Abstract
Despite the demonstrated functional importance of gut microbes, our understanding of how animals regulate their metabolism in response to nutritionally beneficial symbionts remains limited. Here, we elucidate the functional importance of the African cotton stainer's (Dysdercus fasciatus) association with two actinobacterial gut symbionts and subsequently examine the insect's transcriptional response following symbiont elimination. In line with bioassays demonstrating the symbionts' contribution towards host fitness through the supplementation of B vitamins, comparative transcriptomic analyses of genes involved in import and processing of B vitamins revealed an upregulation of gene expression in aposymbiotic (symbiont-free) compared with symbiotic individuals; an expression pattern that is indicative of B vitamin deficiency in animals. Normal expression levels of these genes, however, can be restored by either artificial supplementation of B vitamins into the insect's diet or reinfection with the actinobacterial symbionts. Furthermore, the functional characterization of the differentially expressed thiamine transporter 2 through heterologous expression in Xenopus laevis oocytes confirms its role in cellular uptake of vitamin B1. These findings demonstrate that despite an extracellular localization, beneficial gut microbes can be integral to the host's metabolic homeostasis, reminiscent of bacteriome-localized intracellular mutualists.
Keywords: nutritional symbiosis, mutualism, symbiont transmission, vitamin supplementation, host–microbe metabolic integration
1. Introduction
Insects derive a range of nutritional benefits from their microbial partners. These can include the supply of essential nutrients, as well as aiding in the detoxification and digestion of ingested food [1]. As a result, many nutritional partnerships are considered to have played a significant role in the ecological diversification of insects by mediating the host's subsistence on novel, and often nutritionally deficient, food sources [2].
Like other animals, insects lack the metabolic pathways for the synthesis of several vital nutritional compounds, including 10 essential amino acids, as well as most vitamins [2,3]. Hence, insects can achieve complete nourishment only by acquiring the missing nutrients either from ingested food sources or through symbiotically mediated supplementation involving microbial partners [4–6].
In bugs of the family Pyrrhocoridae (Hemiptera), recent characterization of the microbial community revealed a consistent and conserved microbiota, with the notable co-occurrence of two actinobacterial taxa belonging to the Coriobacteriaceae family (Coriobacterium glomerans and Gordonibacter sp.) [7,8]. Similar to other heteropteran insects [9,10], firebugs rely on an extracellular post-hatch mechanism for the vertical transmission of their actinobacterial symbionts. This involves the deposition of symbiont-containing droplets by adult females over newly laid eggs, and the subsequent probing and uptake of the symbionts by the hatched nymphs [11]. In the European firebug (Pyrrhocoris apterus) and the African cotton stainer (Dysdercus fasciatus) (both Pyrrhocoridae), egg surface sterilization specifically results in Coriobacteriaceae-free individuals (aposymbionts) [11] that, relative to their symbiotic counterparts, exhibit slower growth rates, higher mortality and lower reproductive success [8], suggesting an essential function of the actinobacterial symbionts towards their insect hosts. Re-infection of previously sterilized eggs with the Coriobacteriaceae symbionts (via gut suspensions) completely rescues firebug survivorship and reproductive potential [8].
Dysdercus species, like many pyrrhocorids, are specialized feeders that exhibit a strong preference for seeds of the plant order Malvales, particularly cottonseeds [12]. Given the rich protein content of many Malvales seeds, the putative nutritional provisions by the symbionts have been suggested to lie not in the supply of essential amino acids—as with aphids and other sap-feeding insects—but rather in the supplementation of B vitamins [7,8], which were described as limiting in many seed-based diets, including those of the Malvales plant order (e.g. cotton) [13]. Here, cottonseed hulls were found to be an especially poor source of B vitamins [13].
In this study, we used an integrative approach to study both host and symbiont contributions towards vitamin production, transport and processing in cotton stainers, in order to gain comprehensive insights into the metabolic integration of extracellular gut symbionts in an insect host. To this aim, we first investigate the possible nutritional role of the Coriobacteriaceae symbionts towards D. fasciatus using a defined artificial diet that allows for omission of specific nutrients. Through bioassays that combine symbiont elimination with direct manipulations of the host's diet, we test the hypothesis that the actinobacterial symbionts provide D. fasciatus with B vitamins. These experiments are complemented by genomic analyses of C. glomerans' potential for vitamin production. Furthermore, we examine the effect of symbiont elimination on the midgut transcriptome of cotton stainers, specifically with respect to the expression of B vitamin transporters and downstream processing genes, with the expectation that these candidate genes would be differentially regulated in case of vitamin deficiency. Our findings provide a detailed description for symbiotic contributions of the Coriobacteriaceae towards pyrrhocorid bugs and shed light on a highly dependent and integrated nutritional partnership featuring extracellularly localized gut symbionts.
2. Results
(a). Success of symbiont elimination procedure
In order to manipulate symbiont infection status, D. fasciatus eggs were surface-sterilized by ethanol and bleach treatment [8]. This procedure consistently resulted in aposymbiotic firebugs that were free of both Coriobacteriaceae symbionts, as confirmed by C. glomerans- and Gordonibacter-specific diagnostic PCRs. Conversely, symbiotic bugs tested positive for both bacterial species.
(b). Fitness of symbiotic and aposymbiotic Dysdercus fasciatus on complete and B vitamin-deficient artificial diets
To investigate the possibility of symbiont-mediated supplementation of B vitamins, we examined the survivorship of aposymbiotic and symbiotic bugs reared on complete and B vitamin-deficient artificial diets, respectively, resulting in four treatment groups. When reared on a complete artificial diet containing all B vitamins in sufficient quantities, firebug survivorship until adulthood was consistently high, irrespective of symbiont presence or absence (figure 1a). Equivalent survivorship rates were observed for symbiotic bugs reared on a B vitamin-deficient diet (figure 1a). However, aposymbiotic bugs reared on a vitamin-deficient diet suffered significantly higher mortality compared with all other treatments (Friedman test, p = 0.012, with Wilcoxon–Wilcox post hoc measures, p < 0.05) (figure 1a). While similar trends were also observed for the developmental rates across treatments, with vitamin omission from the artificial diet increasing the time needed to reach adulthood of aposymbiotic individuals, the differences were not significant (Friedman test, p = 0.129) (figure 1b).
Figure 1.
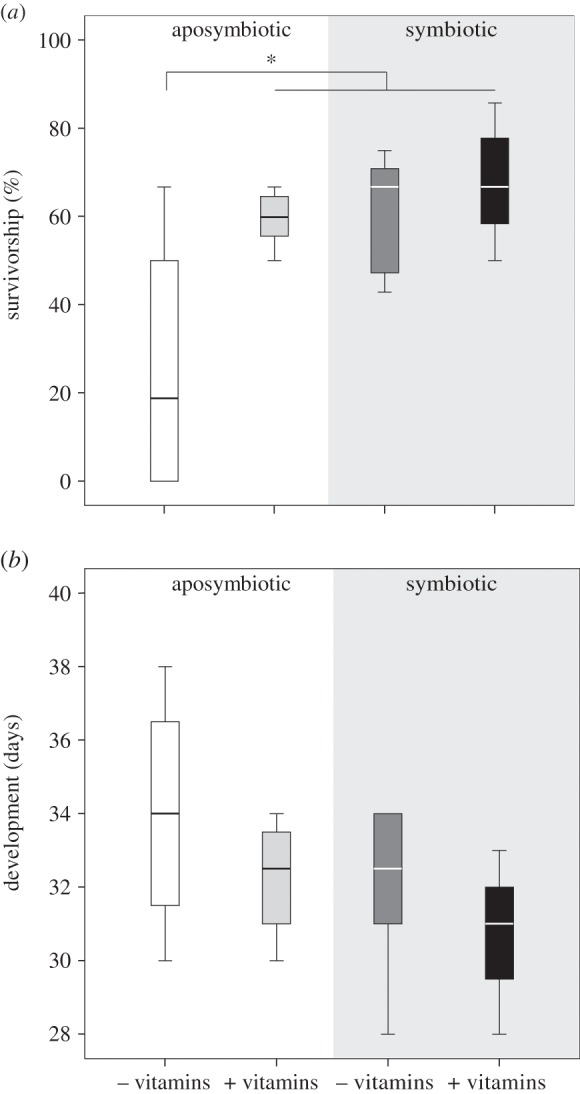
Fitness of control and aposymbiotic D. fasciatus on an artificial diet with and without B vitamins, respectively. (a,b) Survivorship and developmental time from egg hatching to adulthood, respectively. Shading of boxes signifies the experimental treatments. Lines represent medians, boxes comprise the 25–75 percentiles and whiskers denote the range. Significant differences (Friedman test with Wilcoxon–Wilcox post hoc measures, p < 0.05) are marked by an asterisk.
(c). Midgut transcriptome of Dysdercus fasciatus reared on the natural diet of cottonseeds
To examine the metabolic response to symbiont elimination under natural feeding conditions, we conducted a comparative transcriptome analysis of the midguts of aposymbiotic and symbiotic D. fasciatus that had been reared on their natural diet of cottonseeds (electronic supplementary material, figure S1).
Among the 55 222 assembled contigs, 20 174 received a BLAST annotation (electronic supplementary material, figure S1), and 3362 contigs were assigned to functional categories according to gene ontology (electronic supplementary material, figure S1). However, we focus here on (i) gene candidates involved in the transport and processing of B vitamins and (ii) genes whose expression patterns are known to be indicative of a stress response due to nutrient limitation.
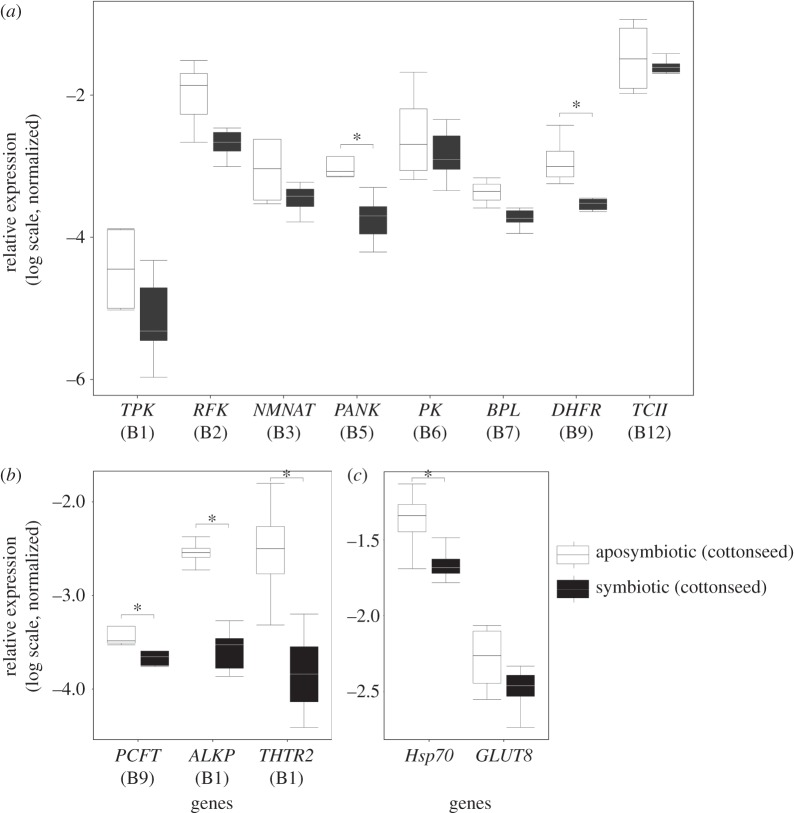
In total, 11 transcripts of proteins putatively involved in B vitamin metabolism and transport were detected in the D. fasciatus midgut transcriptome (figure 2; electronic supplementary material, table S1), including eight enzymes involved in the intracellular activation of these cofactors (figure 3a), i.e. thiamine pyrophosphokinase (TPK) for vitamin B1, riboflavin kinase (RFK) for vitamin B2, nicotinamide mononucleotide adenylyltransferase (NMNAT) for vitamin B3, pantothenate kinase (PANK) for vitamin B5, pyridoxal kinase (PK) for vitamin B6, biotin-protein lyase (BPL) for vitamin B7, dihydrofolate reductase (DHFR) for vitamin B9 and transcobalamine 2 (TCII) for vitamin B12. Assessing relative expression levels across individuals by quantitative PCR (qPCR) (normalized to the expression of the 60S ribosomal protein L13a) revealed that PANK and DHFR were differentially upregulated in the aposymbiotic treatment relative to symbiotic bugs (figure 3a). While similar trends were also observed for TPK, RFK, NMNAT, PK, BPL and TCII (figure 3a), the effects were not significant.
Figure 2.
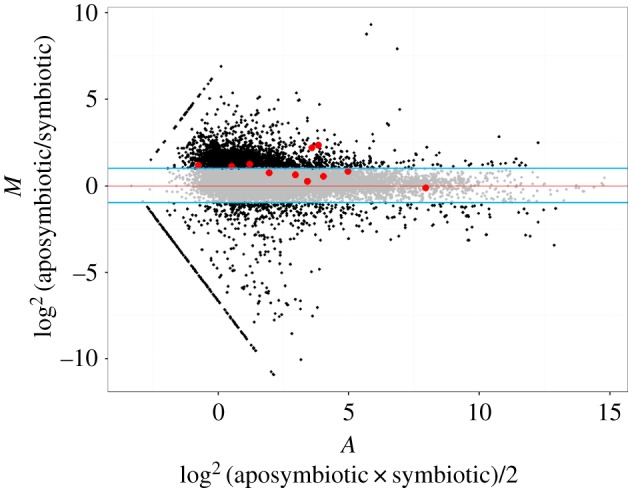
Pairwise MA plots for expressed genes among aposymbiotic and symbiotic D. fasciatus reared on their natural diet of cottonseeds. The scatter plot depicts the distribution of aposymbiotic/symbiotic log2 intensity ratio (M-value) versus the log2 average intensity (A-value). Each dot represents a single gene in comparison. The red line indicates M = 0. The upper and lower blue lines represent expression fold changes of 2 and 0.5, respectively. Grey dots represent constitutively expressed genes, black dots depict differentially expressed genes (more than twofold). B vitamin genes are designated by red dots. The lines of dots on the left side signify genes that were only observed in a single sample.
Figure 3.
Differential expression of B-vitamin metabolism and stress-related host genes by qPCR for symbiotic and aposymbiotic D. fasciatus reared on cottonseeds (normalized to the 60S ribosomal protein L13a). (a) Intracellular B vitamin processing genes: thiamine pyrophosphokinase (TPK), riboflavin kinase (RFK), nicotinamide mononucleotide adenylyltransferase (NMNAT), pantothenate kinase (PANK), pyridoxal kinase (PK), biotin-protein lyase (BPL), dihydrofolate reductase (DHFR), and transcobalamine 2 (TCII). (b) B vitamin transport and extracellular processing genes: thiamine transporter 2 (THTR2), proton-coupled folate transporter (PCFT) and thiamine alkaline phosphatase (ALKP). (c) A stress indicator; heat shock protein (Hsp) 70 and a glucose transporter (GLUT8). Shading of boxes signifies the experimental treatments (see legend). Lines represent medians, boxes comprise the 25–75 percentiles and whiskers denote the range. Significant differences were assessed based on the normalized expression in reference to the 60S ribosomal protein L13a with Mann–Whitney U-tests. Differentially expressed transcripts at p < 0.05 are marked by asterisks (*).
B vitamin transport genes included the thiamine transporter 2 (THTR2) and a proton-coupled folate transporter (PCFT) for the epithelial transport of thiamine [14] and folate [15], respectively. Additionally, an extracellular thiamine alkaline phosphatase (ALKP) gene was annotated and described to putatively catalyse the transphosphorylation of thiamin (B1) to thiamin monophosphate, thereby facilitating its epithelial transport [14]. Similar to the aforementioned B vitamin activation genes, all three genes were found by qPCR to be significantly upregulated in the aposymbiotic treatment as compared with symbiont-containing control individuals (figure 3b).
A stress response was additionally ascribed to the aposymbiotic condition, based on the upregulation of many stress-related genes in the transcriptome of aposymbiotic bugs—including the highly expressed heat shock protein Hsp70 and the glucose transporter 8 (GLUT8)—relative to the symbiotic treatment (electronic supplementary material, table S2). While Hsp70 has previously been found to be upregulated in firebugs under conditions of temperature stress [16], in other animal groups, the elevated expression of Hsp70 has also been reported under conditions of nutrient limitation [17]. Given that symbiotic and aposymbiotic bugs were reared under the same constant temperatures, a climate-induced stress response can be ruled out. Additionally, a similar trend was also observed according to the host's transcriptome for GLUT8—which has also been described as a stress indicator under conditions of nutrient limitation [18]—however, the effects were not significant in a subsequent qPCR (figure 3c).
(d). Vitamin transport and processing gene expression for symbiotic and aposymbiotic Dysdercus fasciatus reared on complete and vitamin-deficient artificial diets
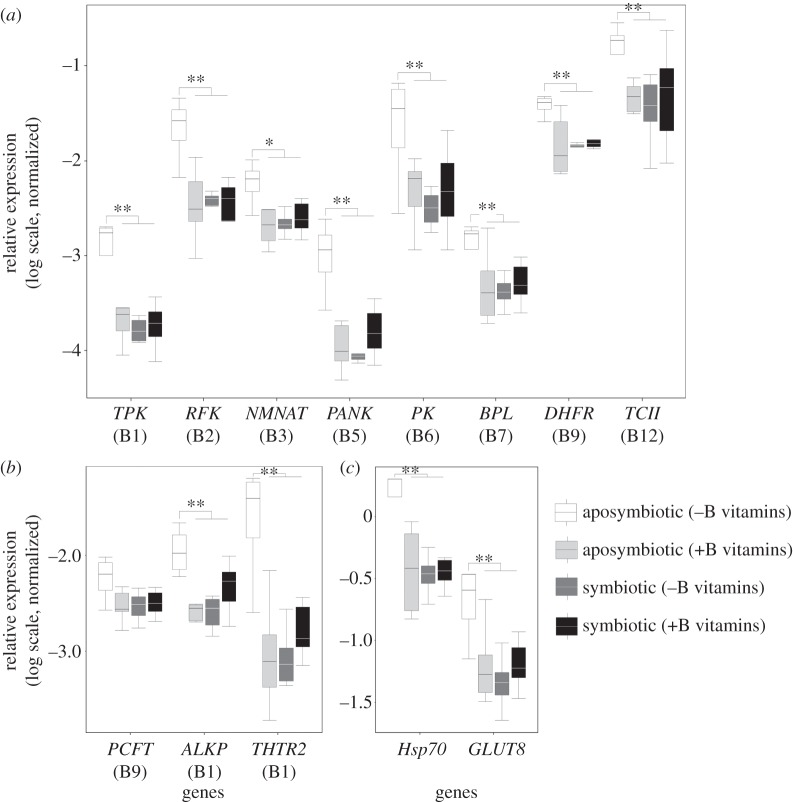
qPCRs for the candidate host genes involved in B vitamin metabolism were conducted across the four experimental treatments of aposymbiotic and symbiotic bugs reared on a complete and vitamin-deficient artificial diet to examine whether the upregulation of vitamin-transport and -processing genes (figures 2 and 3a,b) substantiates under controlled conditions with vitamin availability as the only nutritional difference between dietary treatments.
For most B vitamin-related genes annotated in this study, a consistent differential expression pattern was observed across the four experimental treatments, with aposymbiotic bugs reared on a vitamin-deficient diet exhibiting significantly higher relative expression levels of vitamin-transport and -processing genes than aposymbiotic bugs on a vitamin-deficient diet or symbiotic D. fasciatus reared on either complete or vitamin-deficient diets. This was the case for 10 out of the 11 predicted B vitamin-related genes (figure 4a,b), the heat shock protein (Hsp70), as well as the glucose transporter (GLUT8) (figure 4c). While the same trend was observed for the PCFT, the differences were not significant.
Figure 4.
Differential expression of B-vitamin metabolism and stress-related host genes by qPCR for symbiotic and aposymbiotic D. fasciatus reared on a complete or vitamin-deficient artificial diet (normalized to the 60S ribosomal protein L13a). (a) Intracellular B vitamin processing genes: TPK, RFK, NMNAT, PANK, PK, BPL, DHFR and TCII. (b) B vitamin transport and extracellular processing genes: THTR2, PCFT and ALKP. (c) A stress indicator Hsp70 and a glucose transporter GLUT8. For gene abbreviations, see figure 2. Shading of boxes signifies the experimental treatments (see legend). Lines represent medians, boxes comprise the 25–75 percentiles and whiskers denote the range. Significant differences were assessed based on the normalized expression in reference to the 60S ribosomal protein L13a with the Kruskal–Wallis test and Dunn post hoc tests. Differentially expressed transcripts at p < 0.05 and p < 0.01 are marked by single (*) and double (**) asterisks, respectively.
(e). Gene expression of other nutritional transporters for symbiotic and aposymbiotic Dysdercus fasciatus reared on complete and vitamin-deficient artificial diets
qPCRs for candidate host genes involved in amino acid transport, as well as phosphate transport were conducted across the four experimental treatments of aposymbiotic and symbiotic bugs reared on a complete and vitamin-deficient artificial diet to examine whether other genes that are not involved in the B vitamin metabolism exhibit a similar differential expression pattern or, instead, remain constitutively expressed (electronic supplementary material, figure S2). For both transporters, no differences in expression levels were observed across treatment groups.
(f). Functional characterization of the thiamine transporter THTR2
Of the two B vitamin transporters detected in D. fasciatus' transcriptome (PCFT and THTR2), only THTR2's sequence could be completely assembled from the retrieved transcripts, and in turn, functionally analysed by heterologous expression in Xenopus laevis oocytes. The functional assessment of THTR2 through radiotracer experiments revealed that the transporter is in fact capable of transporting thiamine (B1). This was demonstrated through the significant uptake of labelled thiamine in frog oocytes injected with THTR2 cRNA relative to control oocytes injected with cRNA for the human glucose transporter 1 (hGLUT1) or water (p < 0.001, ANOVA; figure 5a). Conversely, no discernible transport activity of labelled deoxyglucose could be reported for oocytes expressing THTR2 relative to hGLUT1-injected oocytes (p < 0.001, ANOVA; figure 5b). The specific and elevated transport activity reported for thiamine or deoxyglucose in oocytes injected with THTR2 or hGLUT1 cRNA, respectively, demonstrates that the injection procedure itself was not responsible for the uptake of radiolabelled compounds.
Figure 5.
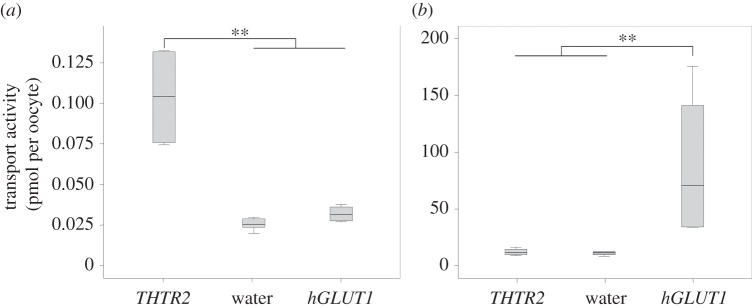
THTR2 mediated uptake of (a) 50 pM Thiamin and (b) 100 µM 2-deoxy-d-glucose into Xenopus oocytes. Transport activity was determined by quantifying the radiolabelled substrate uptake in oocytes injected with THTR2-cRNA, water or hGLUT1-cRNA, respectively. Lines represent medians, boxes comprise the 25–75 percentiles and whiskers denote the range. Significant differences at p < 0.01 are marked by double asterisks (**) (ANOVA).
(g). Coriobacterium glomerans' genomic potential for B vitamin supplementation
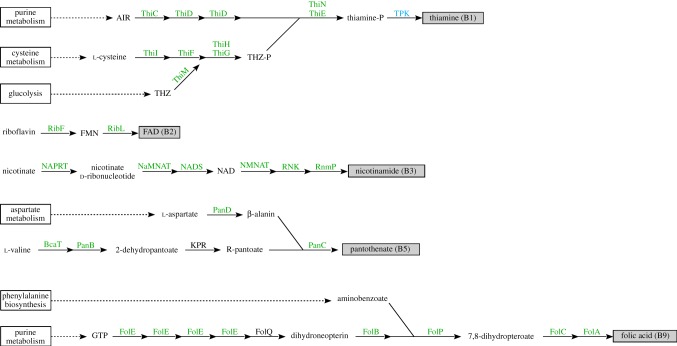
From the recently sequenced genome of C. glomerans [19], we report on the presence of pathways for the biosynthesis of five B vitamins (figure 6). For thiamine (B1), the complete pathway was discovered that incorporates derivatives of purine, cysteine and thiazole to form thiamine phosphate. The gene coding for TPK—the enzyme responsible for catalysing the cleavage of the final phosphate group to form thiamine (B1)—is nonetheless missing from the genome. However, the host itself possesses the TPK gene (figures 3 and 4), thereby providing a metabolic complement to C. glomerans' synthesis pathway for thiamine phosphate. For riboflavin (B2), genes encoding its conversion to flavin adenine dinucleotide (FAD) were detected, and the pathway for nicotinamide (B3) contained all the necessary genes for its conversion from nicotinate. For the complete biosynthesis of pantothenate (B5), a pathway was discovered to form the cofactor from aspartate and valine. Almost all necessary genes were accounted for, except for ketopantoate reductase which catalyses the reduction of dehydropantoate to pantoate. Lastly, the pathway leading to the biosynthesis of folic acid (B9) from purine and aminobenzoate was nearly complete, except for the gene FolQ. However, FolQ is absent from the genomes of many bacteria that are known to produce folic acid [20], therefore it has been proposed that alternate (but in most cases currently unknown) genes can substitute for this gene.
Figure 6.
Metabolic pathways for B vitamin biosynthesis in the genome of C. glomerans. Each arrow represents one step in the biosynthetic pathway. Enzyme names in green font indicate that a candidate gene for this step was detected in the annotated genome sequence. Enzyme names in blue indicate that a candidate gene for this step was detected in the host's genome (see figures 3a and 4a).
For Gordonibacter sp., the absence of a sequenced genome (or that of a closely related species) and its refractoriness to in vitro cultivation prevented us from making inferences regarding its genomic potential for B vitamin synthesis.
3. Discussion
Insects, depending on their feeding ecology, require vitamins of the B complex in varying amounts [21], and early work by Fraenkel & Blewett [22] demonstrated that the variation in the vitamin requirements of similarly oligophagous insects can often be attributed to the presence (or absence) of vitamin-supplementing microbial symbionts.
In contrast to their performance on Malvales seeds [8], aposymbiotic D. fasciatus that were exclusively fed on a complete artificial diet were found to exhibit near identical survivorship values relative to symbiotic individuals. Upon omission of B vitamins from the artificial diet, however, survivorship of aposymbiotic individuals decreased significantly compared with the symbiotic treatment on the same diet, as well as compared with symbiotic and aposymbiotic firebugs fed on a complete artificial diet (figure 1), thereby implicating the Coriobacteriaceae symbionts in supplementing these compounds to the host.
Haematophagous (blood-feeding) insects like the assassin bug Rhodnius prolixus, the tsetse fly Glossina morsitans and the bedbug Cimex lectularius have thus far presented us with excellent systems that illustrate the host's nutritional dependency on beneficial microbes for the supply of B vitamins that are consistently lacking in their vertebrate blood meals [4,5,23]. Symbiont-mediated B vitamin supplementation has also been demonstrated in non-haematophagous insects, such as the anobiid beetles Lasioderma serricorne and Stegobium paniceum, which specialize on a range of oilseeds, cereals and dried fruit [24].
In an effort to understand the host's response to this nutritional partnership, we assessed how symbiont elimination affects gene expression in the midgut of D. fasciatus when the bugs are reared on their natural diet of cottonseeds, as well as on complete and vitamin-deficient artificial diets. We specifically considered the expression patterns of host genes involved in the B vitamin metabolism. We initially observed that symbiont elimination in cottonseed-fed firebugs generally induces the upregulation of genes involved in the transport and processing of B vitamins (figures 2 and 3a,b). This pattern of differential gene expression was even more evident in aposymbiotic firebugs reared on an artificial diet deficient in B vitamins (figure 4a,b). We propose that the variation in the degree of differential expression across both sets of experiments can be attributed to the presence of some (albeit limiting) amounts of B vitamins in cottonseeds compared with the vitamin-deficient artificial diet. The gene expression patterns reported for D. fasciatus in this study are consistent with previous findings in mammals and yeast, highlighting the negative regulatory control of genes involved in the transport and activation of B vitamins, including thiamine, riboflavin, nicotinamide, panthothenate, pyradoxin, biotin, folate and B12 [14,15,25–29]. For example, the expression of thiamine-related uptake and processing genes is upregulated under conditions of thiamine deficiency [14]. Regulation in this system is mediated by transcriptional activators whose binding appears to be dependent on the vitamin's absence, as evidenced by the transcriptional repression of thiamine transporter genes following the supplementation of the vitamin to a previously deficient cell culture [14]. Similarly, the mouse homologue of the PCFT investigated in this study was also shown to undergo similar patterns of regulation, where a 13-fold increase in expression was recorded in the small intestine of mice fed on a folate-deficient diet relative to mice reared on a complete diet [15]. This is also the case for intracellular processing and activation genes. For example, PANK, the enzyme catalysing the phosphorylation of pantothenate (B5) to form phosphopantothenate, is regulated through a two-step negative feedback inhibition, both by pantothenate, as well as the downstream product acetyl-CoA [30]. Thus, despite the extracellular localization of the Coriobacteriaceae in the gut, our findings are reminiscent of highly derived mutualisms (e.g. aphid–Buchnera) where the host differentially regulates nutrient transport to and from the bacteriome in response to the symbiont's synthesized end product [31].
Gene expression patterns also seem to suggest that symbiont elimination in firebugs induces a general stress response, as evidenced by the elevated expression of Hsp70 and GLUT8, as well as several other stress-related genes in cottonseed-fed aposymbiotic firebugs relative to their symbiotic counterparts (figure 4c; electronic supplementary material, table S1), possibly as a product of nutrient limitation. This hypothesis was confirmed by the differential upregulation of both genes in aposymbiotic firebugs reared on a vitamin-deficient diet. The stress response could be rescued by artificial B vitamin supplementation or by the presence of the actinobacterial symbionts (figure 4c), demonstrating that vitamin omission is only stressful for firebugs in the absence of their Coriobacteriaceae symbionts. Additionally, constitutive expression of other transporter genes (e.g. phosphate and amino acid transporters; electronic supplementary material, figure S2) excluded the possibility that all genes are consistently upregulated in aposymbiotic insects reared on a vitamin-deficient diet. Collectively, these findings demonstrate that under B vitamin-limiting conditions, symbiont elimination initiates a general nutritional stress response in cotton stainers that results in the upregulation of vitamin transport and activation genes, which may represent an adaptive metabolic regulatory mechanism to cope with vitamin deficiency as a result of symbiont absence.
The case for symbiont-mediated nutritional supplementation in firebugs is further supported by the maintenance of many genes involved in the metabolism of B vitamins as predicted by C. glomerans' recently sequenced genome [19], including pathways for the synthesis of thiamine (B1), FAD (B2), nicotinamide (B3), pantothenate (B5) and folic acid (B9) (figure 6). While the exact mechanism for deriving B vitamins from the gut symbionts in firebugs is currently unknown, preliminary findings suggest that the active lysis of symbiont cells by the host through the expression of lysozyme plays an important role (Bauer et al. [32]), not unlike how R. prolixus [23], Drosophila melanogaster [33] or Riptortus pedestris [34] have been suggested to acquire important nutrients by harvesting their microbial gut associates.
In contrast to the aforementioned associations involving putative vitamin-supplementing symbionts that are housed intracellularly in specialized bacteriocytes, firebugs derive their vitamins from extracellular symbionts residing in their midgut lumen. In bacteriocyte-based mutualisms, the complementary partnerships are highly integrated from a metabolic standpoint [35]. Aphids—in their reliance on the essential amino acids provided by their primary endosymbiont Buchnera—differentially regulate gene expression to accommodate and facilitate the beneficial mutualism [36]. This includes the upregulation of host genes involved in the transport of essential amino acids in Buchnera-harbouring bacteriocytes compared with the rest of the aphid's body. Additionally, bacteriocytes also exhibit an upregulation of genes involved in the synthesis of non-essential amino acids [36,37], an expression pattern that is consistent with the hypothesis that Buchnera derives these compounds from the host, given the extensive loss affecting the bacterium's ability to produce non-essential amino acids [36,37]. Additionally, by comparing the aphid's gene expression between bacteriocytes and other body tissues using RNA sequencing and coupling it with comparative pathway analysis of amino acid synthesis between host and symbiont, Hansen & Moran [37] demonstrate that among the host genes that were upregulated in bacteriocytes, seven were found to fill in the gaps of Buchnera's essential amino acid pathways. Thus, despite the extracellular symbiont localization in cotton stainers, the differential expression of genes involved in the transport and processing of symbiont-provided nutritional compounds is reminiscent of bacteriome-associated intracellular symbioses, indicating a high degree of intimacy in the Pyrrhocoridae–Coriobacteriaceae association.
Our results offer novel insights into the metabolic integration between an animal host and its gut microbes. We demonstrate that despite an extracellular localization in D. fasciatus' midgut, the Coriobacteriaceae symbionts influence a range of metabolic pathways in firebugs including the differential expression of genes related to the transport and processing of B vitamins, thereby highlighting an important interface for the exchange of symbiont-provided nutritional supplements. Such findings enhance our understanding of the mechanisms involved in symbiont-mediated nutritional supplementation between animals and their extracellularly localized gut symbionts.
4. Material and methods
(a). Insect sampling and rearing
Live specimens of D. fasciatus were originally collected from the Comoé National Park, Cote d'Ivoire, but have since been bred in the laboratory at the University of Würzburg, Germany, and a sub-culture was later established at the Max Planck Institute for Chemical Ecology, Jena, Germany. The insects were reared in plastic containers (20 × 35 × 22 cm) at a constant temperature of 28°C and exposed to long light regimes (16 L : 8 D cycles). The bugs were provided ad libitum with previously autoclaved water and dry cottonseeds (Gossypium hirsutum) or different variations of the artificial diet (see below).
(b). Development and manipulation of a defined artificial diet
The artificial diet was modified for our study system from Panizzi et al. [38]. A single batch was produced for every experimental treatment and used throughout the course of the study to ensure consistency. Composition of the nutritionally complete diet is listed in the electronic supplementary material, table S3. A second variation of this diet was developed with the complete omission of B vitamins, i.e. thiamine (B1), riboflavin (B2), nicotinamide (B3), calcium pantothenate (B5), pyridoxine (B6), biotin (B7), folic acid (B9) and vitamin B12. Both diets were stored at −20°C, and small cubes (1 cm3) were thawed and supplied every other day to cages of the various experimental treatments.
(c). Experimental set-up
The generation of aposymbiotic bugs involved the sterilization of egg surfaces following the procedure used by Salem et al. [8]. Briefly, the eggs were submerged in ethanol for 5 min, followed by bleach (12% NaOCl) for 45 s. Residual bleach was removed by washing in sterile autoclaved water. To investigate symbiont-mediated supplementation of B vitamins, eight egg clutches (approx. 35 eggs each) from different females of D. fasciatus were harvested 3 days following oviposition and kept separately. We then split each of the collected egg clutches into four experimental treatments: (i) aposymbiotic bugs reared on a vitamin-deficient diet (aposymbiotic −B vitamins), (ii) aposymbiotic bugs reared on a complete diet (aposymbiotic +B vitamins), (iii) symbiotic bugs reared on a vitamin-deficient diet (symbiotic −B vitamins) and (iv) symbiotic bugs reared on a complete diet (symbiotic +B vitamins). The four treatments were provided with autoclaved water and the two variations of the artificial diet ad libitum. To examine the differential pattern of host gene expression in response to symbiont elimination on the natural diet of cottonseeds, five additional egg clutches of D. fasciatus were harvested, and each clutch was separated into two experimental treatments: (i) symbiotic and (ii) aposymbiotic bugs reared on cottonseeds.
(d). Fitness measurements
Individuals across all experimental treatments were observed on a daily basis for the assessment of fitness effects across the different groups. For the bugs reared on the different variations of the artificial diet, growth rate (days until moulting to the next stage) and survival until adulthood (%) were recorded.
(e). RNA extraction and reverse transcription
Three days following adult emergence, a single individual was collected from every experimental treatment replicate, and through dissection, its midgut region (M1–M4) was harvested. Once dissected, the midgut region was stabilized in RNAlater solution (Qiagen) and stored at −20°C. Total RNA isolations were performed using the RNeasy Micro Kit (Qiagen) following the manufacturer's guidelines. Integrity and quality of the RNA samples were determined using the RNA 6000 Nano LabChip kit (Agilent Technologies) on an Agilent 2100 Bioanalyzer (Agilent Technologies) according to the manufacturer's instructions. cDNA was then generated according to the manufacturer's guidelines (QuantiTect Reverse Transcription kit, Qiagen). To account for possible shortcomings during the RNA extraction procedure and to test for the success of the symbiont elimination procedure, diagnostic PCR screens targeting the host's 18S rRNA gene and the Coriobacteriaceae symbionts' 16S rRNA genes were conducted using the generated cDNA according to previously described procedures [8].
(f). Illumina-based transcriptome sequencing and quantitative PCRs
RNA was extracted from dissected whole midgut regions (M1–M4) of five symbiotic and aposymbiotic bugs, respectively, fed on their natural diet of cottonseeds, resulting in two pooled samples. Transcriptome sequencing of poly-A enriched mRNAs was done commercially (Fasteris; http://www.fasteris.com) on the HiSeq 2000 Sequencing System from Illumina (http://www.illumina.com), using the paired read 100 bp technology. For details on assembly and annotation procedures, see the electronic supplementary material, Methods.
qPCRs for candidate host genes involved in B vitamin metabolism, metabolic stress response, as well as non-B vitamin transporters were conducted across (i) aposymbiotic and symbiotic bugs reared on their natural diet of cottonseeds (to confirm the transcriptome sequencing results) and (ii) across the four experimental treatments of aposymbiotic and symbiotic bugs reared on a complete and vitamin-deficient artificial diet to examine whether such patterns substantiate under controlled conditions with vitamin availability as the only nutritional difference between dietary treatments. Primers were designed based on the candidate gene sequences available from the transcriptome, checked for specificity in vitro using capillary sequencing of amplified PCR products (electronic supplementary material, table S4) and used for qPCRs on a RotorGene-Q cycler (Qiagen, Hilden, Germany) (see the electronic supplementary material, Methods).
(g). Functional characterization of transporter activity using Xenopus laevis oocytes
THTR2 and hGLUT1 (control) cRNA were generated by in vitro transcription using mMESSAGE mMACHINE kit (Ambion, Inc.). Xenopus laevis oocytes were kindly provided by Stefan Heinemann (University of Jena). cRNA (100–125 ng) was injected per oocyte, and RNase-free water was used as a negative control. All oocytes were maintained at 17.5°C in modified Barth's saline (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.33 Ca(NO3)2, 0.41 CaC12, TRIS-HCl, pH 7.4) with 10 µg ml−1 penicillin, 10 µg ml−1 streptomycin and 4 µg ml−1 cefuroxim solution for 3 days.
Functional zero-trans-influx studies were carried out using radiolabelled thiamine hydrochloride and deoxyglucose as substrates in X. laevis oocytes. Three days after cRNA injection, groups of 5–10 oocytes were transferred into tubes containing 200 μl Barth's saline and thiamine (4 µCi ml−1 thiamine-[3H]-hydrochloride; American Radiolabelled Chemicals) or 2-deoxy-d-glucose (2 µCi ml−1 2-Deoxy-d-[1-3H]-glucose; Amersham Biosciences). After incubation at room temperature, cells were transferred to ice-cold Barth's saline, washed three times, solubilized with 100 μl 1% (w/v) SDS and measured individually (five replicates) as previously described by Strauss and co-workers [39].
(h). B vitamin pathways in the genome of Coriobacterium glomerans
To examine the metabolic potential of C. glomerans for B vitamin supplementation, we analysed the genomic pathways using the symbiont's recently sequenced genome (NC_015389) [17]. This was accomplished using the pathway analysis platform available in the Kyoto Encyclopaedia of Genes and Genomes [40]. Complete pathways with all necessary genes were recorded; however, a number of missing genes were additionally supplemented by manual curation. In this process, each missing gene was searched in closely related bacterial species for potential homologues. The homologues were then searched with blastn and blastx (using an e-value cut-off score of 10−5) against a custom created database of all predicted genes in the genome of C. glomerans. The additional genes discovered by this method were then included in the respective pathways.
(i). Statistical analysis
Growth and survival rates until adulthood across the four experimental treatments of symbiotic and aposymbiotic groups fed on a complete and vitamin-deficient diet were compared using Friedman tests with Wilcoxon–Wilcox post hoc measures using BiaS v. 7.40 (Epsilon-Verlag; Hochheim-Darmstadt, Germany). To assess differential expression patterns of host genes from aposymbiotic and symbiotic bugs reared on cottonseeds, as well as on the two variations of the artificial diet, gene copy numbers estimated from qPCRs were first normalized against the 60S ribosomal protein L13a (electronic supplementary material, table S3), then compared using the Mann–Whitney U-test and Kruskal–Wallis tests with Dunn post hoc tests, respectively, to asses levels of significance (p < 0.05) across treatments. The absence of a significant differential expression pattern in a second housekeeping gene, elongation factor 1 alpha (EF1α), across treatments following normalization with 60S ribosomal protein L13a (electronic supplementary material, figure S2b) confirmed the suitability of L13a as a reference with stable expression.
To compare the differential uptake in labelled compounds in X. laevis oocytes, ANOVA was used, after confirming that the data were normally distributed (p < 0.05, Kolmogorov–Smirnov test). Both tests were performed using the SPSS v. 17.0 software package (SPSS, Chicago, IL, USA).
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Benjamin Weiss for his assistance in caring for the bugs, Jonathan Gershenzon for insightful discussions on the experimental set-up and Angela Douglas for helpful comments on an earlier version of the manuscript.
Funding statement
Financial support from the Max Planck Society and the DFG (MA5082-1/1) is gratefully acknowledged.
Conflict of interests
The authors declare that they have no competing interests.
References
- 1.Douglas AE. 2009. The microbial dimension in insect nutritional ecology. Funct. Ecol. 23, 38–47. ( 10.1111/j.1365-2435.2008.01442.x) [DOI] [Google Scholar]
- 2.Moran NA. 2007. Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl Acad. Sci. USA 104, 8627–8633. ( 10.1073/pnas.0611659104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payne SH, Loomis WF. 2006. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences . Eukaryot. Cell 5, 272–276. ( 10.1128/Ec.5.2.272-276.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl Acad. Sci. USA 107, 769–774. ( 10.1073/pnas.0911476107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32, 402–407. ( 10.1038/ng986) [DOI] [PubMed] [Google Scholar]
- 6.Feldhaar H, Straka J, Krischke M, Berthold K, Stoll S, Mueller MJ, Gross R. 2007. Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol. 5, 48 ( 10.1186/1741-7007-5-48) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudakaran S, Salem H, Kost C, Kaltenpoth M. 2012. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae). Mol. Ecol. 21, 6134–6151. ( 10.1111/Mec.12027) [DOI] [PubMed] [Google Scholar]
- 8.Salem H, Kreutzer E, Sudakaran S, Kaltenpoth M. 2013. Actinobacteria as essential symbionts in firebugs and cotton stainers (Hemiptera, Pyrrhocoridae). Environ. Microbiol. 15, 1956–1968. ( 10.1111/1462-2920.12001) [DOI] [PubMed] [Google Scholar]
- 9.Fukatsu T, Hosokawa T. 2002. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl. Environ. Microbiol. 68, 389–396. ( 10.1128/Aem.68.1.389-396.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi Y, Hosokawa T, Fukatsu T. 2007. Insect–microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73, 4308–4316. ( 10.1128/Aem.00067-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaltenpoth M, Winter SA, Kleinhammer A. 2009. Localization and transmission route of Coriobacterium glomerans, the endosymbiont of pyrrhocorid bugs. FEMS Microbiol. Ecol. 69, 373–383. ( 10.1111/j.1574-6941.2009.00722.x) [DOI] [PubMed] [Google Scholar]
- 12.Ahmad IS, Schaeffer C. 1987. Food plants and feeding biology of the Pyrrhocoroidea (Hemiptera). Phytophaga 1, 75–92. [Google Scholar]
- 13.Whitsitt M. 1933. Vitamin B (B1) and G (B2) Content of cotton-seed products. Ind. Eng. Chem. 25, 1169–1171. ( 10.1021/ie50286a027) [DOI] [Google Scholar]
- 14.Hohmann S, Meacock PA. 1998. Thiamin metabolism and thiamin diphosphate-dependent enzymes in the yeast Saccharomyces cerevisiae: genetic regulation. Biochem. Biophys. Acta Protein Struct. Mol. Enzymol. 1385, 201–219. ( 10.1016/S0167-4838(98)00069-7) [DOI] [PubMed] [Google Scholar]
- 15.Qiu A, et al. 2007. Rodent intestinal folate transporters (SLC46A1): secondary structure, functional properties, and response to dietary folate restriction. Am. J. Physiol. Cell Physiol. 293, C1669–C1678. ( 10.1152/ajpcell.00202.2007) [DOI] [PubMed] [Google Scholar]
- 16.Kostal V, Tollarova-Borovanska M. 2009. The 70 kDa heat shock protein assists during the repair of chilling injury in the insect, Pyrrhocoris apterus. PLoS ONE 4, e4546 ( 10.1371/Journal.Pone.0004546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribeil JA, et al. 2007. Hsp70 regulates erythropoiesis by preventing caspase-3-mediated cleavage of GATA-1. Nature 445, 102–105. ( 10.1038/nature05378) [DOI] [PubMed] [Google Scholar]
- 18.Piroli GG, Grillo CA, Charron MJ, McEwen BS, Reagan LP. 2004. Biphasic effects of stress upon GLUT8 glucose transporter expression and trafficking in the diabetic rat hippocampus. Brain Res. 1006, 28–35. ( 10.1016/j.brainres.2004.01.044) [DOI] [PubMed] [Google Scholar]
- 19.Stackebrandt E, et al. 2013. Complete genome sequence of Coriobacterium glomerans type strain (PW2(T) from the midgut of Pyrrhocoris apterus L. (red soldier bug). Stand. Genomic Sci. 8, 15–25. ( 10.4056/sigs.3507020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Crecy-Lagard V, El Yacoubi B, de la Garza RD, Noiriel A, Hanson AD. 2007. Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genomics 8, 245 ( 10.1186/1471-2164-8-245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraenkel G, Blewett M. 1943. Vitamins of the B-group required by insects. Nature 151, 703–704. ( 10.1038/151703a0) [DOI] [Google Scholar]
- 22.Fraenkel G, Blewett M. 1943. Intracellular symbionts of insects as a source of vitamins. Nature 152, 506–507. ( 10.1038/152506a0) [DOI] [Google Scholar]
- 23.Beard CB, Cordon-Rosales C, Durvasula RV. 2002. Bacterial symbionts of the triatominae and their potential use in control of Chagas disease transmission. Annu. Rev. Entomol. 47, 123–141. ( 10.1146/annurev.ento.47.091201.145144) [DOI] [PubMed] [Google Scholar]
- 24.Blewett M, Fraenkel G. 1944. Intracellular symbiosis and vitamin requirements of two insects, Lasioderma serricorne and Sitodrepa panacea. Proc. R. Soc. Lond. B 132, 212–221. ( 10.1098/rspb.1944.0013) [DOI] [Google Scholar]
- 25.Abbas CA, Sibirny AA. 2011. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. R. 75, 321–330. ( 10.1128/Mmbr.00030-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Said HM, Ma TY, Kamanna VS. 1994. Uptake of biotin by human hepatoma cell line, Hep G2: a carrier-mediated process similar to that of normal liver. J. Cell Physiol. 161, 483–489. ( 10.1002/jcp.1041610311) [DOI] [PubMed] [Google Scholar]
- 27.Ink SL, Henderson LM. 1984. Vitamin B6 metabolism. Annu. Rev. Nutr. 4, 455–470. ( 10.1146/annurev.nu.04.070184.002323) [DOI] [PubMed] [Google Scholar]
- 28.Said HM, Nabokina SM, Balamurugan K, Mohammed ZM, Urbina C, Kashyap ML. 2007. Mechanism of nicotinic acid transport in human liver cells: experiments with HepG2 cells and primary hepatocytes. Am. J. Physiol. Cell Physiol. 293, C1773–C1778. ( 10.1152/ajpcell.00409.2007) [DOI] [PubMed] [Google Scholar]
- 29.Zhao R, Diop-Bove N, Visentin M, Goldman ID. 2011. Mechanisms of membrane transport of folates into cells and across epithelia. Annu. Rev. Nutr. 31, 177–201. ( 10.1146/annurev-nutr-072610-145133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackowski S, Rock CO. 1981. Regulation of coenzyme A biosynthesis. J. Bacteriol. 148, 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price DR, Feng H, Baker JD, Bavan S, Luetje CW, Wilson AC. 2014. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc. Natl Acad. Sci. USA 111, 320–325. ( 10.1073/pnas.1306068111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer M, Salem H, Vogel H, Marz M, Kaltenpoth M. In press. Transcriptomic immune response of the cotton stainer Dysdercus fasciatus to experimental elimination of vitamin-supplementing intestinal symbionts. PLoS ONE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daffre S, Kylsten P, Samakovlis C, Hultmark D. 1994. The lysozyme locus in Drosophila melanogaster: an expanded gene family adapted for expression in the digestive tract. Mol. Gen. Genet. 242, 152–162. ( 10.1007/BF00391008) [DOI] [PubMed] [Google Scholar]
- 34.Kim JK, Kim NH, Jang HA, Kikuchi Y, Kim CH, Fukatsu T, Lee BL. 2013. Specific midgut region controlling the symbiont population in an insect–microbe gut symbiotic association. Appl. Environ. Microb. 79, 7229–7233. ( 10.1128/AEM.02152-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douglas AE. 2006. Phloem-sap feeding by animals: problems and solutions. J. Exp. Bot. 57, 747–754. ( 10.1093/jxb/erj067) [DOI] [PubMed] [Google Scholar]
- 36.Nakabachi A, Shigenobu S, Sakazume N, Shiraki T, Hayashizaki Y, Carninci P, Ishikawa H, Kudo T, Fukatsu T. 2005. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc. Natl Acad. Sci. USA 102, 5477–5482. ( 10.1073/pnas.0409034102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen A, Moran N. 2011. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc. Natl Acad. Sci. USA 108, 2849–2854. ( 10.1073/pnas.1013465108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panizzi AR, Parra JRP, Santos CH, Carvalho DR. 2000. Rearing the southern green stinkbug using an artificial dry diet and an artificial plant. Pesq. Agropec. Bras. 35, 1709–1715. ( 10.1590/S0100-204X2000000900002) [DOI] [Google Scholar]
- 39.Strauss A, Peters S, Boland W, Burse A. 2013. ABC transporter functions as a pacemaker for the sequestration of plant glucosides in leaf beetles. eLife 2, e01096 ( 10.7554/eLife.01096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114. ( 10.1093/nar/gkr988) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.