Abstract
Histone modifications on major transcription factor target genes are one of the major regulatory mechanisms controlling adipogenesis. Plant homeodomain finger 2 (PHF2) is a Jumonji domain-containing protein and is known to demethylate the histone H3K9, a repressive gene marker. To better understand the function of PHF2 in adipocyte differentiation, we constructed stable PHF2 knock-down cells by using the mouse pre-adipocyte cell line 3T3-L1. When induced with adipogenic media, PHF2 knock-down cells showed reduced lipid accumulation compared to control cells. Differential expression using a cDNA microarray revealed significant reduction of metabolic pathway genes in the PHF2 knock-down cell line after differentiation. The reduced expression of major transcription factors and adipokines was confirmed with reverse transcription- quantitative polymerase chain reaction and Western blotting. We further performed co-immunoprecipitation analysis of PHF2 with four major adipogenic transcription factors, and we found that CCATT/enhancer binding protein (C/EBP)α and C/EBPδ physically interact with PHF2. In addition, PHF2 binding to target gene promoters was confirmed with a chromatin immunoprecipitation experiment. Finally, histone H3K9 methylation markers on the PHF2-binding sequences were increased in PHF2 knock-down cells after differentiation. Together, these results demonstrate that PHF2 histone demethylase controls adipogenic gene expression during differentiation.
Keywords: adipogenesis, epigenetic regulation, histone demethylase, PHF2, transcriptional control
INTRODUCTION
Research related to adipose tissue has revealed important insight demonstrating that this organ is not only a source of spare energy but also a vital endocrine organ that plays a central role in regulation of the energy balance of the organism. The main dysfunctions of adipose tissue, obesity and lipodystrophy, are related to the development of many geriatric diseases, including diabetes, hypertension, and atherosclerosis (Friedman, 2000). Therefore, better understanding of the controlling mechanism of fat cell differentiation would facilitate development of novel approaches for treating or relieving symptoms of these diseases. Adipocyte differentiation is tightly controlled through hierarchic control of many transcriptional factors. In the 3T3-L1 mouse preadipocyte model system, hormonal stimuli of cAMP, insulin, and glucocorticoids are known to induce CCATT/enhancer-binding protein β (C/EBPβ) and C/EBPδ followed by peroxisome proliferator-activated receptor γ (PPARγ) and C/EBPα expression during differentiation (Rosen and Spiegelman, 2000). PPARγ is the terminal factor that controls adipogenesis and the ultimate expression of many adipogenic genes, including fatty acid-binding protein (FABP), glycerol kinase, lipoprotein receptor, and phosphoenolpyruvate carboxykinase (Rosen et al., 1999).
Eukaryotic transcriptional regulation is tightly controlled by various mechanisms. One such mechanism is histone modification, which has been widely and intensively studied in the past few decades (Margueron et al., 2005). The nucleosome consists of 6 different histones, each of which has a unique site for post-translational modification, including phosphorylation, acetylation, and methylation (Zhang and Reinberg, 2001). Histone methylations are related to both gene activation and repression depending on the amino acid position in which they reside. For example, H3K4 and H3K36 methylations are associated with active transcription, whereas H3K9, H3K27, and H4K20 methylations are associated with gene repression (Cao et al., 2002). Specific enzymes with methyl-transferase or demethylase activity have been identified to be involved in methylation and demethylation at each amino acid position.
As a transcriptionally controlled process, adipogenesis is accompanied by several histone modifications. In particular, histone H3 and H4 acetylation is associated with the activation of adipogenic gene expression (Debril et al., 2004). The methyltransferase enzyme PR-Set7/Setd8 is known to act on PPARγ for gene-targeted histone regulation during adipogenesis (Wakabayashi et al., 2009). The H3K27 methyl transferase enhancer of zeste homolog 2 (EZH2) induces adipogenesis by silencing genes involved in the WNT pathway (Wang et al., 2010). Knock-down of the H3K4/K9 demethylase LSD1 repressed adipogenesis, and H3K9 methyl-transferase Setdb1 knock-down promoted adipogenesis in 3T3-L1 and 10T1/2 cell lines (Musri et al., 2010).
PHF2 is a member of the KDM7 histone demethylase family. KDM7 family proteins harbor a plant homeo domain (PHD) in the N-terminal and the Jumonji-C domain, which has demethylase enzymatic activity (Fortschegger and Shiekhattar, 2011). PHF2 was first identified as a candidate gene for hereditary sensory neuropathy type I because of its high expression in the neural tube and dorsal root ganglia (Fortschegger and Shiekhattar, 2011). Biochemical analysis showed that PHF2 acts as an H3K9 demethylase upon physical interaction with H3K4 (Wen et al., 2010). In hepatocytes, PHF2 was found to be phosphorylated by protein kinase A and interacted with ARID5B to activate targeting promoters (Baba et al., 2011); in addition, several reports have demonstrated an association between PHF2 and potential oncogenic processes (Ghosh et al., 2012; Sun et al., 2013). PHF2 is also involved in pro-inflammatory gene expression in macrophages by demethylating the H4K20 methyl group at a nuclear factor-kappa B (NF-κB)-binding site (Stender et al., 2012).
In this study, we analyzed the role of PHF2 in adipocyte differentiation using PHF2 knock-down cell lines. We found that PHF2 interacts with C/EBPα and C/EBPδ transcription factors during adipogenesis and positively regulates adipogenesis.
MATERIALS AND METHODS
Materials
DMEM cell culture media, trypsine, anti-biotics, Trizol and Lipofectamine 2000 were purchased from Invitrogen (USA), fetal bovine serum and culture media were obtained from Hyclone Laboratories Inc. (USA). Antibodies against PPARγ, C/EBPα, C/EBPβ, C/EBPδ, FABP4, HA and beta-tubulin were purchased from SantaCruz Biotech. (USA), and antibody against PHF2 and H3K9-Me2 were from Cell Signaling (USA). Insulin, dexamethasone, 3-isobutyl-1-methylxanthine (IBMX) and other chemicals were purchased form Sigma-Aldrich (USA).
Cell lines, plasmids, virus production and siRNA transfection
3T3-L1 cell line was purchased from American Type Culture Collection (USA). The cell lines were cultured in DMEM supplemented with 10% bovine serum at 37°C in 5% CO2. Full-length PHF2 cDNAs were cloned by RT-PCR with specific primers (Supplementary Table S1), and amplified using a pfu DNA polymerase, which were inserted into Flag/Streptavidin binding protein (F/S)-tagged vector. PHF2-H249A mutant was made with QuikChange™ mutagenesis kit (stratagene, USA). HA tagged C/EBPα, C/EBPβ, C/EBPδ and PPARγ cDNAs were kind gift from Dr. JB Kim (Seoul National University). For gene silencing, the pLKO.1-puro vector was purchased from Sigma-Aldrich and oligonucleotides for control shRNA or PHF2 shRNA (sequences are listed in Supplementary Table S1) were inserted into the vector using AgeI and EcoRI restriction enzymes. 3T3-L1 cells were infected overnight with virus. For siRNA transfection, control and target siRNAs (sequences listed on Supplementary Table S2) were transfected as 50nM concentration on confluent 3T3-L1 cells and adipogenesis were started after two days.
3T3-L1 cell adipogenesis induction and Oil Red O staining
3T3-L1 stable cell lines were grown to confluent and induced with adipogenesis media (DMEM with 10% FBS, 10 μg/ml insulin, 0.5 mM IBMX and 1 μM dexamethasone). Media were changed with DMEM with 10% FBS and 10 μg/ml insulin after two days and maintained with it. After 8 days of differentiation, cells were fixed with 10% Formaldehyde solution for 20 min at room temperature. Fixed cells were incubated 1 h with freshly prepared 0.3% Oil Red-O solution in 60% isopropanol. After removal of staining solution, immobilized Oil Red-O dye was extracted with 100% isopropanol and OD520 were measured.
RNA isolation and RT-qPCR
Total RNA was extracted using Trizol according to the instructions of the manufacturer (Ambion, USA). For each reverse transcription, 1 μg of total RNA was used for cDNA synthesis using MultiScribe Reverse Transcription kit (Applied Biosystems, USA). Real time PCR was done with the Evagreen qPCR mastermix from Applied Biological Materials Inc. (Canada) using StepOne™ Real-Time PCR System (Applied Biosystems). Quantity of 18S ribosomal RNA was used for an internal control. PCR primer sequences are listed in Supplementary Table S1.
Microarray analysis and data acquisition
3T3-L1 stable cell lines expressing control shRNA or PHF2 shRNA were differentiated with adipogenic media for 4 days. Total RNAs after differentiation were extracted, and the synthesis of target cDNA probes and hybridization were performed using Agilent’s Low RNA Input Linear Amplification kit (Agilent Technology, USA) according to the manufacturer’s instructions. The hybridized images were scanned using Agilent’s DNA microarray scanner and quantified with Feature Extraction Software (Agilent Technology). All data normalization and selection of fold-changed genes were performed using GeneSpringGX 7.3 (Agilent Technology). The averages of normalized ratios were calculated by dividing the average of normalized signal channel intensity by the average of normalized control channel intensity. Functional annotation of genes was performed according to DAVID (http://david.abcc.ncifcrf.gov/), and Medline databases (http://www.ncbi.nlm.nih.gov/).
Western blotting
Cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.2, 0.5% NP-40, 1% Triton X-100, 1% sodium deoxycholate) containing a protease inhibitor cocktail. Cell lysates were separated on SDS-polyacrylamide gels, and transferred to an Immobilon-P membrane (Millipore, USA). Membranes were blocked with 5% skim milk and 0.1% Tween-20 for 1 h, and then incubated overnight at 4°C with a primary antibody diluted 1:1000. Membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (1:5000) for 1 h, and developed using the ECL-plus kit (Thermo Scientific, USA).
Streptavidin-binding peptide (SBP) pull-down analysis and Co-immunoprecipitations
For SBP pull-down, cell lysates (1 mg of protein) were applied to 10 μl of Streptavidin Sepharose™ High Performance bead from GE Healthcare (England) overnight at 4°C. After extensive wash with binding buffer, bead-bound proteins were eluted with biotin, and then subjected to SDS-PAGE and Western blotting with indicated antibodies. For immunoprecipitation, cell lysates (1 mg of protein) were incubated with 5 μl of a primary antibody at 4°C for 16 h, and then with 10 μl of protein A/G-Sepharose beads from GE Healthcare for 2 h at 4°C. Bead-bound proteins were eluted with SDS sample buffer and then subjected to SDS-PAGE and Western blotting with indicated antibodies
Chromatin Immunoprecipitation
Lysates from 1 × 107 cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and glycine (finally 125 mM) was added to quench the cross-linking reaction. Lysed cell extracts were subjected to sonication until the DNA fragments become less than 500 base pair. Chromatin complexes were precipitated with indicated antibodies overnight at 4°C. Protein A/G-sepharose beads were added and incubated 2 more hours. After washing with saline, DNA-protein complexes were eluted with elution buffer (1% SDS, 0.1 M NaHCO3) and incubated overnight at 65°C to reverse cross-links. The eluted DNAs were purified with Mega-quick spin DNA purification kit (Intron, Korea). The precipitated DNAs were amplified and quantified on StepOne™ Real-Time PCR System using the Evagreen qPCR master mix. PCR primer sequences used for ChIP are listed in Supplementary Table S1.
Statistical analyses
All data were analyzed using Microsoft Excel 2007 software, otherwise stated. Continuous variables were analyzed using Student’s t-test if the data were normally distributed. Otherwise, the Mann Whitney U-test was used as a non-parametric analysis. All statistical tests were two-sided. Significance was considered when p is < 0.05.
RESULTS
PHF2 knock-down reduced the adipogenesis capacity of 3T3-L1 pre-adipocytes and induced expression changes in metabolic pathway genes
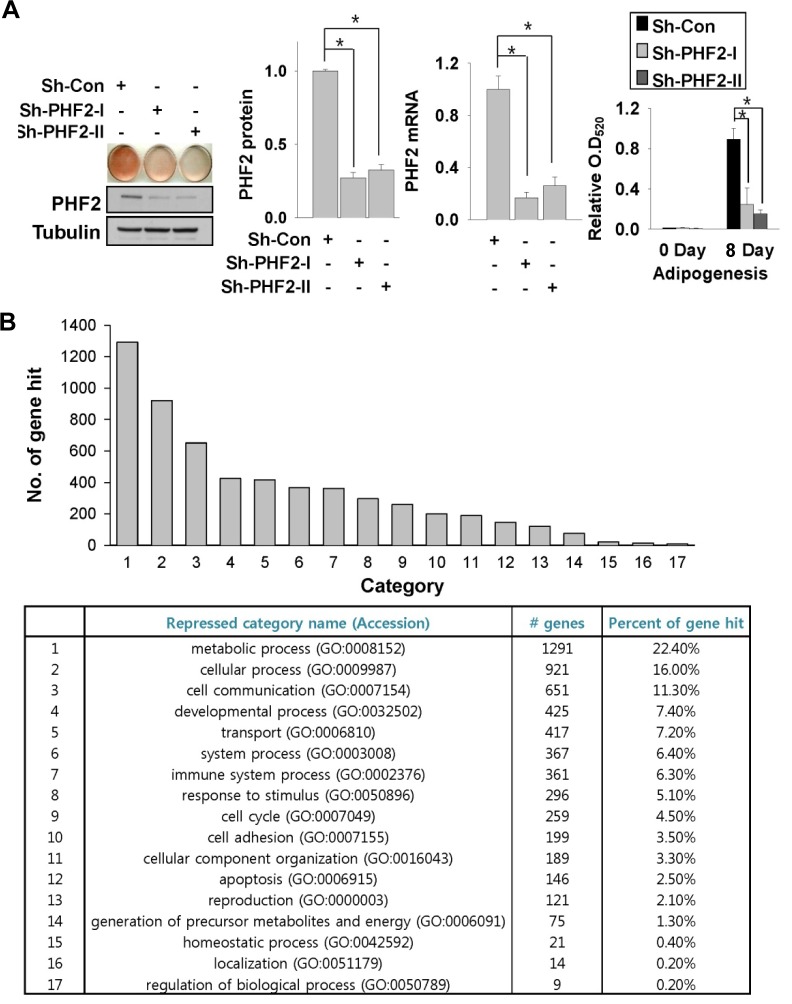
To better understand the biological role of the histone demethylase PHF2 in the adipocyte differentiation process, we established stable knock-down cell lines using a lentiviral shRNA expression system in 3T3-L1 pre-adipocytes. After selection with appropriate antibiotics, we induced adipogenesis using control and PHF2 shRNA-expressing cell lines. Western blotting using a PHF2 antibody and qRT-PCR (Fig. 1A) showed a significant reduction in PHF2 protein and mRNA levels, respectively, in. PHF2 shRNA-expressing cell lines compared to the control. After eight days of adipogenic induction, there was a significant decrease in lipid accumulation in PHF2 knock-down cell lines compared to the control. cell line, as determined by Oil Red-O staining (Fig. 1A, left); isopropanol extraction of Oil Red-O showed a significant reduction in dye quantity (Fig. 1A, right). To evaluate the genome-wide effect of PHF2 knock-down during the adipogenesis process, we induced both control and PHF2 knock-down 3T3-L1 stable cell lines with adipogenic media and compared their mRNA expression profiles using a mouse cDNA micro-array. We compared the most dramatically down-regulated genes (< 0.5-fold, p-value < 0.05) between control and PHF2 knock-down cell lines after adipogenesis using the Panther 9.0 program (www.pantherdb.org). Most of the genes of representative cellular pathways in the microarray data showed reduced expression in PHF2 knock-down stable cells (Fig. 1B). Genes related to metabolic processes showed the most substantial repression, corresponding to 22.4% of all genes that were significantly repressed in PHF2 knock-down cell lines after differentiation.
Fig. 1.
PHF2 knock-down reduced the adipogenesis capacity of 3T3-L1 pre-adipocytes. (A) Oil Red-O staining of stable 3T3-L1 cell lines expressing control or PHF2 shRNA. Cell extracts were analyzed with Western blotting using anti-PHF2 and tubulin antibodies (left). PHF2 mRNA and protein levels of indicated stable cell lines were measured before adipogenesis and plotted (middle panels). Oil Red-O dye was extracted with isopropanol and the optical density at 520 nm (O. D520) was measured (left). Bars represents the means ± SDs of three independent experiments, and “*” denotes P < 0.01 versus the control shRNA (sh-con) group. (B) Number of hits for genes involved in cellular pathways showing reduced expression after adipogenesis in the PHF2 knock-down cell line. Genes repressed by more than 0.5-fold compared to the control cell line and had a p-value of less than 0.05 were selected. A total of 4069 genes were analyzed using the DAVID (http://david.abcc.ncifcrf.gov/) database.
Adipogenic genes were down-regulated in PHF2 knockdown stable cell lines
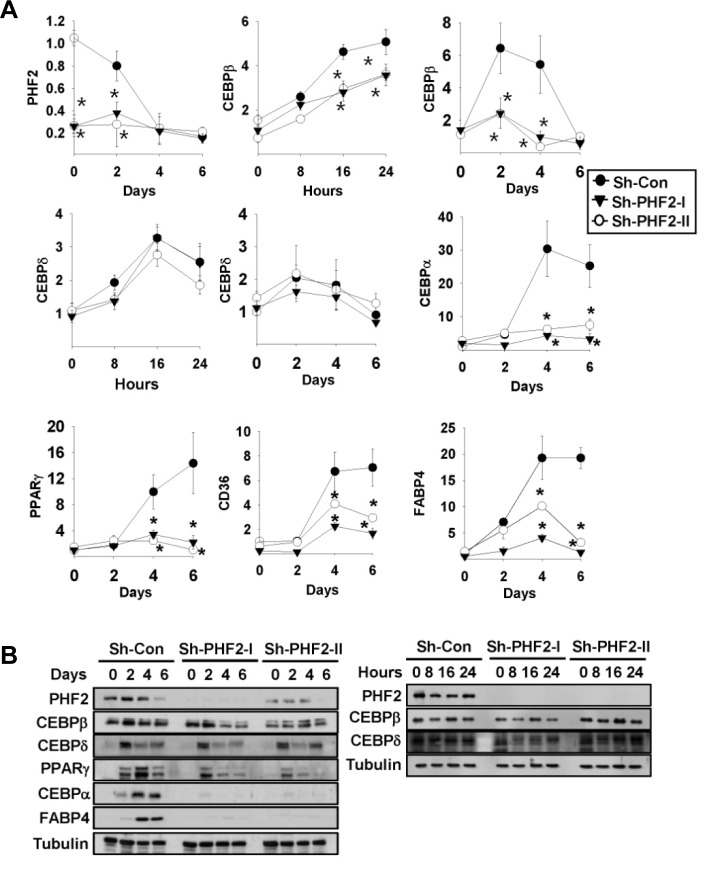
To confirm the expression changes observed in the cDNA microarray analysis, we evaluated the mRNA levels of representative adipogenic genes, including CEBPA, CEBPB, CEBPD, PPARG, CD36, and FABP4, by using qRT-PCR. We induced adipogenesis for six days and harvested cells as indicated in the “Materials and Methods”. We found the PHF2 mRNA of control shRNA was reduced during adipogenesis (Fig. 2A). The expression of all tested adipogenic genes from control shRNA-expressing cells was significantly induced after adipogenic stimulation, whereas PHF2 shRNA-expressing cells did not show induction of these genes, except for the CEBPD gene (Fig. 2A). Next, we measured protein levels of major adipogenic markers from the same samples. Under the same conditions, the two most upstream adipogenic transcription factors C/EBPβ and C/EBPδ showed a slight reduction and no change in protein levels, respectively, in PHF2 knock-down cell lines. However, protein levels of downstream transcription factors, PPARγ and C/EBPα were dramatically reduced in PHF2 knockdown cells. Finally, expression of the adipogenic marker FABP4 was not induced during adipogenesis of the PHF2 shRNA-expressing cell line (Fig. 2B). These results demonstrated that PHF2 may function as a major regulator during the adipogenic process. Furthermore, the protein level of PHF2 was gradually decreased during adipogenesis, as shown in the control shRNA sample in Fig. 2B, which is consistent with the mRNA data shown in Fig. 2A.
Fig. 2.
PHF2 knock-down reduced mRNA and protein levels of adipogenesis marker genes. (A) 3T3-L1 stable cells were differentiated with adipogenesis-inducing media and then lysed at the indicated times. Relative mRNA levels of the indicated genes were determined by qRT-PCR. Bars represents the means ± SDs of three independent experiments, and “*” denotes p < 0.01 versus the control shRNA (sh-con) group. (B) Control and PHF2 shRNA-expressing stable 3T3-L1 cell lines were treated with adipogenic rea-gent for the indicated number of days (left) or hours (right), and cell lysates were analyzed with Western blotting with the indicated antibodies.
PHF2 physically interacts with C/EBPα and C/EBPδ
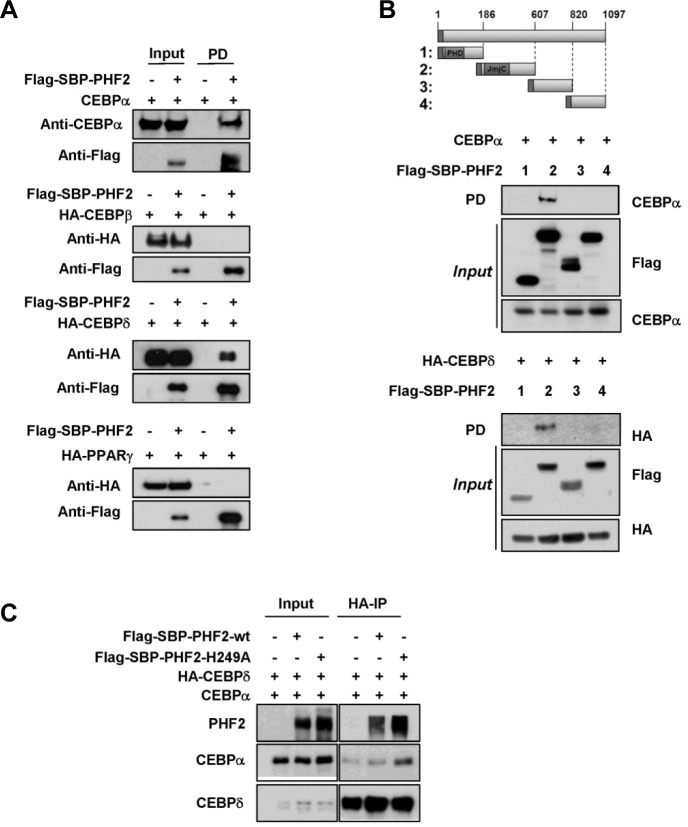
As PHF2 is a well-known transcriptional activator via regulation of epigenetic histone H3K9 methylation interacting with NF-κB or ARID5 (Baba et al., 2011; Stender et al., 2012), we hypothesized that PHF2 might interact with one of the transcription factors involved in fat cell differentiation for recruitment to the transcriptional complex. Therefore, we conducted a co-immunoprecipitation analysis of PHF2 with ectopically expressed transcriptional factors for adipogenesis, including C/EBPα, C/EBPβ, C/EBPδ and PPARγ. PHF2 was found to interact with C/EBPα and C/EBPδ (Fig. 3A), but not with C/EBPβ or PPARγ. We next evaluated the domain of PHF2 that is involved in the interaction with C/EBPα and C/EBPδ. We ectopically expressed PHF2 domains with C/EBPα and C/EBPδ, and found that the Jumonji domain of PHF2 interacted with C/EBPα and C/EBPδ (Fig. 3B). C/EBPα and C/EBPδ can combine to make a heterodimer complex; thus, we further evaluated the possibility that PHF2 contributes to the formation of this complex. The co-immunoprecipitation of C/EBPα and C/EBPδ was confirmed in ectopically expressed proteins, but the interaction was not affected by the presence of PHF2 or a PHF2 non-functional mutant (Fig. 3C).
Fig. 3.
PHF2 physically interacts with C/EBPα and C/EBPδ (A) Flag-SBPPHF2 and HA-tagged C/EBPβ, C/EBPδ, PPARγ and non-tagged C/EBPα were ectopically expressed in HEK293T cells, and cell lysates were incubated with streptavidin beads. The bound proteins were eluted with biotin. Input and streptavidin-bound proteins were detected by western blotting with the indicated antibodies. (B) F/S-tagged PHF2 fragments (1–4) were co-expressed with C/EBPα or HA-C/EBPδ plasmid in HEK293T cells. After F/S-PHF2 fragments were precipitated using streptavidin beads, co-precipitated proteins were detected by Western blotting using the indicated antibodies. (C) C/EBPα and C/EBPδ were co-expressed with wild-type PHF2 or the PHF2-H249A mutant. After HAC/EBPδ was precipitated using an HA antibody, co-precipitated proteins were detected by western blotting using the indicated antibodies.
PHF2 is recruited to C/EBP response element (CEBPRE) of the PPARG, CEBPA and FABP4 promoters during adipogenesis together with C/EBPδ or C/EBPα
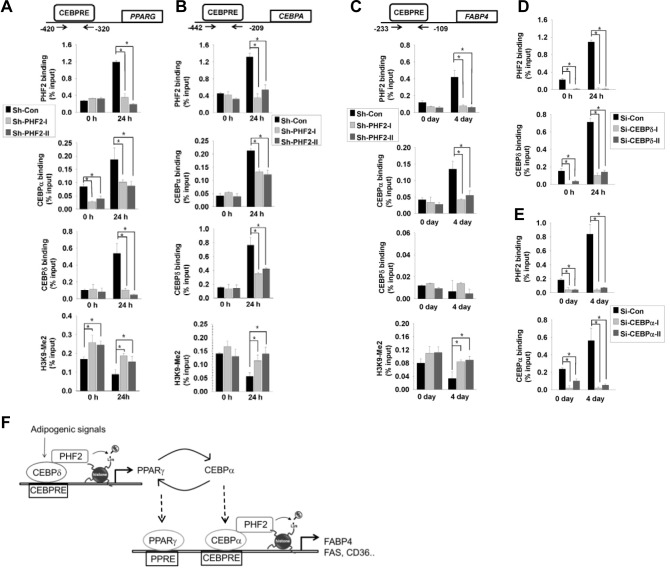
As C/EBPα and C/EBPδ are known to regulate the PPARG gene, we performed chromatin immunoprecipitation analysis with PHF2, C/EBPα and C/EBPδ antibodies and confirmed the binding of these proteins on the PPARG promoter. The binding of PHF2 on the PPARG promoter was induced after adipogenesis induction and was abolished in PHF2 shRNA stable cell lines (Fig. 4A, first row). The binding of C/EBPα and C/EBPδ on the PPARG promoter also increased after adipogenesis induction in control cells, and this induction was lost in PHF2 shRNA stable cell lines (Fig. 4A, second and third rows). Finally, PHF2 knockdown cell lines showed increased dimethyl H3K9 modification on the PPARG promoter compared to the control cell line after adipogenesis (Fig. 4A, fourth row). Next, we measured PHF2, C/EBPα and C/EBPδ binding on the CEBPA (Fig. 4B) and FABP4 (Fig. 4C) promoters. The binding of PHF2, C/EBPα, and C/EBPδ was observed on the CEBPA promoter after adipogenic induction, and these bindings were abolished with PHF2 knockdown (Fig. 4B). The binding of PHF2 and C/EBPα to the FABP4 promoter was observed and PHF2 knock-down abolished this binding (Fig. 4C, first row and second row), but C/EBPδ was not associated with the FABP4 promoter during adipogenesis (Fig. 4C, third row). Finally, similar to the PPARG promoter, H3K9 dimethylation of the CEBPA and FABP4 promoter was induced in PHF2 knock-down cells (Figs. 4B and 4C; fourth row). To address whether the binding of PHF2 on PPARG is C/EBPα dependent and the binding on the FABP4 promoter is C/EBPδ dependent, we transfected 3T3L1 cells with siRNAs targeting C/EBPα or C/EBPδ to knock them down. When we immunoprecipitated PHF2 from C/EBPδ knock-down adipocytes after 24 h of induction, the binding of PHF2 was dramatically reduced (Fig. 4D). Moreover, PHF2 binding on the FABP4 promoter was abolished when C/EBPα was repressed (Fig. 4E). These results showed that the binding of PHF2 on each CEBPRE is dependent on transcription factors binding together.
Fig. 4.
PHF2 was recruited to PPARG and FABP4 promoters with C/EBPα and C/EBPδ. 3T3-L1 cells expressing control shRNA or PHF2 shRNA were stimulated with an adipogenesis cocktail for the indicated times 24th, and chromatin immunoprecipitation was performed with anti-PHF2, anti-C/EBPα anti-C/EBPδ or histone H3K9-Me2 antibodies. The resulting immuno-precipitated DNA was analyzed by qPCR using primers for (A) the PPARG promoter sequence, (B) the CEBPA promoter sequence, or (C) the FABP4 promoter sequence. The positions of primers relative to the transcriptional start site are indicated. 3T3L1 pre-adipocytes transfected with (D) C/EBPδ or (E) C/EBPα siRNAs (sequences listed in Supplement Table S2) were induced with adipogenesis cocktail for the indicated times. Immunoprecipitated DNAs were PCR-amplified with primers targeting the (D) PPARG promoter or (E) FABP4 promoter CEBP response element. Bars represents the means ± SDs of three independent experiments, and “*” denotes p < 0.01 versus the control group. (F) Schematic illustrating the role of PHF2 in C/EBPδ and C/EBPα transcriptional activation.
DISCUSSION
C/EBPβ and C/EBPδ expression occur early in the adipogenesis process, which results in the transcriptional activation of CEBPA and PPARG expression. PPARγ then activates its own promoter and CEBPA expression. PPARγ and C/EBPα cooperate to activate the expression of most adipocyte genes, including FABP4, FAS, and CD36. In this study, we identified the histone demethylase PHF2 as an epigenetic co-regulator of the adipogenic transcription factors C/EBPα and C/EBPδ In the 3T3-L1 pre-adipocyte model system, PHF2 knock-down dramatically repressed adipogenesis (Fig. 1A). The microarray and subsequent qRT-PCR data suggested that PHF2 knock-down resulted in reduction in the expression of major genes related to fat cell differentiation, including CEBPA and PPARG. These results indicate that the transcriptional activity of the upstream factors of CEBPA and PPARG had changed. Although it was previously reported that PHF2 interacts with C/EBPα and demethylates its target promoter to activate adipogenesis (Okuno et al., 2012), their result does not sufficiently explain why the CEBPA mRNA level changed in PHF2 knock-down cells (Fig. 2A). In this study, we not only confirmed the interaction of PHF2 with C/EBPα but also demonstrated that PHF2 interacts with C/EBPδ, an upstream regulator of C/EBPα and PPARγ. This indicates that PHF2 acts at a much earlier stage of adipogenesis than previously expected. We also demonstrated the physical interaction of PHF2 with C/EBPδ, an upstream transcriptional factor of C/EBPα and PPARγ, and confirmed the binding of PHF2 on the CEBPA and PPARG promoters after adipogenic induction (Figs. 4A and 4B). The repression of adipogenesis after PHF2 knock-down in Okuno’s (2013) study was rather moderate compared to the dramatic repression of adipogenesis observed in our study. This may reflect a knock-down efficiency difference between the shRNA vectors used in the different studies or might simply be due to the adipogenic capacity difference of the 3T3L1 cells used for stable cell lines.
As a key regulator of the adipogenic process, C/EBPα and PPARγ expression is a major target for studies related to histone modification. The H3K9 methyltransferase G9a inhibits adipogenesis by inhibiting PPARG expression and facilitating Wnt10a expression (Wang et al., 2013). The methylation regulator PTIP, which associates with the histone methyl transferase MLL4, is required for PPARG and CEBPA expression during adipogenesis (Cho et al., 2009). Our study shows that yet another histone demethylase, PHF2, is also involved in the change in CEBPA and PPARG expression by interacting with C/EBPδ and modifying histone methylation on the CEBPA and PPARG gene promoters.
PPARγ is a critical regulator of adipogenesis and a target of the anti-diabetic drug thiazolidinedione. Several transcription factors, including C/EBPs, GATAs and CREB, have been reported to regulate the gene expression of PPARG in various cell types (Fox et al., 2006). Specifically, C/EBPδ was reported to be responsible for the induction of PPARγ expression in 3T3-L1 mouse pre-adipocytes through dexamethasone-mediated stimulation (Shi et al., 2000) or TNFα-mediated inhibition (Kudo et al., 2004). In the human hepatocyte cell line HepG2, C/EBPδ was found to bind to the PPARG promoter after lipogenic stimulation, and this activation was accompanied by HDAC1 and HDAC3 binding (Lai et al., 2008). Considering our observation that C/EBPδ can epigenetically regulate PPARG expression in adipogenic process, it would be interesting to next investigate whether PHF2 has a biological function in the regulation of the PPARG promoter activation described earlier.
In summary, we found that the histone demethylase PHF2 controls adipogenesis through binding with the transcription factors C/EBPδ and C/EBPα. We demonstrated a significant increase in C/EBPδ binding on the CEBPA and PPARG promoters and in C/EBPα binding on the FABP4 promoter in the 3T3-L1 cell line after adipogenic stimulation, which were abolished in PHF2 knock-down cell lines. Our findings suggest that C/EBPδ and C/EBPα mediate PHF2 binding to their target promoters during adipogenesis to demethylate the repressive H3K9 histone methyl marker. As we identified the upstream regulator of PPARG, the most important transcription factor for fat cell differentiation and a diabetic drug target, it would be useful to next identify the target molecule of PHF2 activity for regulating its activity, which might help to improve metabolic and diabetic disease symptoms.
Acknowledgments
This work was supported by the National Research Foundation of Korea funded by the Korean Government (MESF) (NRF-2011-0008634) and the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A121106).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Baba A., Ohtake F., Okuno Y., Yokota K., Okada M., Imai Y., Ni M., Meyer C.A., Igarashi K., Kanno J., et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5. B. Nat. Cell Biol. 2011;13:668–675. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cho Y.W., Hong S., Jin Q., Wang L., Lee J.E., Gavrilova O., Ge K. Histone methylation regulator PTIP is required for PPARγ and C/EBPalpha expression and adipogenesis. Cell Metabol. 2009;10:27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debril M.B., Gelman L., Fayard E., Annicotte J.S., Rocchi S., Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J. Biol. Chem. 2004;279:16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- Fortschegger K., Shiekhattar R. Plant homeodomain fingers form a helping hand for transcription. Epigenetics. 2011;6:4–8. doi: 10.4161/epi.6.1.13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K.E., Fankell D.M., Erickson P.F., Majka S.M., Crossno J.T., Klemm D.J. Depletion of cAMP-response element-binding protein/ATF1 inhibits adipogenic conversion of 3T3-L1 cells ectopically expressing CCAAT/Enhancer-binding protein (C/EBP)α, C/EBPβ, or PPARγ2. J. Biol. Chem. 2006;281:40341–40353. doi: 10.1074/jbc.M605077200. [DOI] [PubMed] [Google Scholar]
- Friedman J.M. Obesity in the new millennium. Nature. 2000;404:632–634. doi: 10.1038/35007504. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Ghosh S., Maiti G.P., Mukherjee S., Mukherjee N., Chakraborty J., Roy A., Roychoudhury S., Panda C.K. Association of FANCC and PTCH1 with the development of early dysplastic lesions of the head and neck. Ann. Surg. Oncol. 2012;19(Suppl 3):S528–538. doi: 10.1245/s10434-011-1991-x. [DOI] [PubMed] [Google Scholar]
- Kudo M., Sugawara A., Uruno A., Takeuchi K., Ito S. Transcription suppression of peroxisome proliferator-activated receptor γ2 gene expression by tumor necrosis factor α via an inhibition of CCAAT/enhancer-binding protein δ during the early stage of adipocyte differentiation. Endocrinology. 2004;145:4948–4956. doi: 10.1210/en.2004-0180. [DOI] [PubMed] [Google Scholar]
- Lai P.H., Wang W.L., Ko C.Y., Lee Y.C., Yang W.M., Shen T.W., Chang W.C., Wang J.M. HDAC1/HDAC3 modulates PPARG2 transcription through the sumoylated CEBPD in hepatic lipogenesis. Biochim. Biophy. Acta. 2008;1783:1803–1814. doi: 10.1016/j.bbamcr.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Margueron R., Trojer P., Reinberg D. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Musri M.M., Carmona M.C., Hanzu F.A., Kaliman P., Gomis R., Parrizas M. Histone demethylase LSD1 regulates adipogenesis. J. Biol. Chem. 2010;285:30034–30041. doi: 10.1074/jbc.M110.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y., Ohtake F., Igarashi K., Kanno J., Matsumoto T., Takada I., Kato S., Imai Y. Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes. 2013;62:1426–1434. doi: 10.2337/db12-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E.D., Spiegelman B.M. MOLECULAR REGULATION OF ADIPOGENESIS. Ann. Rev. Cell Dev. Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Sarraf P., Troy A.E., Bradwin G., Moore K., Milstone D.S., Spiegelman B.M., Mortensen R.M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Shi X.M., Blair H.C., Yang X., McDonald J.M., Cao X. Tandem repeat of C/EBP binding sites mediates PPARγ2 gene transcription in glucocorticoid-induced adipocyte differentiation. J. Cell. Biochem. 2000;76:518–527. doi: 10.1002/(sici)1097-4644(20000301)76:3<518::aid-jcb18>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Stender J.D., Pascual G., Liu W., Kaikkonen M.U., Do K., Spann N.J., Boutros M., Perrimon N., Rosenfeld M.G., Glass C.K. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol. Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.L., Sun X.X., Xu X.E., Zhu M.X., Wu Z.Y., Shen J.H., Wu J.Y., Huang Q., Li E.M., Xu L.Y. Overexpression of Jumonji AT-rich interactive domain 1B and PHD finger protein 2 is involved in the progression of esophageal squamous cell carcinoma. Acta Histochem. 2013;115:56–62. doi: 10.1016/j.acthis.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K., Okamura M., Tsutsumi S., Nishikawa N.S., Tanaka T., Sakakibara I., Kitakami J., Ihara S., Hashimoto Y., Hamakubo T., et al. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol. Cell. Biol. 2009;29:3544–3555. doi: 10.1128/MCB.01856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Jin Q., Lee J.E., Su I.H., Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xu S., Lee J.E., Baldridge A., Grullon S., Peng W., Ge K. Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J. 2013;32:45–59. doi: 10.1038/emboj.2012.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H., Li J., Song T., Lu M., Kan P.-Y., Lee M.G., Sha B., Shi X. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J. Biol. Chem. 2010;285:9322–9326. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.