Abstract
The genes of the major histocompatibility complex (MHC) are a key component of the adaptive immune system and among the most variable loci in the vertebrate genome. Pathogen-mediated natural selection and MHC-based disassortative mating are both thought to structure MHC polymorphism, but their effects have proven difficult to discriminate in natural systems. Using the first model of MHC dynamics incorporating both survival and reproduction, we demonstrate that natural and sexual selection produce distinctive signatures of MHC allelic diversity with critical implications for understanding host–pathogen dynamics. While natural selection produces the Red Queen dynamics characteristic of host–parasite interactions, disassortative mating stabilizes allele frequencies, damping major fluctuations in dominant alleles and protecting functional variants against drift. This subtle difference generates a complex interaction between MHC allelic diversity and population size. In small populations, the stabilizing effects of sexual selection moderate the effects of drift, whereas pathogen-mediated selection accelerates the loss of functionally important genetic diversity. Natural selection enhances MHC allelic variation in larger populations, with the highest levels of diversity generated by the combined action of pathogen-mediated selection and disassortative mating. MHC-based sexual selection may help to explain how functionally important genetic variation can be maintained in populations of conservation concern.
Keywords: adaptive immunity, disassortative mating, genetic drift, natural selection, pathogen-mediated selection, sexual selection
1. Introduction
The major histocompatibility complex (MHC) includes the most variable loci in the vertebrate genome [1]. Classical MHC genes are an essential component of the adaptive immune system, responsible for the recognition and presentation of foreign antigens [2]. The peptide-binding regions (PBRs) of MHC loci are involved in antigen recognition, and the unique distribution of genetic variation at PBR sites is one of the textbook examples of balancing selection [3]. The MHC also shows considerable variation in genomic structure [1], with some species carrying dozens of copies of classical MHC genes [4], whereas others maintain a minimal MHC system consisting of individual loci [5,6]. The recent discovery of species that naturally lack key components of the MHC [7,8] suggests the possible existence of alternative genetic pathways associated with adaptive immune function.
Haldane [9] was the first to recognize the potential importance of host–parasite interactions in the evolutionary process, noting that individuals carrying rare allelic variants associated with pathogen resistance have a fitness advantage relative to conspecifics. This advantage is expected to decline as resistant alleles increase in frequency, owing to the concomitant increase of opportunistic pathogens, an idea that has been used to explain the Red Queen dynamics characteristic of host–parasite interactions [10]. As the front line in the adaptive immune process, MHC loci are subjected to strong selection by local pathogen communities, and negative-frequency dependence is thought to be the primary driver of MHC diversity [11]. Individuals heterozygous at MHC loci are thought to be able to recognize a broader array of pathogens, and may also be favoured by selection [12]. Selection dynamics may be even more complex when pathogens are spatially segregated [11,13], or if hosts carry multiple copies of MHC loci, where individual-level MHC diversity is thought to reflect a trade-off between immune protection and self-incompatibility [14,15].
There is a growing appreciation that sexual selection can also contribute to the unique patterns of variation observed at MHC loci [16], and may, in some cases, be more important than pathogen-mediated selection for the maintenance of functional diversity [17]. The fact that MHC loci are subjected to both natural and sexual selection may help to explain why MHC allelic diversity is so much greater than that observed at other immune loci [18]. Studies of congenic populations of laboratory mice provided the first evidence that individuals could discriminate MHC-based odour cues and use this information when making reproductive decisions [19,20], potentially enhancing genetic diversity via MHC-based disassortative mating. Selection may also extend after mating, via sperm competition and/or cryptic choice prior to fertilization [21,22] and/or sperm–egg incompatibility associated with MHC genotype (reviewed in [23]).
Although the mechanisms of MHC discrimination during reproduction remain poorly understood [24], evidence of the importance of MHC-based signals during mating is now available for a wide range of terrestrial and aquatic vertebrates [25]. The bulk of empirical evidence is consistent with preferences for MHC-dissimilar mates, which may limit inbreeding and increase MHC heterozygosity of progeny (reviewed in [16,26]), but preferences for MHC-diverse mating partners [27] and/or individuals carrying specific MHC alleles conveying parasite resistance [28] have also been found. Recent studies of the three-spine stickleback suggest that mate choice behaviour may be even more complex in species that carry multiple copies of MHC loci, where individual variation in mating preferences maintains intermediate levels of diversity in offspring (reviewed in [15]).
Unfortunately, while both natural and sexual selection probably contribute to the generation and maintenance of MHC diversity, the two mechanisms are expected to generate qualitatively similar patterns of variation, enhancing both allelic diversity and heterozygosity, something which has complicated efforts to discriminate their relative importance in wild populations [11,29]. Previous efforts to model the effects of selection on patterns of MHC variation have investigated the effects of natural and sexual selection in isolation, and have been unable to address the key question of how these two forms of selection are likely to interact in natural systems [30–33]. Such interactions may not only determine levels of MHC polymorphism maintained in populations, but also may impact the resistance of host populations to prevalent pathogen genotypes, for example when alleles conferring resistance to infection are disfavoured during mate choice.
The recent development of biologically realistic models of MHC dynamics has provided unique insights into the importance of selection and drift in maintaining MHC diversity. These models have consistently demonstrated the importance of negative-frequency-dependent selection, but indicate that heterozygote advantage alone is insufficient to explain the levels of genetic variation observed in natural systems [30,33,34], supporting earlier analytical work [35]. Here, we present the results of the first model of MHC evolution incorporating both survival and reproduction, modelling the combined effects of natural selection and MHC-based disassortative mating on patterns of MHC allelic diversity. Our results demonstrate that these two forms of selection produce distinct allele frequency spectra with important implications for understanding the relationship between population size and MHC diversity in natural systems.
2. Material and methods
(a). The model
Our model is based on the theoretical framework of Borghans et al. [30] and adapted from Ejsmond et al. [33,34], considering a population of sexually reproducing hosts (Ne = 50–10 000), with non-overlapping generations and a 1 : 1 sex ratio. Each host carried two MHC alleles represented as 16-bit binary strings, with bits corresponding to the amino acids involved in antigen binding and T cell presentation (approx. 18–20 peptide-binding sites have been identified at human MHC class I and II loci [2]). Mutations in host MHC molecules were simulated by micro-recombination (10−5 mutations per allele, per host generation), the dominant mode of MHC mutation [36]. The parameter settings of our model are based on extensive sensitivity analyses of earlier simulation models [30,33,34], and are designed to explore the full range of host–pathogen dynamics predicted under such conditions. Our model explicitly incorporates the two major explanations for the high genetic diversity observed at MHC loci: pathogen-mediated natural selection and disassortative mating during reproduction.
(b). Pathogen-mediated selection
Pathogen-mediated selection followed the design of earlier models by Ejsmond et al. [33,34]. Host populations coevolved with 25 haploid pathogen species, each with population size N equal to host Ne and a generation time 1/10 that of hosts. Each pathogen individual produced twenty 16-bit antigens. In each pathogen generation, each host was attacked by a randomly drawn individual of each of the 25 pathogen species. When at least 7/16 adjacent bits of at least one of the host's MHC alleles matched one or more of the 20 antigen sequences produced by a pathogen, the pathogen was presented, and the host mounted a successful defence. Consequently, a random MHC molecule had a 0.043 probability of recognizing a randomly generated antigen [33], a value in line with empirical estimates [37]. The number of antigens simultaneously presented did not affect the strength of the immune response.
Each new generation of pathogens was produced using pathogen haplotype frequencies proportional to the number of hosts infected in the previous generation. Pathogens reproduced asexually, with genetic variation generated by point mutations, which allowed each antigen bit to change to its reverse in each generation. Pathogen populations were derived under three mutation rates: 5 × 10−3, 2 × 10−3 or 10−3 per antigen, per pathogen generation. Previous analyses have demonstrated that model results are robust to alternative forms of host and pathogen mutation, including point mutation, recombination and/or randomly constituted alleles [30]. Pathogen mutation rates directly influence the potential for host–pathogen coevolution: at low mutation rates, pathogens may not evolve quickly enough to adapt to hosts, whereas at higher rates, random rather than host-adapted pathogen genotypes may appear [33,34]. Similar dynamics can be produced by manipulating host generation time relative to pathogens (data not shown).
The probability of host survival until reproduction was set to equal the proportion of pathogens successfully presented, and the immune response of a homozygote host bearing an allele capable of pathogen recognition was equal to that of a heterozygote carrying a single copy of the allele (i.e. codominance), such that heterozygote hosts were twice as likely to respond to a random pathogen (i.e. heterozygote advantage). The population of reproductive hosts was determined by drawing individuals with a sampling probability proportional to the fraction of presented pathogens, maintaining a constant effective population size. The sex of reproductive individuals was set randomly, so that the sex ratio remained at 1 : 1 throughout each generation of the simulation.
(c). Disassortative mating
Each surviving individual had K = 10 mating attempts. Sensitivity analyses varying this parameter indicated that the majority of individuals could find a suitable mating partner within their first 10 attempts, with the exception of small populations, where fewer potential mating partners were available for reproduction. During each round of mating, females and males were randomly paired and allowed to mate. Three scenarios of partner choice were implemented (random mating, soft assortative mating, hard assortative mating), according to the preference function developed by Hedrick [31]. Randomly mating individuals mated independent of MHC genotype, whereas the soft (h = 0) and hard (h = 1) forms of disassortative mating required that mating partners differed at one or both MHC alleles, respectively. Individuals that mated successfully were removed from the breeding pool. If individuals failed to find a preferred mate during their first nine pairings, they mated randomly during their final mating attempt.
Each mated pair produced four offspring genotypes according to Mendelian expectations, each with the same probability of recruitment to the host population in the next generation. The recruitment probability of offspring genotypes into the F1 population was determined by the timing of reproduction by their parents, according to a linearly decreasing function γ(i), where γ(i) indicates the recruitment probability of offspring conceived at ith mating occasion, a function which can be interpreted biologically as the relative decrease in litter size or offspring reproductive value at birth due to delayed reproduction [38]. γ(K) was set to 10−6 in our model, a biologically realistic form of disassortative mating that penalizes delayed reproduction, but avoids the possibility of population extinction if all individuals fail to find a preferred partner during mating trials.
Host populations evolved under three scenarios of partner choice performed with and without coevolving pathogens (denoted as Pon and Poff, respectively) for each combination of host/pathogen population size and pathogen mutation rates. All scenarios were initiated with a random population of hosts and pathogens, and evolved until they reached mutation–selection equilibrium (within 2500 host generations, i.e. 25 000 pathogen generations), after which allelic richness and the effective number of alleles (1/(1 − He)) [39] were calculated for each generation. Simulations were performed with Matlab 7.9 (MathWorks, Natick, MA). Summary data for all simulation conditions are provided in the electronic supplementary material.
3. Results
(a). Non-additive interactions of natural and sexual selection
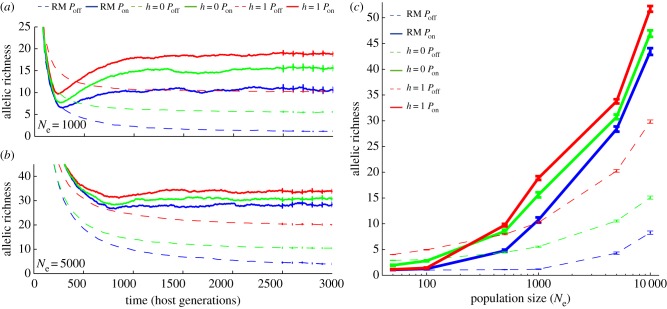
Simulated populations reached mutation–selection equilibrium after approximately 2500 host generations, after which allelic richness fluctuated around a stable mean (figure 1; electronic supplementary material, figure S1). The lowest levels of MHC variation were observed under a neutral model (random mating without pathogens, RM Poff; 1–8 alleles for Ne = 50–10 000; figure 1 and table 1), and whereas both natural selection (with pathogens, Pon) and disassortative mating (soft selection, h = 0; hard selection, h = 1; see Material and methods) enhanced MHC allelic richness (figure 1 and table 1), their combined effects were non-additive (figure 1).
Figure 1.
Natural and sexual selection act together to structure patterns of MHC diversity. MHC allelic richness under disassortative mating (RM, blue; h = 0, green; h = 1, red) and host–pathogen coevolution (Pon, solid; Poff, dashed) for (a) Ne = 1000 and (b) Ne = 5000 hosts. Error bars denote bootstrapped confidence limits (100 replicates) for the mean, calculated every 100 generations after simulations reached mutation–selection equilibrium. (c) The relative importance of natural and sexual selection is dependent on population size. Plots show mean allelic richness after equilibrium for host populations of variable size. Error bars denote bootstrapped confidence limits (100 replicates) for the mean, calculated for the final 500 generations after simulations reached mutation–selection equilibrium. The pathogen mutation rate was fixed to 2 × 10−3 per antigen per generation for all simulations shown.
Table 1.
The effect of pathogen-mediated selection and disassortative mating on allelic richness of MHC loci (n = 100 simulations; mean ±95% CI).
| random mating | |||||||
|---|---|---|---|---|---|---|---|
| pathogen mutation rate | Ne | 50 | 100 | 500 | 1000 | 5000 | 10 000 |
| no pathogens | 1.00 ± 0.00 | 1.01 ± 0.00 | 1.08 ± 0.03 | 1.18 ± 0.06 | 4.29 ± 0.22 | 8.27 ± 0.32 | |
| 10−3 | 1.05 ± 0.02 | 1.17 ± 0.05 | 2.48 ± 0.09 | 3.69 ± 0.14 | 11.78 ± 0.25 | 23.72 ± 0.55 | |
| 2 × 10−3 | 1.06 ± 0.04 | 1.24 ± 0.07 | 4.75 ± 0.24 | 10.23 ± 0.16 | 28.38 ± 0.52 | 43.44 ± 0.68 | |
| 5 × 10−3 | 1.33 ± 0.08 | 2.21 ± 0.15 | 15.69 ± 0.33 | 26.60 ± 0.43 | 54.85 ± 0.55 | 88.00 ± 0.64 | |
| soft disassortative mating (h = 0) | |||||||
|---|---|---|---|---|---|---|---|
| pathogen mutation rate | Ne | 50 | 100 | 500 | 1000 | 5000 | 10 000 |
| no pathogens | 2.90 ± 0.06 | 3.04 ± 0.02 | 4.44 ± 0.10 | 5.55 ± 0.12 | 10.55 ± 0.19 | 15.07 ± 0.32 | |
| 10−3 | 1.41 ± 0.10 | 2.52 ± 0.08 | 4.72 ± 0.18 | 7.08 ± 0.39 | 13.87 ± 0.32 | 26.20 ± 0.53 | |
| 2 × 10−3 | 1.94 ± 0.09 | 2.80 ± 0.09 | 8.53 ± 0.29 | 15.63 ± 0.53 | 30.74 ± 0.46 | 46.96 ± 0.63 | |
| 5 × 10−3 | 2.40 ± 0.09 | 3.96 ± 0.13 | 17.30 ± 0.26 | 27.90 ± 0.36 | 56.23 ± 0.47 | 88.05 ± 0.62 | |
| hard disassortative mating (h = 1) | |||||||
|---|---|---|---|---|---|---|---|
| pathogen mutation rate | Ne | 50 | 100 | 500 | 1000 | 5000 | 10 000 |
| no pathogens | 4.03 ± 0.02 | 4.96 ± 0.06 | 7.96 ± 0.12 | 10.23 ± 0.16 | 20.26 ± 0.26 | 29.81 ± 0.32 | |
| 10−3 | 1.06 ± 0.03 | 1.24 ± 0.08 | 4.29 ± 0.27 | 10.06 ± 0.33 | 17.36 ± 0.38 | 31.15 ± 0.59 | |
| 2 × 10−3 | 1.15 ± 0.06 | 1.41 ± 0.08 | 9.74 ± 0.26 | 10.74 ± 0.51 | 33.77 ± 0.41 | 51.70 ± 0.57 | |
| 5 × 10−3 | 1.66 ± 0.12 | 3.09 ± 0.22 | 19.12 ± 0.23 | 29.76 ± 0.26 | 58.07 ± 0.38 | 88.65 ± 0.51 | |
(b). Natural selection accelerates the loss of MHC diversity in small populations
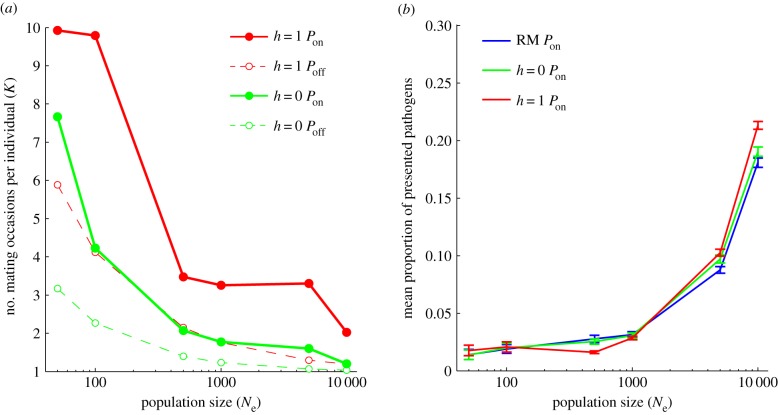
The relative importance of natural and sexual selection depends critically on Ne (figure 1c). While the number of mating attempts required to find a suitable partner increased at low Ne (figure 2a), the highest levels of MHC allelic richness in small populations were maintained by disassortative mating (h = 0 Poff and h = 1 Poff; figure 1c). Natural selection, in contrast, did not have an appreciable effect on allelic diversity in small populations, and was antagonistic to the effects of sexual selection (figure 1c), a pattern evident for all three pathogen mutation rates tested (electronic supplementary material, figure S2; table 1).
Figure 2.
The effectiveness of natural (Pon, Poff) and sexual selection (RM, h = 0, h = 1) is influenced by effective population size. (a) The intensity of sexual selection as measured by the mean number of mating attempts necessary for successful mating under soft (h = 0, green) and hard disassortative mating (h = 1, red) with (Pon, solid) and without pathogens (Poff, dashed). (b) The effect of disassortative mating (RM, blue; h = 0, green; h = 1, red) on the mean proportion of presented pathogens. Mean ± 95% CI values presented for the final 500 generations of simulated data. The pathogen mutation rate was set to 2 × 10–3 per antigen per generation.
Pathogen-mediated selection was the primary driver of MHC diversity in large populations (figure 1c and table 1). However, despite the increasing importance of pathogen-mediated selection at large Ne, disassortative mating significantly enhanced the immune response in larger populations (figure 2b), a pattern most evident at low to intermediate pathogen mutation rates, where negative-frequency-dependent selection was most efficient (electronic supplementary material, figure S3). As a result, the highest levels of MHC variation were always achieved via the combined effects of natural and sexual selection (table 1).
(c). Sexual selection and the Red Queen
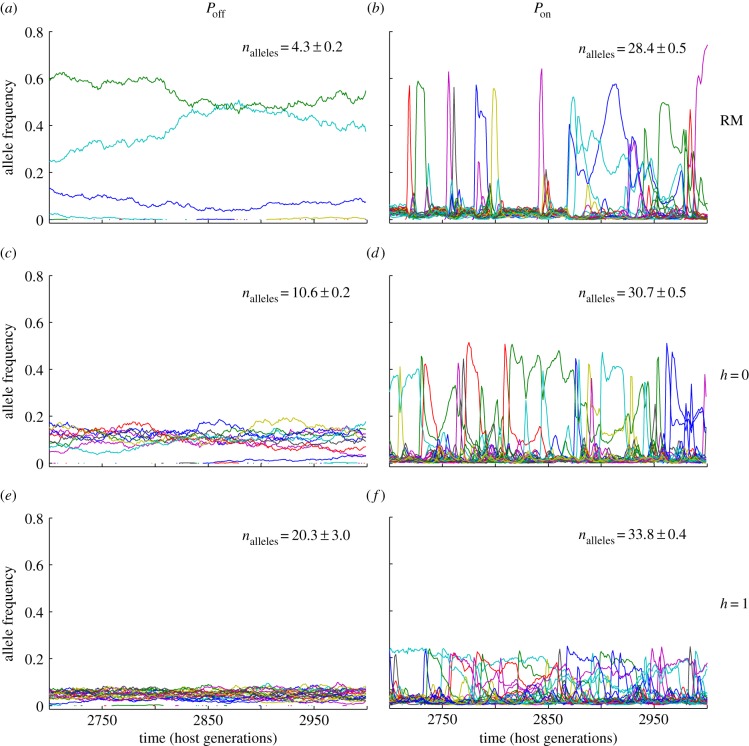
An analysis of the allele frequency spectrum under selection suggests an explanation for how natural and sexual selection act to shape MHC diversity (figure 3; electronic supplementary material, figure S4). While allele frequencies in the absence of selection followed a random walk, consistent with genetic drift (figure 3a), the presence of pathogen-mediated selection (RM Pon) produced the Red Queen dynamics characteristic of parasite–host interactions (figure 3b). The majority of host alleles were held at low frequencies by natural selection, but new MHC variants rapidly increased in frequency owing to the absence of effective pathogens (more than 68% of new alleles reached their maximum frequency within the first 50 generations after their origin; electronic supplementary material, figures S5 and S6). This dominance was short-lived, however, as opportunistic pathogens invaded the population, reducing the fitness advantages of the formerly dominant allele [33]. This boom–bust cycle was repeated over time, with new host alleles spiking in frequency in response to the pathogen community (figure 3b), significantly reducing the effective number of alleles [39] present at any point in time (electronic supplementary material, table S1).
Figure 3.
(a–f) Natural and sexual selection generate distinctive signatures on the allele frequency spectrum. Allele frequency variation under six scenarios of natural (Pon, Poff) and sexual selection (RM, h = 0, h = 1), for a host population of Ne = 5000 individuals. The pathogen mutation rate for all simulations was set to 2 × 10−3 per antigen per generation.
Sexual selection had a dramatically different effect on the allelic dynamics of host populations. In the absence of natural selection, disassortative mating selectively benefitted individuals carrying rare allelic variants, maintaining higher levels of allelic variation and stabilizing allele frequencies (figure 3c,e). This effect was also evident to a lesser extent when natural and sexual selection acted together (figure 3d,f). While the typical Red Queen dynamics associated with parasite–host interactions were still evident under such conditions (figure 3; electronic supplementary material, figure S4), major fluctuations in allele frequencies were buffered by the effects of disassortative mating, reducing the variability of allele frequencies over time.
4. Discussion
Our results indicate that disassortative mating with respect to MHC variation can be a potent evolutionary force, capable of maintaining levels of genetic variation in excess of those expected under pathogen-mediated selection alone. Disassortative mating is a form of balancing selection akin to heterozygote advantage, but one which selectively favours rare alleles, regardless of their fitness in host–pathogen interactions. By stabilizing allele frequency variation at equilibrium, disassortative mating creates the optimal conditions for the maintenance of functional genetic variation in small populations [40]. Empirical studies indicate that MHC variation is often eroded during population bottlenecks at a rate that equals or exceeds that of neutral loci [29,41], but exceptional cases have been documented in which MHC variation has been maintained in the face of severe declines in population size [42,43]. Analyses of the intensity of sexual selection in such populations could help to explain whether non-random mating during periods of population decline and recovery can contribute to the maintenance of MHC diversity.
While MHC-based disassortative mating produces a distinctive distribution of functionally important genetic variation at the population level (figure 3), it does not negatively impact average host resistance to pathogens, except in the cases of strong disassortative mating at intermediate population size (figure 2b). Instead, disassortative mating appears to often result in an even more effective immune response, an effect that is particularly evident in populations of large effective size (figure 2b). Recent work on sticklebacks [28] is consistent with a good-genes sexual selection mechanism, in which individuals exhibit preferences for specific MHC alleles conveying pathogen resistance [44]. Good-genes sexual selection has the potential to reinforce the Red Queen dynamics of host : pathogen coevolution, and the investigation of alternative forms of sexual selection would be an important area for future research.
Our model simulates disassortative mate choice, the best-documented form of MHC-based sexual selection, but it is important to note that the genetic consequences of disassortative mating should be identical whether non-random fertilization results from events that occur before or after copulation. Post-copulatory sexual selection may have an important influence on MHC allelic dynamics [21], even in species that show no MHC-based preferences during mate choice.
The distinct allele frequency spectra produced by natural and sexual selection suggest that it should, in principle, be possible to evaluate the relative importance of these two selective forces in natural systems through the examination of the dispersion of allele frequencies in empirical data. Disassortative mating produces a relatively flat distribution of allele frequencies, whereas pathogen-mediated selection produces a highly skewed allele frequency spectrum dominated by a small number of abundant alleles (figure 3). In practice, the application of this approach in a natural setting would require recurrent sampling and a stable pattern of selection over time, but such an analysis could yield important insights into the factors responsible for the exceptionally high diversities maintained at MHC loci. Long-term datasets including detailed pedigree information [27] may be particularly useful in testing model predictions and exploring the interaction of natural and sexual selection in shaping MHC diversity in wild populations.
Our model is the first to address how discrete phases of survival and reproduction can interact to shape patterns of MHC variation, but models only one form of sexual selection, and does not yet incorporate higher-level factors, which may also contribute to immune gene variation in natural systems. MHC loci are thought to evolve according to a birth–death process in which individuals gain and lose loci through homologous recombination and gene conversion [45], resulting in copy number variation both within [46] and among species [1]. Species that carry multiple copies of MHC loci must balance the selective advantages of allelic diversity in host–pathogen interactions with the potential costs of MHC diversity during T cell maturation [14]. Recent empirical work has demonstrated that species carrying multiple MHC loci show mating behaviour consistent with the advantages of intermediate levels of MHC diversity (reviewed in [15]). The incorporation of multiple loci into future models would help to explore the generality of our results in more complex genetic systems. There is now clear evidence that the composition of pathogen communities can vary substantially over both space and time, and that pathogen-mediated selection may be an unappreciated driver of local adaptation [11,13]. While our results are based on a temporally dynamic coevolution of hosts and pathogens, the development of spatially explicit models will be essential to explore the extent to which spatial segregation of pathogens can structure populations of their hosts, and contribute novel MHC variants through host migration and/or introgression [47].
The stabilizing effects of sexual selection may also help to explain the existence of trans-species MHC polymorphism, the apparent maintenance of ancient MHC variants over evolutionary time [48]. Despite the computational challenges associated with simulating spatially dynamic communities of hosts and pathogens, the modular nature of the present model offers a useful starting point for the testing of more complex hypotheses of MHC evolution. Ultimately, future models should allow natural and sexual selection to dynamically interact and change depending on the demographic structure of hosts and pathogens.
Finally, it has not escaped our attention that the combined effects of natural and sexual selection on MHC variants may help to explain how diverse communities of hosts and pathogens can be maintained over evolutionary time, even in the face of strong frequency-dependent selection favouring particular MHC genotypes. As the form of sexual selection described here favours rare alleles, regardless of their fitness in host–pathogen interactions, it reduces the probability of the loss of host genetic diversity by drift and/or directional selection, and is expected to be particularly important when hosts encounter pathogens presenting a higher diversity of antigens (data not shown). The maintenance of a genetically diverse population of hosts has effects that probably extend to local pathogen communities, and we are presently investigating how variation in selection pressures in host populations may influence the long-term dynamics of pathogen communities, an underexplored facet of host–pathogen evolution.
Supplementary Material
Acknowledgements
We thank Philip Hedrick and Filip Kapustka for comments on an earlier draft of the manuscript.
Data accessibility
The dataset used in the analyses presented here is available on Dryad (doi:10.5061/dryad.26k11), with a 12-month embargo period in place. The data and Matlab code used in the simulations are also available from the authors upon request.
Funding statement
This research was supported in part with funding from the Foundation for Polish Science ‘START’ and Jagiellonian University (DS/WBiNoZ/INoS 757/13) (M.J.E.) and the University of Zurich and City University of New York (A.B.W.), and by the Zurich HPC (C. Bollinger) and cluster computing (R. Murri) facilities.
References
- 1.Kelley J, Walter L, Trowsdale J. 2005. Comparative genomics of major histocompatibility complexes. Immunogenetics 56, 683–695. ( 10.1007/s00251-004-0717-7) [DOI] [PubMed] [Google Scholar]
- 2.Reche PA, Reinherz EL. 2003. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 331, 623–641. ( 10.1016/S0022-2836(03)00750-2) [DOI] [PubMed] [Google Scholar]
- 3.Hughes AL. 2007. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity 99, 364–373. ( 10.1038/sj.hdy.6801031) [DOI] [PubMed] [Google Scholar]
- 4.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. 2004. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 14, 1501–1515. ( 10.1101/gr.2134504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes CR, Miles S, Walbroehl JM. 2008. Support for the minimal essential MHC hypothesis: a parrot with a single, highly polymorphic MHC class IIB gene. Immunogenetics 60, 219–231. ( 10.1007/s00251-008-0287-1) [DOI] [PubMed] [Google Scholar]
- 6.Bahr A, Wilson AB. 2012. The evolution of MHC diversity: evidence of intralocus gene conversion and recombination in a single-locus system. Gene 497, 52–57. ( 10.1016/j.gene.2012.01.017) [DOI] [PubMed] [Google Scholar]
- 7.Star B, et al. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nat. Lond. 477, 207–210. ( 10.1038/nature10342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haase D, Roth O, Kalbe M, Schmiedeskamp G, Scharsack JP, Rosenstiel P, Reusch TBH. 2013. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: evidence from deep transcriptome sequencing. Biol. Lett. 9, 20130044 ( 10.1098/rsbl.2013.0044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haldane JBS. 1949. Disease and evolution. Suppl. Ric. Sci. 1949, 68–76. [Google Scholar]
- 10.Lively CM, Dybdahl MF. 2000. Parasite adaptation to locally common host genotypes. Nat. Lond. 405, 679–681. ( 10.1038/35015069) [DOI] [PubMed] [Google Scholar]
- 11.Spurgin LG, Richardson DS. 2010. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B 277, 979–988. ( 10.1098/rspb.2009.2084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penn DJ, Damjanovich K, Potts WK. 2002. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA 99, 11 260–11 264. ( 10.1073/pnas.162006499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eizaguirre C, Lenz TL, Traulsen A, Milinski M. 2009. Speciation accelerated and stabilized by pleiotropic major histocompatibility complex immunogenes. Ecol. Lett. 12, 5–12. ( 10.1111/j.1461-0248.2008.01247.x) [DOI] [PubMed] [Google Scholar]
- 14.Nowak MA, Tarczyhornoch K, Austyn JM. 1992. The optimal number of major histocompatibility complex molecules in an individual. Proc. Natl Acad. Sci. USA 89, 10 896–10 899. ( 10.1073/pnas.89.22.10896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woelfing B, Traulsen A, Milinski M, Boehm T. 2009. Does intra-individual major histocompatibility complex diversity keep a golden mean? Phil. Trans. R. Soc. B 364, 117–128. ( 10.1098/rstb.2008.0174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milinski M. 2006. The major histocompatibility complex, sexual selection, and mate choice. Annu. Rev. Ecol. Evol. Syst. 37, 159–186. ( 10.1146/annurev.ecolsys.37.091305.110242) [DOI] [Google Scholar]
- 17.Winternitz JC, Minchey SG, Garamszegi LZ, Huang S, Stephens PR, Altizer S. 2013. Sexual selection explains more functional variation in the mammalian major histocompatibility complex than parasitism. Proc. R. Soc. B 280, 20131605 ( 10.1098/rspb.2013.1605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penn DJ, Potts WK. 1999. The evolution of mating preferences and major histocompatibility complex genes. Am. Nat. 153, 145–164. ( 10.1086/303166) [DOI] [PubMed] [Google Scholar]
- 19.Yamazaki K, Boyse EA, Miké V, Thaler HT, Mathieson BJ, Abbott J, Boyse J, Zayas ZA, Thomas L. 1976. Control of mating preferences in mice by genes in the major histocompatibility complex. J. Exp. Med. 144, 1324–1335. ( 10.1084/jem.144.5.1324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki K, Yamaguchi M, Baranoski L, Bard J, Boyse EA, Thomas L. 1979. Recognition among mice. Evidence from the use of a y-maze differentially scented by congenic mice of different major histocompatibility types. J. Exp. Med. 150, 755–760. ( 10.1084/jem.150.4.755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Løvlie H, Gillingham MAF, Worley K, Pizzari T, Richardson DS. 2013. Cryptic female choice favours sperm from major histocompatibility complex-dissimilar males. Proc. R. Soc. B 280, 20131296 ( 10.1098/rspb.2013.1296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rülicke T, Chapuisat M, Homberger FR, Macas E, Wedekind C. 1998. MHC-genotype of progeny influenced by parental infection. Proc. R. Soc. Lond. B 265, 711–716. ( 10.1098/rspb.1998.0351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez N, Cooper J, Sprinks M, Abdelrahman M, Fiszer D, Kurpisz M, Dealtry G. 1999. A critical review of the role of the major histocompatibility complex in fertilization, preimplantation development and feto-maternal interactions. Hum. Reprod. Update 5, 234–248. ( 10.1093/humupd/5.3.234) [DOI] [PubMed] [Google Scholar]
- 24.Kwak J, Willse A, Preti G, Yamazaki K, Beauchamp GK. 2010. In search of the chemical basis for MHC odourtypes. Proc. R. Soc. B 277, 2417–2425. ( 10.1098/rspb.2010.0162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piertney SB, Oliver MK. 2006. The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21. [DOI] [PubMed] [Google Scholar]
- 26.Penn DJ. 2002. The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology 108, 1–21. ( 10.1046/j.1439-0310.2002.00768.x) [DOI] [Google Scholar]
- 27.Richardson DS, Komdeur J, Burke T, von Schantz T. 2005. MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc. R. Soc. B 272, 759–767. ( 10.1098/rspb.2004.3028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eizaguirre C, Yeates SE, Lenz TL, Kalbe M, Milinski M. 2009. MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol. Ecol. 18, 3316–3329. ( 10.1111/j.1365-294X.2009.04243.x) [DOI] [PubMed] [Google Scholar]
- 29.Sutton JT, Nakagawa S, Robertson BC, Jamieson IG. 2011. Disentangling the roles of natural selection and genetic drift in shaping variation at MHC immunity genes. Mol. Ecol. 20, 4408–4420. ( 10.1111/j.1365-294X.2011.05292.x) [DOI] [PubMed] [Google Scholar]
- 30.Borghans JAM, Beltman JB, De Boer RJ. 2004. MHC polymorphism under host–pathogen coevolution. Immunogenetics 55, 732–739. ( 10.1007/s00251-003-0630-5) [DOI] [PubMed] [Google Scholar]
- 31.Hedrick PW. 1992. Female choice and variation in the major histocompatibility complex. Genetics 132, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahata N, Nei M. 1990. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124, 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ejsmond MJ, Babik W, Radwan J. 2010. MHC allele frequency distributions under parasite-driven selection: a simulation model. BMC Evol. Biol. 10, 332 ( 10.1186/1471-2148-10-332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ejsmond MJ, Radwan J. 2011. MHC diversity in bottlenecked populations: a simulation model. Conserv. Genet. 12, 129–137. ( 10.1007/s10592-009-9998-6) [DOI] [Google Scholar]
- 35.Lewontin RC, Ginzburg LR, Tuljapurkar SD. 1978. Heterosis as an explanation for large amounts of genic polymorphism. Genetics 88, 149–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinsohn JT, Sousa AB, Guethlein LA, Howard JC. 1999. The gene conversion hypothesis of MHC evolution: a review. Immunogenetics 50, 168–200. ( 10.1007/s002510050593) [DOI] [PubMed] [Google Scholar]
- 37.Kast WM, Brandt RM, Sidney J, Drijfhout JW, Kubo RT, Grey HM, Melief CJ, Sette A. 1994. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J. Immunol. 152, 3904–3912. [PubMed] [Google Scholar]
- 38.Ejsmond MJ, Czarnołęski M, Kapustka F, Kozłowski J. 2010. How to time growth and reproduction during the vegetative season: an evolutionary choice for indeterminate growers in seasonal environments. Am. Nat. 175, 551–563. ( 10.1086/651589) [DOI] [PubMed] [Google Scholar]
- 39.Kimura M, Crow JF. 1964. The number of alleles that can be maintained in a finite population. Genetics 49, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson A. 1962. Selection for heterozygotes in small populations. Genetics 47, 1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radwan J, Biedrzycka A, Babik W. 2010. Does reduced MHC diversity decrease viability of vertebrate populations? Biol. Conserv. 143, 537–544. ( 10.1016/j.biocon.2009.07.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilar A, Roemer G, Debenham S, Binns M, Garcelon D, Wayne RK. 2004. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl Acad. Sci. USA 101, 3490–3494. ( 10.1073/pnas.0306582101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliver MK, Piertney SB. 2012. Selection maintains MHC diversity through a natural population bottleneck. Mol. Biol. Evol. 29, 1713–1720. ( 10.1093/molbev/mss063) [DOI] [PubMed] [Google Scholar]
- 44.Hamilton WD, Zuk M. 1982. Heritable true fitness and bright birds: a role for parasites? Science 218, 384–387. ( 10.1126/science.7123238) [DOI] [PubMed] [Google Scholar]
- 45.Nei M, Rooney AP. 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39, 121–152. ( 10.1146/annurev.genet.39.073003.112240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lighten J, van Oosterhout C, Paterson IG, McMullan M, Bentzen P. 2014. Ultra-deep Illumina sequencing accurately identifies MHC class IIb alleles and provides evidence for copy number variation in the guppy (Poecilia reticulata). Mol. Ecol. Resour. 14, 753–767. ( 10.1111/1755-0998.12225) [DOI] [PubMed] [Google Scholar]
- 47.Nadachowska-Brzyska K, Zieliński P, Radwan J, Babik W. 2012. Interspecific hybridization increases MHC class II diversity in two sister species of newts. Mol. Ecol. 21, 887–906. ( 10.1111/j.1365-294X.2011.05347.x) [DOI] [PubMed] [Google Scholar]
- 48.Klein J, Sato A, Nikolaidis N. 2007. MHC, TSP, and the origin of species: from immunogenetics to evolutionary genetics. Annu. Rev. Genet. 41, 281–304. ( 10.1146/annurev.genet.41.110306.130137) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset used in the analyses presented here is available on Dryad (doi:10.5061/dryad.26k11), with a 12-month embargo period in place. The data and Matlab code used in the simulations are also available from the authors upon request.