Abstract
Estrogen and its receptors are essential hormones for normal reproductive function in males and females during developmental stage. To better understand the effect of estrogen receptor (ER) gene in yak (Bos grunniens), reverse transcription-polymerase chain reaction (PCR) was carried out to clone ERα and ERβ genes. Bioinformatics methods were used to analyze the evolutionary relationship between yaks and other species, and real-time PCR was performed to identify the mRNA expression of ERα and ERβ. Sequence analysis showed that the ER open reading frames (ORFs) encoded 596 and 527 amino acid proteins. The yak ERα and ERβ shared 45.3% to 99.5% and 53.9% to 99.1% protein sequence identities with other species homologs, respectively. Real-time PCR analysis revealed that ERα and ERβ were expressed in a variety of tissues, but the expression level of ERα was higher than that of ERβ in all tissues, except testis. The mRNA expression of ERα was highest in the mammary gland, followed by uterus, oviduct, and ovary, and lowest in the liver, kidney, lung, testis, spleen, and heart. The ERβ mRNA level was highest in the ovary; intermediary in the uterus and oviduct; and lowest in the heart, liver, spleen, lung, kidney, mammary gland, and testis. The identification and tissue distribution of ER genes in yaks provides a foundation for the further study on their biological functions.
Keywords: Yak, Estrogen Receptor Gene, Gene Expression, Tissue Distribution
INTRODUCTION
Estrogen, which is a steroid hormone primarily synthesized in ovary and testis (Katsu et al., 2010), regulates a variety of functions in vertebrates, including reproductive immune, and central nervous systems (Bakker and Brock, 2010; McCarthy, 2010; Vasudevan and Pfaff, 2008). At present, a large amount of research has been done on the biological roles of estrogen in vertebrates, especially in reproductive performance (Hewitt and Korach, 2003; Wang, 2005). Wu et al. found treatment of in vitro mouse embryo cultures with the anti-estrogen CI 628 could block embryo development (Sengupta et al., 1982), and this type of blockage could be alleviated by the co-administration of E2, indicating a direct effect of estrogens on embryo development. In primates, it also has been shown that near-term fetuses deprived of estrogen in utero reduced the number of primordial follicles in the ovaries, and the phenomenon can be restored to normal in animals administered E2 (Billiar et al., 2003). These researches indicated the key role of estrogen during human primordial follicle formation. Moreover, a number of studies showed that endogenous estradiol-17β acted as a natural inducer of ovarian differentiation in non-mammalian vertebrates (Devlin and Nagahama, 2002; Sinclair et al., 2002).
Although estrogen has an important effect on fetal development, its extensive physiological functions are mediated by specific cell surface receptors, i.e., the estrogen receptors (ERs) (Beyer et al., 2003; Mermelstein and Micevych, 2008). Accordingly, it is important to analyze the ERs to understand their physiological role. ERs belong to a superfamily of nuclear hormone receptors that include other steroid hormone receptors, such as progestogen, androgen, glucocorticoid, and mineralocorticoid receptors (Blumberg and Evans, 1998). The members of this superfamily have a number of common features and their proteins can be divided into six distinct domains. The N terminal of the A/B domain has a transactivation function, and the C domain contains two zinc finger motifs, which is formed by a number of cysteine residues and necessary for DNA binding. The D area is the hinge region, which enables the protein to change its conformation. In addition, the E domain is possibly the ligand binding domain. The function of the F domain is not fully understood (Todo et al., 1996). Two types of ER, ERβ, and ERβ, arising from two distinct genes, have been isolated in vertebrates. DNAs encoding ERs have been cloned from a variety of vertebrate species including mammals (Green et al., 1986; White et al., 1987), bird (Krust et al., 1986), reptiles (Sumida et al., 2001; Katsu et al., 2004), amphibian (Weiler et al., 1987), and teleost fish (Pakdel et al., 1990). However, no information is available on the sequence and the expression pattern of ERs mRNA in yaks.
Yak (Bos grunniens), living in the Tibetan Plateau, has successfully adapted to the chronic cold and low-oxygen environment of high altitude (~3,500 to 5,500 m). As a key species in Tibetan Plateau, yaks play an important role in Tibetan life by providing meat and milk where few other animals can survive. Unfortunately, female yaks usually have a low reproductive rate (40% to 60%) compared with other bovines (Zi, 2003; Sarkar and Prakash, 2005). Therefore, it’s of significance to study the reproductive biology of this species to meet an increasing demand in Tibet. To date, there is little information concerning the reproductive endocrinology of yak. Considering this, we isolated cDNA clones encoding yak ERs and detected their expression pattern in order to provide some data on their phylogenic relationship with other known vertebrate ERs and further investigated the special reproductive endocrine system of yak.
MATERIALS AND METHODS
Sample collection and preservation
The yaks were obtained from the Songpan Bovine Breeding Farm in Sichuan, China. These animals were killed, and their tissues, including heart, liver, spleen, lung, kidney, mammary gland, uterus, oviduct, ovary, and testis (male = 3, female = 3), were collected and immediately frozen in liquid nitrogen until use.
RNA isolation and cDNA cloning
Total RNA was extracted from tissues (11 difference tissues) using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The primers for amplification of yak ERα (ERα01 and ERα02) and ERβ (ERβ01 and ERβ02) gene coding sequences were based on the full-length sequences of bovine ERs, designed by Bacon designer (Table 1). cDNA synthesis was performed using PrimeScript reagent kit (TakaRa, Dalian, China) in a total volume of 10 μL of reaction mixture, containing 2 μL of 5× PrimeScript Buffer, 0.5 μL of PrimeScript RT enzyme Mix I, 0.5 μL of Oligo dT primer, 0.5 μL of random 6 mers, and 6.5 μL of total RNA. This cDNA was used as a template in reverse transcription-polymerse chain reaction (RT-PCR). The PCR reaction was performed in 25 μL volumes of the reaction mixture, containing 12.5 μL of PCR Mastermix, 1 μL of cDNA, 9.5 μL of ddH2O, and 1 μL of each primer. The PCR procedure consisted of first denaturation step at 94°C for 5 min, followed by 35 amplification cycles (denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min 30 s), with a final elongation step at 72°C for 5 min. The expected fragment ERα 01 (820 bp), fragment ERα 02 (1,280 bp), fragment ERβ 01 (783 bp), and fragment ERβ 02 (875 bp) were extracted from the EZNA gel extraction kit (OMEGA, USA). The fragments were cloned into pMD19-T Vector (TakaRa, Dalian, China).
Table 1.
Primers used for molecular cloning and Q-PCR in this study
| Gene name | Primer sequence | bp | Accession number |
|---|---|---|---|
| For molecular clone | |||
| ERα 01F | ACTGTCTCAGCCCTTGACTTCTA | 820 | NM_001001443.1 |
| ERα 01R | GCTCTTCCTCCTGTTTTTATCAA | ||
| ERα 02F | ACGATTGATAAAAACAGGAGGAA | 1,280 | NM_001001443.1 |
| ERα 02R | ACTGAGTGAGCGAATGAATGG | ||
| ERβ01F | GCTGTTACCTACTCAAGACATGG | 783 | NM_174051.3 |
| ERβ01R | AGCTCTTTCACTCGGGTCAT | ||
| ERβ02F | ACTGCCTGAGCAAAACCAA | 875 | NM_174051.3 |
| ERβ02R | TCACTGAGCCTGGGGTTTC | ||
| For real-time PCR | |||
| ERα QF | TCAGGCTACCATTACGGAGT | 230 | AC_000166.1 |
| ERα QR | CGCTTGTGCTTCAACATTCT | ||
| ERβ QF | CAGCCGTCAGTTCTGTATGC | 191 | AC_000167.1 |
| ERβ QR | GGCACAACTGCTCCCACTA | ||
| GAPDHF | TGCTGGTGCTGAGTAGTTGGTG | 290 | AC_000162.1 |
| GAPDHR | TCTTCTGGGTGGCAGTGATGG | ||
Q-PCR, quantity polymerase chain reaction; F, forward primer; R, reverse primer; QF, quantity forward primer; QR, quantity reverse primer; GAPDHF, GAPDH forward primer; GAPDHR, GAPDH reverse primer.
Sequence analysis
The nucleotide and deduced amino acid sequence identity was performed using LaserGene software package (DNASTAR, London, UK). The sequences of yak ERα and ERβ were aligned using the Multiple Sequence Alignment option in Clustal W. The neighbor-joining phylogenetic tree of ERα and ERβ was constructed using molecular evolutionary genetics analysis 5.
The tissue distributions of ERα and ERβ mRNA were examined in the heart, liver, spleen, lung, kidney, testis, mammary, oviduct, uterus, and ovary by quantitative RT-PCR using the SYBR Premix Ex Taq (TakaRa, China) with Bio-Rad Connect (iCycler iQ5 Real-time Detection System) in a 15 μL of reaction volume. GlycERαldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control gene. The primers ERαQ, ERβQ, and GAPDH were designed using Bacon designer (Table 1). The reaction mixture for quantitative RT-PCR contained 7.5 μL of Green premix (TakaRa), 1.0 μL of cDNA, and 0.5 μL of each gene-specific primer, and ddH2O was used to adjust the total volume to 15 μL. Melting curve was performed to detect the specificity. PCR parameters were based on a three-step method: 94°C for 30 s; 40 cycle of amplification step (denaturation at 94°C for 5 s, annealing at 58°C for 25 s, and extension at 72°C for 25 s); and dissociation curve analysis at 95°C/10 s, 65°C to 95°C in 0.5°C intervals. Each sample was tested in triplicate. Serial dilutions of pooled cDNA samples of each tissue were used to generate the standard curves. The amplification efficiency between the target gene and reference gene is 97% to 99%. The expression of the target gene was compiled relative to the expression of GADPH by the relative quantification method 2−ΔΔCt (Schmittgen and Livak, 2008).
Statistical analysis
The statistical significance of the variation was analyzed by one-way analysis of variance or Student’s t-test, followed by Tukey’s multiple comparison test. All the quantitative RT-PCR data were expressed as mean±SE, and significance was set at p<0.05.
RESULTS
Molecular cloning and characterization of ERα and ERβ cDNA
The ERα PCR products were 820 and 1,280 bp, which were composed of 1,791 bp of open reading frame (ORF) coding 596 amino acids. The predicted molecular mass of yak ERα was 66.5 kDa. Multiple alignments were carried out based on amino acid sequences of ERα from Bos taurus, Gallus gallus, Homo sapiens, Mus musculus, Rattus norvegicus, alligator, Xenopus laevis, and zebrafish. ERα showed 99.5% and 91.1% amino acid identities with B. taurus and H. sapiens, whereas only 45.3% amino acid identity with zebrafish (Table 2). The ERβ PCR products were 783 and 875 bp, which contained 1,584 bp of ORF with a coding potential for 527 amino acid residues. The molecular mass of yak ERβ was 59.0 kDa. The protein sequence showed 99.0%, 70.5%, 78.8%, 80.9%, 96.5%, 87.8%, 79.6%, and 91.9% similarities to B. taurus, G. gallus, H. sapiens, M. musculus, R. norvegicus, alligator, X. laevis, and zebrafish, respectively (Table 2). Similar to most nuclear receptors, both ERα and ERβ contained six important domains that are labeled A though F (Figure 1). C and E domains of yak were highly conserved from fish to mammals between ERα and ERβ (C domain, 94% to 99% amino acid identity; E domain, 58% to 94%) in the six bindings (Figures 2 and 3). Both ERα and ERβ contained eight cysteine residues (Figure 4).
Table 2.
Similarities of amino acid of ERα and ERβ to other species
| Species | Similarity | |
|---|---|---|
|
| ||
| ERα | ERβ | |
| Bos taurus | 99.5 | 99.1 |
| Gallus gallus | 76.1 | 78.0 |
| Homo sapiens | 91.1 | 84.0 |
| Mus musculus | 88.1 | 87.3 |
| Alligator | 75.5 | 77.8 |
| Xenopus laevis | 68.0 | 70.0 |
| Rattus norvegicus | 87.8 | 86.8 |
| Zebrafish | 45.3 | 53.9 |
ER, estrogen receptor.
Figure 1.
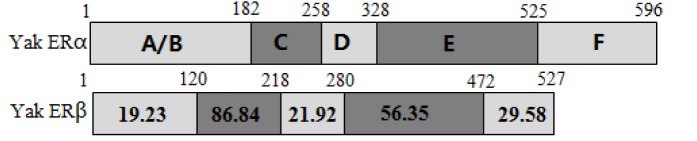
Domain structures of ERα and ERβ. The numbers within each box indicate the percentage identity of the domain between yak ERα and ERβ. ER, estrogen receptor.
Figure 2.

Domain structures of yak ERα and identities with ERα of alligator, Xenopus laevis, Gallus gallus, and zebrafish. The numbers within each box indicate the percentage identity of the domain relative to yak ERα. ER, estrogen receptor.
Figure 3.
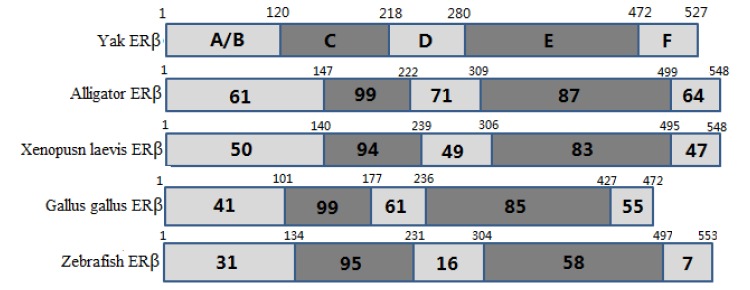
Domain structures of ERβ and identities with ERβ of alligator, Xenopus laevis, Gallus gallus, and zebrafish. The numbers within each box indicate the percentage identity of the domain relative to yak ERβ. ER, estrogen receptor.
Figure 4.

Position of eight cysteines of yak ERα and ERβ. ER, estrogen receptor.
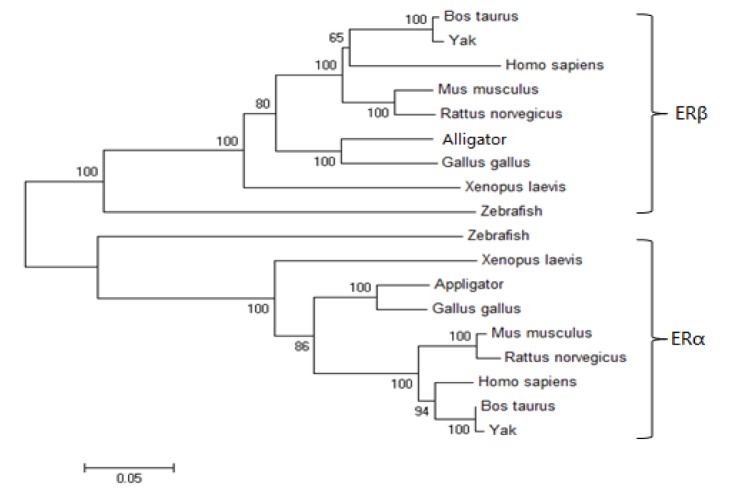
Phylogenetic analysis
Based on the amino acid sequences of ERα and ERβ of other species, a phylogenetic tree was constructed using the neighbor-joining method (Figure 5). The phylogenetic analysis showed that all mammalian ERs formed an independent branch, whereas birds, reptile, amphibian, and fish formed another branch.
Figure 5.
Phylogenetic tree of yak ERs. The tree was constructed based on the amino acid sequences by neighbor-joining method in ClustalX and MEGA 5.0 software with 1,000 bootstrap replications. ERs, estrogen receptors.
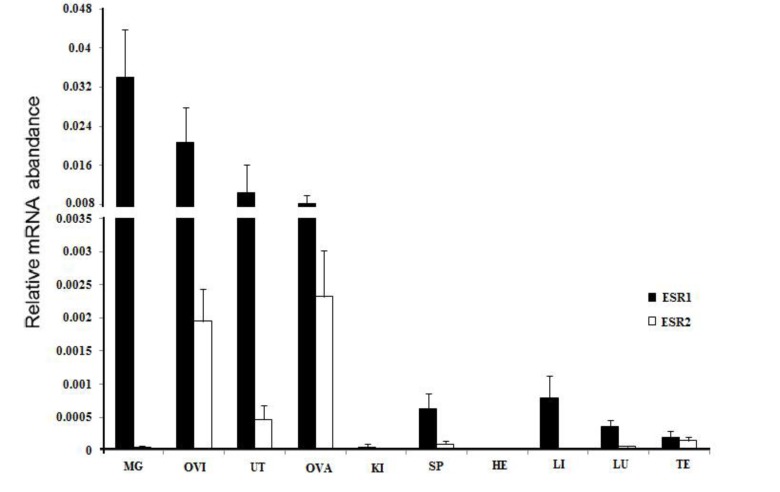
Tissue expression of estrogen receptor α and estrogen receptor β in yak
The two transcripts were expressed in a variety of tissues, but their expression levels varied (Figure 6). The expression of ERα was highest in the oviduct, followed by the uterus, mammary gland, spleen, stomach, and ovary, and lowest in the liver, lung, testis, spleen, and heart. The expression of ERβ was highest in the oviduct; intermediary in the uterus, testis, mammary gland, and ovary; and lowest in the spleen, mammary gland, heart, spleen, and liver. Moreover, in the same reproductive organs, the yERα mRNA level was higher than yERβ, except in the ovary.
Figure 6.
Real-time quantification of ERα and ERβ gene expression in various tissues in MaiWa yak. Different uppercase letters indicate significant difference within the groups (p<0.05), and different lowercase letters indicate significant difference between the groups (p<0.05). ER, estrogen receptor.
DISCUSSION
Estrogen and its receptors are essential hormones for normal reproductive function in males and females during developmental stage. In this study, we cloned the ORFs of ERs from yak and examined the expression pattern of mRNA in 10 tissues to elucidate the evolution, structure, and function of the genes.
The results of the phylogenetic analysis showed that yak ERα and ERβ belonged to the ERα and ERβ clusters, respectively. Both also shared high identities of amino acid sequences with other ER of mammals. These results suggest that ERs were quite conserved in mammal molecular evolution.
ERs are divided into six domains designated A-F by the deletion and point mutation technology similarity to other steroid hormone receptors. The results of the alignment showed that the A/B domain was hyper-variable between ERα and ERβ. In this study, we found that yak ERα had a considerably longer A/B domain than ERβ (182 compared with 120 amino acids), which is similar with previous reports of other species (Ma et al., 2000; Choi and Habibi, 2003). hERα was significantly different compared with hERβ; the A/B domain of hERα had a ligand-independent transactivation (AF-1) function by the promoter and cell context. Both are a combination of GAL4 DNA binding domain (GAL4-DBD) fusion protein; the N-terminal region of ERα possessed an autonomous and ligand-independent activity in HeLa cells, but not ERβ. After deletion, the N-terminal region of hERβ showed higher activity than the whole hERβ. This result indicates that the A/B domain of ERβ could repress a target function. Therefore, we speculate that the length of yak A/B domains between the ERα and ERβ may be related to the activity of the A/B domain.
The distribution of the eight cysteine residues was the same in both ERα and ERβ, which is composed of two zinc fingers (Menue et al., 2002). These structures were necessary for combining with sequences of the target gene. (Kumar et al., 1987). Moreover, the C and E domains of the yak ERs were highly conserved from fish to mammals in both ERα and ERβ (C domain, 94% to 99%; E domain, 58% to 94%) in the six bindings, which agreed with previous results (Katsu et al., 2010). The aforementioned evidence indicated that these two domains were the core of the estrogen and are essential for estrogen actions. Therefore, the basic functions of ERs have been conserved during evolution. The D domain had a less conserved fragment than other domains, whereas an arginine residue, which is surrounded by other residues, was conserved in ERs. The present pattern of arginine showed that it has function in the ER secondary structure. Moreover, this pattern contributes to consolidating the structure of DNA binding.
This study showed that both ERα and ERβ were expressed in a variety of tissues in yak, which is similar to the ERs in many species (Socorro et al., 2000; Menuet et al., 2002; Choi and Habibi, 2003). This result further supports the diverse functions of ERs in yak. The mRNA expression levels of yak ERs were predominant in the mammary gland, uterus, and oviduct and showed low expression in the liver, heart, spleen, lung, kidney, and testis. Similarly, Katsu et al. (2010) reported that the reproductive organs are the main sites of ER synthesis. Our results also further demonstrated that ER genes possessed many functions, but the main function was in the regulation of sexual differentiation. We also found that in the same reproductive organ, except testis, the expression of ERα was higher than ERβ. Thus, the present data indicates that ERα is essential for fertility, mammary gland development, and lactation. In addition, ERβ has important functions in normal ovulation, but is not significant in lactation and reproduction, which is similar to Rattus norvegicus (Hiroi et al., 1999).
We reported the identification of ER genes, i.e., ERα (belonging to ERα) and yak ERβ (belonging to ERβ), in domestic yak. The extensive distribution of ER gene product expression in domestic yaks strongly supported that ERs have different functions in yak, and the predominant expression in reproductive organs further showed the evolutionary diversification and physiological function of the mammalian ER gene.
REFERENCES
- Bakker J, Brock O. Early oestrogens in shaping reproductive networks: Evidence for a potential organisational role of oestradiol in female brain development. J Neuroendocrinol. 2010;22:728–735. doi: 10.1111/j.1365-2826.2010.02016.x. [DOI] [PubMed] [Google Scholar]
- Beyer C, Pawlak J, Karolczak M. Membrane receptors for oestrogen in the brain. J Neurochem. 2003;87:545–550. doi: 10.1046/j.1471-4159.2003.02042.x. [DOI] [PubMed] [Google Scholar]
- Billiar RB, Zachos NC, Burch MG, Albrecht ED, Pepe GJ. Up-regulation of alpha-inhibin expression in the fetal ovary of estrogen-suppressed baboons is associated with impaired fetal ovarian folliculogenesis. Biol Reprod. 2003;68:1989–1996. doi: 10.1095/biolreprod.102.011908. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Evans RM. Orphan nuclear receptors—new ligands and new possibilities. Genes Dev. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- Chang XT, Kobayashi T, Todo T, Ikeuchi T, Yoshiura M, Kobayashi HK, Morrey C, Nagahama Y. Molecular cloning of estrogen receptors α and β in the ovary of a teleost fish, the tilapia (Oreochromis niloticus) Zoolog Sci. 1999;16:653–658. [Google Scholar]
- Choi C, Habibi H. Molecular cloning of estrogen receptor α and expression pattern of estrogen receptor subtypes in male and female goldfish. Mol Cell Endocrinol. 2003;204:169–177. doi: 10.1016/s0303-7207(02)00182-x. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-ERβ-A. Nature. 1986;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- Hewitt SC, Korach KS. Oestrogen receptor knockout mice: Roles for oestrogen receptors alpha and beta in reproductive tissues. Reproduction. 2003;125:143–149. doi: 10.1530/rep.0.1250143. [DOI] [PubMed] [Google Scholar]
- Hiroi H, Inoue S, Watanabe T, Goto W, Orimo A, Momoeda M, Tsutsumi O, Taketani Y, Muramatsu M. Differential immunolocalization of estrogen receptor alpha and beta in rat ovary and uterus. J Mol Endocrinol. 1999;22:37–44. doi: 10.1677/jme.0.0220037. [DOI] [PubMed] [Google Scholar]
- Katsu Y, Taniguchi E, Urushitani H, Miyagawa S, Takase M, Kubokawa K, Tooi O, Oka T, Santo N, Myburgh J, Matsuno A, Taisenlguchi Molecular cloning and characterization of ligand- and species-specificity of amphibian estrogen receptors. Gen Comp Endocrinol. 2010;168:220–230. doi: 10.1016/j.ygcen.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Klein SL, Strausberg R, Wagner L, Pontius J, Clifton SW, Richardson P. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- Krust A, Green S, Argos P, Kumar V, Walter P, Bornert JM, Chambon P. The chicken oestrogen receptor sequence: Homology with v-erbA and the human oestrogen and glucocorticoid receptors. EMBO J. 1986;5:891–897. doi: 10.1002/j.1460-2075.1986.tb04300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- Ma CH, Dong KW, Yu KL. cDNA cloning and expression of a novel estrogen receptor β-subtype in goldfish (Carassius auratus) Biochim. Biophys Acta (BBA)-Gene Structure and Expression. 2000;1490:145–152. doi: 10.1016/s0167-4781(99)00235-3. [DOI] [PubMed] [Google Scholar]
- McCarthy M. How it’s made: organisational effects of hormones on the developing brain. J Neuroendocrinol. 2010;22:736–742. doi: 10.1111/j.1365-2826.2010.02021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuet A, Pellegrini E, Anglade I, Blaise O, Laudet V, Kah O, Pakdel F. Molecular characterization of three estrogen receptor forms in zebrafish: binding characteristics, transactivation properties, and tissue distributions. Biol Reprod. 2002;66:1881–1892. doi: 10.1095/biolreprod66.6.1881. [DOI] [PubMed] [Google Scholar]
- Mermelstein PG, Micevych PE. Nervous system physiology regulated by membrane estrogen receptors. Rev Neurosci. 2008;19:413–424. doi: 10.1515/revneuro.2008.19.6.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuet A, Pellegrini E, Anglade I, Blaise O, Laudet V, Kah O, Pakdel F. Molecular characterization of three estrogen receptor forms in zebrafish: Binding characteristics, transactivation properties, and tissue distributions. Biol Reprod. 2002;66:1881–1892. doi: 10.1095/biolreprod66.6.1881. [DOI] [PubMed] [Google Scholar]
- Mosselman S, Polman J, Dijkema R. ERβ: Identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- Pakdel F, Gac FL, Goff PL, Valotaire Y. Full-length sequence and in vitro expression of rainbow trout estrogen receptor cDNA. Mol Cell Endocrinol. 1990;71:195–204. doi: 10.1016/0303-7207(90)90025-4. [DOI] [PubMed] [Google Scholar]
- Sarkar M, Prakash B. Circadian variations in plasma concentrations of melatonin and prolactin during breeding and non-breeding seasons in yak (Poephagus grunniens L.) Anim Reprod Sci. 2005;90:149–162. doi: 10.1016/j.anireprosci.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Scatchard G. The attractions of proteins for small molecules and ions. Ann NY Acad Sci. 1949;51:660–672. [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sengupta J, Roy S, Manchanda S. Effect of an oestrogen synthesis inhibitor, 1, 4, 6-androstatriene-3, 17-dione, on mouse embryo development in vitro. J Reprod Fertil. 1982;66:63–66. doi: 10.1530/jrf.0.0660063. [DOI] [PubMed] [Google Scholar]
- Sinclair A, Smith C, Western P, McClive P. A comparative analysis of vertebrate sex determination. Novartis Foundation Symposium. 2002;244:102–125. [PubMed] [Google Scholar]
- Socorro S, Power DM, Olsson PE, Canario AV. Two estrogen receptors expressed in the teleost fish, Sparus aurata: cDNA cloning, characterization and tissue distribution. J Endocrinol. 2000;166:293–306. doi: 10.1677/joe.0.1660293. [DOI] [PubMed] [Google Scholar]
- Sumida K, Ooe N, Saito K, Kaneko H. Molecular cloning and characterization of reptilian estrogen receptor cDNAs. Mol Cell Endocrinol. 2001;183:33–39. doi: 10.1016/s0303-7207(01)00613-x. [DOI] [PubMed] [Google Scholar]
- Todo T, Adachi S, Yamauchi K. Molecular cloning and characterization of Japanese eel estrogen receptor cDNA. Mol Cell Endocrinol. 1996;119:37–45. doi: 10.1016/0303-7207(96)03792-6. [DOI] [PubMed] [Google Scholar]
- Vasudevan N, Pfaff DW. Non-genomic actions of estrogens and their interaction with genomic actions in the brain. Front Neuroendocrinol. 2008;29:238–257. doi: 10.1016/j.yfrne.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Wang PH. Role of sex hormone receptors in ovulation. Taiwan J Obstet Gynecol. 2005;44:16–25. [Google Scholar]
- Weiler IJ, Lew D, Shapiro DJ. The Xenopus laevis estrogen receptor: Sequence homology with human and avian receptors and identification of multiple estrogen receptor messenger ribonucleic acids. Mol Endocrinol. 1987;1:355–362. doi: 10.1210/mend-1-5-355. [DOI] [PubMed] [Google Scholar]
- Zi X-D. Reproduction in female yaks (Bos grunniens) and opportunities for improvement. Theriogenology. 2003;59:1303–1312. doi: 10.1016/s0093-691x(02)01172-x. [DOI] [PubMed] [Google Scholar]