] [Google Scholar]
- 7.Pagan I, González-Jara P, Moreno-Letelier A, Rodelo-Urrego M, Fraile A, Piñero D, García-Arenal F. 2012. Effect of biodiversity changes in disease risk: exploring disease emergence in a plant-virus system. PLoS Pathog. 8, e1002796 ( 10.1371/journal.ppat.1002796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobson A, Cattadori I, Holt RD, Ostfeld RS, Keesing F, Krichbaum K, Rohr JR, Perkins SE, Hudson PJ. 2006. Sacred cows and sympathetic squirrels: the importance of biological diversity to human health. PLoS Med. 3, 714–718. ( 10.1371/journal.pmed.0030231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keesing F, Holt RD, Ostfeld RS. 2006. Effects of species diversity on disease risk. Ecol. Lett. 9, 485–498. ( 10.1111/j.1461-0248.2006.00885.x) [DOI] [PubMed] [Google Scholar]
- 10.Ostfeld RS, Keesing F. 2012. Effects of host diversity on infectious disease. Annu. Rev. Ecol. Evol. Syst. 43, 157–182. ( 10.1146/annurev-ecolsys-102710-145022) [DOI] [Google Scholar]
- 11.Randolph SE, Dobson AD. 2012. Pangloss revisited: a critique of the dilution effect and the biodiversity-buffers-disease paradigm. Parasitology 139, 847–863. ( 10.1017/S0031182012000200) [DOI] [PubMed] [Google Scholar]
- 12.Wood CL, Lafferty KD. 2013. Biodiversity and disease: a synthesis of ecological perspectives on Lyme disease transmission. Trends Ecol. Evol. 4, 239–247. ( 10.1016/j.tree.2012.10.011) [DOI] [PubMed] [Google Scholar]
- 13.Ostfeld RS, Keesing F. 2013. Straw men don't get Lyme disease: response to Wood and Lafferty. Trends Ecol. Evol. 28, 502–503. ( 10.1016/j.tree.2013.05.009) [DOI] [PubMed] [Google Scholar]
- 14.MacArthur R, Levins R. 1967. The limiting similarity, convergence, and divergence of coexisting species. Am. Nat. 101, 377–385. ( 10.1086/282505) [DOI] [Google Scholar]
- 15.Tilman D, Lehman CL, Thomson KT. 1997. Plant diversity and ecosystem productivity: theoretical considerations. Proc. Natl Acad. Sci. USA 94, 1857–1861. ( 10.1073/pnas.94.5.1857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loreau M, Hector A. 2001. Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76. ( 10.1038/35083573) [DOI] [PubMed] [Google Scholar]
- 17.Cardinale BJ, et al. 2007. Impacts of plant diversity on biomass production increase through time because of species complementarity. Proc. Natl Acad. Sci. USA 104, 18 123–18 128. ( 10.1073/pnas.0709069104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook-Patton SC, McArt SH, Parachnowitsch AL, Thaler JS, Agrawal AA. 2011. A direct comparison of the consequences of plant genotypic and species diversity on communities and ecosystem function. Ecology 92, 915–923. ( 10.1890/10-0999.1) [DOI] [PubMed] [Google Scholar]
- 19.McArt SH, Thaler JS. 2013. Plant genotypic diversity reduces the rate of consumer resource utilization. Proc. R. Soc. B 280, 20130639 ( 10.1098/rspb.2013.0639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M. 2008. Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. ( 10.1111/j.1461-0248.2008.01179.x) [DOI] [PubMed] [Google Scholar]
- 21.Huston MA. 1997. Hidden treatments in ecological experiments: re-evaluating the ecosystem function of biodiversity. Oecologia 4, 449–460. ( 10.1007/s004420050180) [DOI] [PubMed] [Google Scholar]
- 22.Barbosa P, Hines J, Kaplan I, Martinson H, Szczepaniec A, Szendrei Z. 2009. Associational resistance and associational susceptibility: having right or wrong neighbors. Annu. Rev. Ecol. Evol. Syst. 40, 1–20. ( 10.1146/annurev.ecolsys.110308.120242) [DOI] [Google Scholar]
- 23.Olson DH, et al. 2013. Mapping the global emergence of Batrachochytrium dendrobatidis, the amphibian Chytrid fungus. PLoS ONE 8, e56802 ( 10.1371/journal.pone.0056802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger L, et al. 1998. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc. Natl Acad. Sci. USA 95, 9031–9036. ( 10.1073/pnas.95.15.9031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lips KR, Diffendorfer J, Mendelson JR, Sears MW. 2008. Riding the wave: reconciling the roles of disease and climate change in amphibian declines. PLoS Biol. 6, e72 ( 10.1371/journal.pbio.0060072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alford RA, Rowley JJL. 2008. The history of amphibian declines in Australia. In Threatened amphibians of the world (eds Stuart SN, et al.) pp. 72–73. Barcelona, Spain: Lynx Editions. [Google Scholar]
- 27.Wake DB, Vredenburg VT. 2008. Colloquium paper. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl Acad. Sci. USA 105, 11 466–11 473. ( 10.1073/pnas.0801921105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lips KR, Reeve JD, Witters LR. 2003. Ecological traits predict amphibian population declines in Central America. Conserv. Biol. 17, 1078–1088. ( 10.1046/j.1523-1739.2003.01623.x) [DOI] [Google Scholar]
- 29.Kriger KM, Hero JM. 2007. The chytrid fungus Batrachochytrium dendrobatidis is non-randomly distributed across amphibian breeding habitats. Divers. Distrib. 6, 781–788. ( 10.1111/j.1472-4642.2007.00394.x) [DOI] [Google Scholar]
- 30.Lips KR. 2006. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc. Natl Acad. Sci. USA 103, 3165–3170. ( 10.1073/pnas.0506889103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vredenburg VT, Knapp RA, Tunstall T, Briggs CJ. 2010. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc. Natl Acad. Sci. USA 107, 9689–9694. ( 10.1073/pnas.0914111107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker SF, et al. 2010. Factors driving pathogenicity vs. prevalence of amphibian panzootic chytridiomycosis in Iberia. Ecol. Lett. 13, 372–382. ( 10.1111/j.1461-0248.2009.01434.x) [DOI] [PubMed] [Google Scholar]
- 33.Rowley JJL, Alford RA. 2007. Behaviour of Australian rainforest stream frogs may affect the transmission of chytridiomycosis. Dis. Aquat. Organ. 77, 1–9. ( 10.3354/dao01830) [DOI] [PubMed] [Google Scholar]
- 34.Longcore JE, Pessier AP, Nichols DK. 1999. Batrachochytrium dendrobatidis gen et sp nov, a chytrid pathogenic to amphibians. Mycologia 91, 219–227. ( 10.2307/3761366) [DOI] [Google Scholar]
- 35.Schloegel LM, et al. 2012. Novel, panzootic and hybrid genotypes of amphibian chytridiomycosis associated with the bullfrog trade. Mol. Ecol. 21, 5162–5177. ( 10.1111/j.1365-294X.2012.05710.x) [DOI] [PubMed] [Google Scholar]
- 36.Pessier AP, Mendelson JR. 2010. A manual for control of infectious diseases in amphibian survival assurance colonies and reintroduction programs. Apple Valley, MN: IUCN/SSC Conservation breeding specialist group. [Google Scholar]
- 37.Longo AV, Burrowes PA, Joglar RL. 2010. Seasonality of Batrachochytrium dendrobatidis infection in direct-developing frogs suggests a mechanism for persistence. Dis. Aquat. Organ. 92, 253–260. ( 10.3354/dao02054) [DOI] [PubMed] [Google Scholar]
- 38.Savage AE, Zamudio KR. 2011. MHC genotypes associate with resistance to a frog-killing fungus. Proc. Natl Acad. Sci. USA 108, 16 705–16 710. ( 10.1073/pnas.1106893108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. 2004. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Organ. 60, 141–148. ( 10.3354/dao060141) [DOI] [PubMed] [Google Scholar]
- 40.Hyatt AD, et al. 2007. Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis. Dis. Aquat. Organ. 73, 175–192. ( 10.3354/dao073175) [DOI] [PubMed] [Google Scholar]
- 41.Price GR. 1970. Selection and covariance. Nature 227, 520–521. ( 10.1038/227520a0) [DOI] [PubMed] [Google Scholar]
- 42.Sokal RR, Rohlf FJ. 2000. Biometry: the principles and practice of statistics in biological research. New York, NY: W. H. Freeman. [Google Scholar]
- 43.SAS. 2012. JMP, version 10. Cary, NC: SAS Institute Inc. [Google Scholar]
- 44.LoGiudice K, Ostfeld RS, Schmidt K, Keesing F. 2003. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc. Natl Acad. Sci. USA 100, 567–571. ( 10.1073/pnas.0233733100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee KA, Wikelski M, Robinson WD, Robinson TR, Klasing KC. 2008. Constitutive immune defenses correlate with life-history variables in tropical birds. J. Anim. Ecol. 77, 356–363. ( 10.1111/j.1365-2656.2007.01347.x) [DOI] [PubMed] [Google Scholar]
- 46.Allan BF, et al. 2009. Ecological correlates of risk and incidence of West Nile virus in the United States. Oecologia 158, 699–708. ( 10.1007/s00442-008-1169-9) [DOI] [PubMed] [Google Scholar]
- 47.Johnson PTJ, Preston DL, Hoverman JT, Richgels KLD. 2013. Biodiversity decreases disease through predictable changes in host community competence. Nature 494, 230–233. ( 10.1038/nature11883) [DOI] [PubMed] [Google Scholar]
- 48.Dobson A. 2004. Population dynamics of pathogens with multiple host species. Am. Nat. 164, S64–S78. ( 10.1086/424681) [DOI] [PubMed] [Google Scholar]
- 49.Rachowicz LJ, Briggs CJ. 2007. Quantifying the disease transmission function: effects of density on Batrachochytrium dendrobatidis transmission in the mountain yellowlegged frog Rana muscosa. J. Anim. Ecol. 76, 711–721. ( 10.1111/j.1365-2656.2007.01256.x) [DOI] [PubMed] [Google Scholar]
- 50.Pires OR, Sebben A, Schwartz EF, Bloch C, Morales RAV, Schwartz CA. 2003. The occurrence of 11-oxotetrodotoxin, a rare tetrodotoxin analogue, in the Brachycephalidae frog Brachycephalus ephippium. Toxicon 42, 563–566. ( 10.1016/S0041-0101(03)00235-6) [DOI] [PubMed] [Google Scholar]
- 51.Searle CL, Biga LM, Spatafora JW, Blaustein AR. 2011. A dilution effect in the emerging amphibian pathogen Batrachochytrium dendrobatidis. Proc. Natl Acad. Sci. USA 108, 16 322–16 326. ( 10.1073/pnas.1108490108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venesky MD, Liu X, Sauer EL, Rohr JR. 2013. Linking manipulative experiments to field data to test the dilution effect. J. Anim. Ecol. 83, 557–565. ( 10.1111/1365-2656.12159) [DOI] [PubMed] [Google Scholar]
- 53.Becker CG, Zamudio KR. 2011. Tropical amphibian populations experience higher disease risk in natural habitats. Proc. Natl Acad. Sci. USA 108, 9893–9898. ( 10.1073/pnas.1014497108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becker CG, Fonseca CR, Haddad CFB, Batista RF, Prado PI. 2007. Habitat split and the global decline of amphibians. Science 318, 1775–1777. ( 10.1126/science.1149374) [DOI] [PubMed] [Google Scholar]
- 55.Becker CG, Fonseca CR, Haddad CFB, Prado PI. 2010. Habitat split as a cause of local population declines of amphibians with aquatic larvae. Conserv. Biol. 24, 287–294. ( 10.1111/j.1523-1739.2009.01324.x) [DOI] [PubMed] [Google Scholar]
- 56.Schloegel LA, et al. 2006. The decline of the sharp-snouted day frog (Taudactylus acutirostris): the first documented case of extinction by infection in a free-ranging wildlife species? Ecohealth 3, 35–40. ( 10.1007/s10393-005-0012-6) [DOI] [Google Scholar]
- 57.Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, Samuel MD. 2002. Climate warming and disease risks for terrestrial and marine biota. Science 296, 2158–2162. ( 10.1126/science.1063699) [DOI] [PubMed] [Google Scholar]
- 58.Keesing F, et al. 2010. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468, 647–652. ( 10.1038/nature09575) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.s4h7h.


