Abstract
The epithelial cell adhesion molecule (EpCAM, also known as CD326) is a transmembrane glycoprotein that is specifically detected in most adenocarcinomas and cancer stem cells. In this study, we performed a Cell systematic evolution of ligands by exponential enrichment (SELEX) experiment to isolate the aptamers against EpCAM. After seven round of Cell SELEX, we identified several aptamer candidates. Among the selected aptamers, EP166 specifically binds to cells expressing EpCAM with an equilibrium dissociation constant (Kd) in a micromolar range. On the other hand, it did not bind to negative control cells. Moreover, EP166 binds to J1ES cells, a mouse embryonic stem cell line. Therefore, the isolated aptamers against EpCAM could be used as a stem cell marker or in other applications in both stem cell and cancer studies.
Keywords: aptamer, EpCAM, SELEX, stem cell marker
INTRODUCTION
Aptamers are single-stranded DNA, RNA, or nucleic acids isolated via systematic evolution of ligands by exponential enrichment (SELEX) (Breaker, 2004; Ellington and Szostak, 1990; Tuerk and Gold, 1990). Due to their capability to form unique secondary or tertiary structures, aptamers can bind to a variety number of targets, such as small molecules, proteins, cells, and even tissues with high binding affinity and specificity (Fang and Tan, 2010; Kim et al., 2014; Mi et al., 2010; Sefah et al., 2010; Shamah et al., 2008; Shangguan et al., 2006). Additionally, aptamers have great advantages, such as small size, easy synthesis, low batch-to-batch variation, lack of immunogenicity, low toxicity, high stability, and easy functionalization. Therefore, aptamers have been used as valuable alternatives to antibodies in a number of applications, including basic research, clinical diagnosis, and therapeutics (Jayasena, 1999; Lee et al., 2006; Tan et al., 2011). Indeed, several aptamers are already considered to be therapeutic or diagnostic agents and are undergoing clinical trials (Keefe et al., 2010).
The epithelial cell adhesion molecule (EpCAM), a single-pass membrane glycoprotein, is a carcinoma-associated antigen, and it is expressed in several progenitor cell populations and cancers (Dalerba et al., 2007; Dan et al., 2006; Hao et al., 2013; Maetzel et al., 2009; Stingl et al., 2005). It functions as a homotypiccalcium-independent cell adhesion molecule (Litvinov et al., 1997). Thus, EpCAM is generally used as a target for immunotherapy treatment of human carcinomas (Patriarca et al., 2012). In addition, the maintenance of self-renewal of embryonic stem cells depends on the high-level expression of EpCAM. It also plays essential functional roles in the proliferation and differentiation of human stem cells. Therefore, EpCAM can serve as a reliable marker for tumor-initiating cells (cancer stem cells) or embryonic stem cells (Gonzalez et al., 2009; Lu et al., 2010; Ng et al., 2010; Sundberg et al., 2009).
The identification of selectively expressed cell surface markers is important in the purification and characterization of pluripotent stem cells, including induced pluripotent stem cells (iPSCs), and it plays a crucial role in research on stem cells (Zhao et al., 2012). In this study, we isolated the aptamers against EpCAM, and in this paper we present data that demonstrates their potential as stem cell markers.
MATERIALS AND METHODS
EpCAM overexpression using retroviral expression system
To express the EpCAM in HepG2 cells, a retrovirus-mediated infection system was used. The gene encoding EpCAM was inserted into the multi-cloning site of the pRetroX-IRES-Ds red vector (Clontech, USA). The FLAG tag was inserted at the C-terminal end of EpCAM. Then, retroviruses were generated by transiently co-transfecting GP2-293 cells with a retroviral vector and the VSV-G plasmid using Lipofectamine 2000 (Life Technologies, USA). At 48 h after transfection, media containing retroviruses were collected, filtered with 0.45 μm filters, and used to infect cells in the presence of polybrene (8 μg/ml). Infected HepG2 cells were selected by means of fluorescence-activated cell sorting (FACS; FACSAria cell sorter, BD Biosciences, USA) and were further maintained in a growth medium as described previously (Kim et al., 2009; 2011; 2012; 2013). The overexpression of EpCAM was assessed by Western blot analysis using anti-EpCAM antibody.
Aptamer library and primers
A single-stranded DNA (ssDNA) library labeled with a fluoresce-in isothiocyanate (FITC) was synthesized by Integrated DNA Technologies, Inc. (USA). The library contains 40 randomized nucleotides (nt) flanked by two 19-nt primer hybridization sites (5′-FITC-CGCGGAAGCGTGCTGGGCC-N40-CATAACCCAGAGG TCGAT-3′). For the amplification of the selected aptamer pool, a FITC-labeled forward primer (5′-FITC-GGGAATTCGCGGAA GCGTGCTGGGCC-3′) and a reverse primer (5′-GGGGGGATCCAT CGACCTCTGGGTTATG-3′) were used in the PCR process. Another forward primer (5′-GGGGAATTCGCGGAAGCGT GCTGGGCC-3′) was utilized for the cloning of the selected DNA pool. Additional nucleotide sequences (containing restriction enzyme sites) were added in forward and reverse primers for efficient sub-cloning (Kim et al., 2014).
Cell SELEX using FACS
The FITC-labeled ssDNA library (10 nmol; initial pool size 1016) dissolved in 20 ml DW was denatured at 95°C for 5 min and then cooled in ice for 10 min to form a secondary structure. During this step, salmon sperm DNA (0.1 mg/ml; Sigma, USA) dissolved in a binding buffer [4.5 g/L glucose, 5 mM MgCl2 and 10% FBS in Dulbecco’s PBS (Sigma)] was incubated with 1 × 106 HepG2 cells overexpressing EpCAM to inhibit non-specific binding. Then, the cells were incubated with the ssDNA library (10 nmol) and bovine serum albumin (1 mg/ml; Thermo Inc., USA) at 37°C for 30 min. After centrifugation, the supernatant was removed and the cells were washed five times with 300 μl of binding buffer. The ssDNA library-bound cells were enriched using a FACSAria cell sorter (BD Biosciences, USA), and the bound ssDNAs were eluted from the sorted cells by heating at 95°C for 5 min. The eluted DNAs were purified by phenol-chloroform (Sigma-Aldrich, USA) extraction, a Sephadex G-25 column (Sigma-Aldrich, USA), and ethanol precipitation. The purified ssDNAs were amplified by PCR with FITC-labeled primers. For the next round of selection, the single-stranded DNA population was obtained via strand separation of PCR products (heat treatment at 95°C for 10 min). After four rounds of selection, negative selection was also performed using control HepG2 cells. The ssDNA pool of the seventh round was cloned into E. coli DH5α and was sequenced (Enzynomics, Korea). A total of 200 aptamers were sequenced, and among these, 33 aptamers were selected, synthesized, and analyzed.
Secondary structure prediction and Kd determination
The secondary structures of the aptamers were predicted using the mfold program (The RNA Institute) (Kim et al., 2014). We chose aptamers with the most thermodynamically stable predicted structure after the sequencing of each aptamer. Individual aptamers were incubated with negative or positive cells. As the treated aptamers increased in number, the mean fluorescence intensity of the aptamer-coated cells was detected using FACS. Next, the equilibrium dissociation constant (Kd) of aptamer-cell interaction was calculated, as described previously (Kim et al., 2014).
Aptamer binding assay by FACS and confocal microscopic imaging
To assess the enrichment of aptamer pools during SELEX, a 100 pmol ssDNA pool of each round was incubated with 1 × 106 cells in 300 μl of binding buffer at 37°C for 30 min. The cells were washed five times with 300 μl of binding buffer. The binding of EP166 with J1ES cells was also analyzed under the same experimental conditions. In the case of EP166 toward HepG2 (Fig. 3), 10 μM of aptamer was incubated with 5 × 105 cells in 200 μl of binding buffer at 37°C for 15 min. The pellets were suspended in 20 μl of binding buffer, and 10 μl of cells was dropped on a glass slide. The FITC signal of the aptamers was detected with a LSM 510 META confocal microscope (Zeiss, USA). The other half of the cells was resuspended in 0.5 ml of binding buffer, and the fluorescence was determined with a FACS Calibur flow cytometer (BD Biosciences, USA).
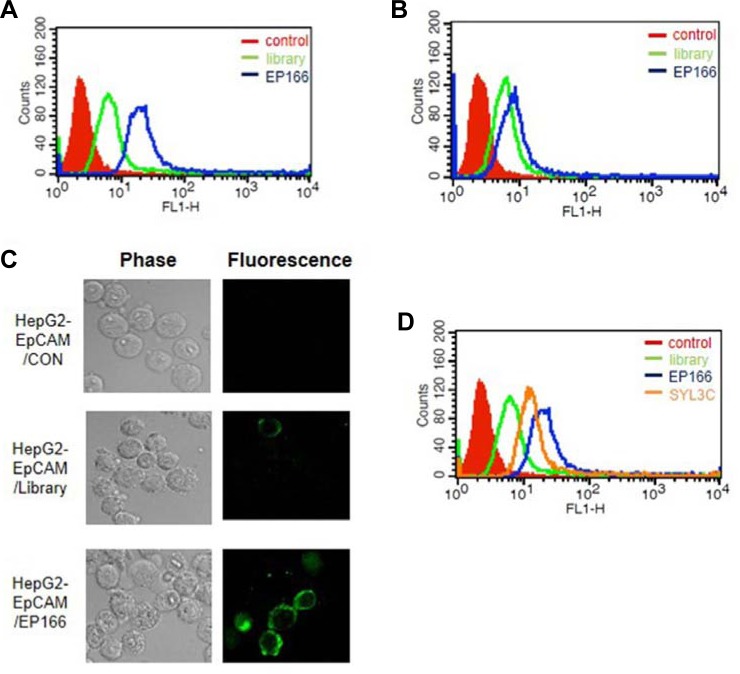
Fig. 3.
The binding affinity and specificity test of EP166 aptamer. (A) The binding affinity of EP166 was measured for HepG2 cells expressing EpCAM. 10 μM of EP166 was incubated with 5 × 105 cells in 200 μl of binding buffer at 37°C for 15 min. (B) The binding affinity of EP166 was measured for HepG2 cells. The 10 μM of EP166 was incubated with 5 × 105 cells in 200 μl of binding buffer at 37°C for 15 min. (C) The binding specificity test of EP166 was measured using HepG2 cells expressing EpCAM by confocal microscopic analysis. (D) The comparison study between EP166 and SYL3C for the binding affinity toward HepG2 expressing EpCAM. The 10 μM of EP166 or SYL3C was incubated with 5 × 105 cells in 200 μl of binding buffer at 37°C for 15 min.
J1ES cell culture and differentiation condition
The mouse embryonic stem cell line, J1ES [American Type Culture Collection (ATCC)], derived from strain 129s4/Jae, was routinely maintained on γ-irradiated mouse embryonic fibroblasts (MEFs) in ESC medium [high-glucose Dulbecco’s modified Eagle’s Medium (DMEM) (Invitrogen, USA) containing 15% ES-tested fetal bovine serum (FBS) (PAA, Colbe, Germany), 1% penicillin-streptomycin (Invitrogen, USA), 2 mM of L-glutamine (Invitrogen, USA), 1% non-essential amino acids (NEAA, Invitrogen, USA), 0.1 mM of β-mercaptoethanol (Invitrogen, USA), and 1 × 103 U/ml of leukemia inhibitory factors (LIF) (Millipore, USA)] at 37°C in a humidified atmosphere with 5% CO2.
For differentiation, the cells were cultured in a medium without LIF (Millipore, USA), as described previously (Bibel et al., 2007). Embryoid bodies (EBs) were generated by plating 3 × 106 cells/cm2 of ES cells onto 90-mm non-cell culture-treated petri dishes (SPL Lifesciences, Korea) in ES medium without LIF (Millipore, USA). The medium was changed every 2 days (2EB, 4EB, 6EB, and 8EB). All trans-RA (10 μM) (Sigma-Aldrich, USA) was added at day 4 (4EB) and day 6 (6EB) of EB formation. At day 8, the EBs were trypsinized, and the cells were plated at a density of 1.5 × 105 cells/cm2 on poly-D-lysine-laminin (Sigma-Aldrich, USA)-coated tissue culture dishes and performed the binding test.
RESULTS AND DISCUSSION
Overexpression of EpCAM and its confirmation in HepG2 cells
As an initial step, we examined the EpCAM expression level of several cell lines by Western blot analysis using anti-EpCAM antibody. Among the cell lines tested, the HepG2 cell line showed the lowest EpCAM expression level. Thus, we determined to ectopically express the EpCAM in HepG2 cells. EpCAM was overexpressed by using a retroviral transduction system (pRetroX-IRES DsRedvector, Clontech). After transduction, we confirmed the EpCAM overexpression by both Western blot and FACS analysis (Figs. 1A and 1B).
Fig. 1.
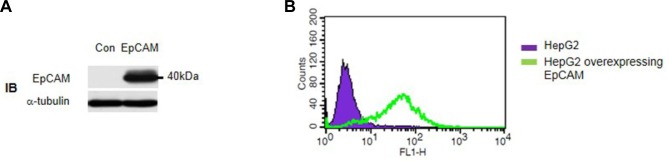
The overexpression of EpCAM in HepG2 cells. (A) The overexpression of EpCAM was confirmed by Western blot analysis using anti-EPCAM antibody. (B) The cell-surface overexpression of EpCAM was confirmed by FACS analysis using anti-EpCAM antibody. The retroviral expression system was used to ectopically express EpCAM. The FLAG tag was inserted at the C-terminal end of EpCAM.
Cell SELEX for identification of aptamers specific for EpCAM
The HepG2 cells and HepG2 cells expressing EpCAM were used for negative and positive selection, respectively. To obtain the aptamers against EpCAM, we applied the Cell SELEX method using flow cytometry (FACS-SELEX). The randomized 40 DNA nucleotide library was used as an initial pool. Eight rounds of SELEX were performed. At the 4th, 6th, 7th and 8th rounds of SELEX, negative selection was applied to drop the non-specific binding of aptamers. After 8 rounds of selection, four representative rounds pools (4th, 6th, 7th and 8th rounds) were measured for their binding affinity against HepG2 cells expressing EpCAM. As shown in Fig. 2A, a gradual increase in the fluorescence intensity against HepG2 cells expressing EpCAM was detected according to the increase in the number of SELEX rounds. However, the peak shift moving back to the library peak was shown in the case of the 8th round pool of SELEX. On the other hand, no peak shift was detected in the case of HepG2 negative control cells. These results clearly indicate that specific aptamers with better binding affinity against EpCAM were enriched as the SELEX rounds continued. Based on these data, approximately 200 aptamers were cloned and sequenced from the 7th round aptamer pool. Unfortunately, we could not obtain the conserved sequences from about 200 aptamer sequences. This may be due to rather insufficient selection processes. After secondary structure prediction of 200 sequenced aptamers using the mfold program (http://mfold.rit.albany.edu), we chose about 33 aptamers showing a distinct secondary structure. Then, their binding affinity toward HepG2 cells expressing EpCAM was assessed by FACS (Supplementary Fig. S1).
Fig. 2.
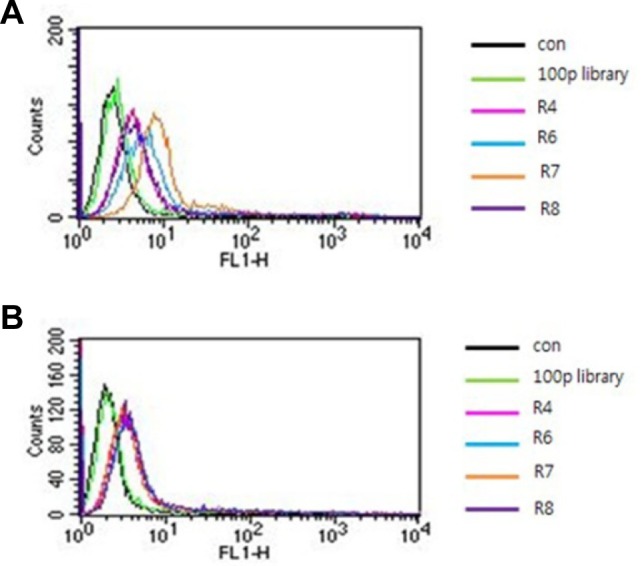
Binding affinity test of each round pool with HepG2 expressing EpCAM or HepG2 only. (A) After a total of 8 rounds of SELEX, the binding affinities of four round pools (4th, 6th, 7th, and 8th rounds) were assessed for HepG2 cells expressing EpCAM. (B) After a total of 8 rounds of SELEX, the binding affinities of four round pools (4th, 6th, 7th and 8th rounds) were assessed for HepG2 cells. The aptamer library (100 pmol) was used as a negative control.
Sequences of aptamers specific for EpCAM and determination of their Kd values
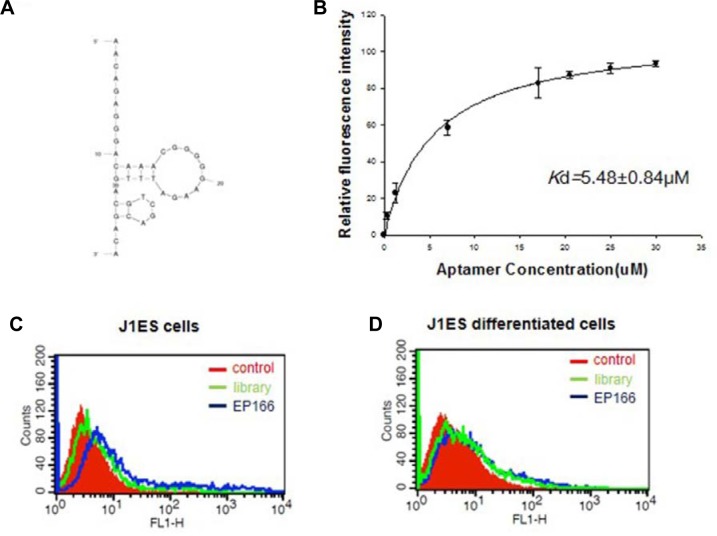
Among the aptamers tested, EP166 (5′-AAC AGA GGG ACA AAC GGG GGA AGA TTT GAC GTC GAC GAC A-3′) showed the highest binding affinity against EpCAM. As shown in Figs. 3A and 3B, EP166 binds with HepG2 cells expressing EpCAM but not HepG2 cells. The confocal microscopic analysis also clearly demonstrated that EP166 binds with HepG2 cells expressing EpCAM (Fig. 3C). The aptamer specific to EpCAM was already identified using recombinant EpCAM protein [29]. This aptamer, named SYL3C, shows high affinity to MDA-MB-231 (breast cancer cell line) and KatoIII (gastric cell line) with nanomolar Kd value (Song et al., 2013). Additionally, various potential applications, such as cancer cell imaging and circulating tumor cell detection, were displayed. Thus, we determined to compare EP166 with the SYL3C aptamer. Surprisingly, EP166 bound more tightly with HepG2 cells expressing EpCAM than SYL3C under our experimental conditions (Fig. 3D). The secondary structure prediction data using the mfold program demonstrated that EP166 has two hairpin structures located at very close range (Fig. 4A). They seem to have an important role in binding with EpCAM, but more experiments are necessary to verify this hypothesis. The EP166 aptamer showed tight binding toward HepG2 cells expressing EpCAM with a Kd value of 5.48 ± 0.84 μM. On the other hand, EP166 binds very weakly with HepG2 cells. This may due to the low expression of EpCAM in HepG2 cells.
Fig. 4.
The characterization of EP166 was carried out. (A) The predicted secondary structure of EP166 was obtained using the mfold program. (B) Along with rising aptamer concentration, the binding affinity of EP166 towards HepG2 cells expressing EpCAM was examined by FACS. (C) The binding affinity of EP166 was measured for J1ES cells. (D) The binding affinity of EP166 was measured for differentiated J1ES cells. The 100 pmol of EP166 was incubated with 1 × 106 cells in 300 μl of binding buffer at 37°C for 30 min.
Binding of aptamers toward J1ES mouse embryonic stem cells
Next, we tested the possibility that EP166 could be used as an embryonic stem cell marker. A wide range of cell surface markers and genetic molecular markers have been reported to be indicative of undifferentiated embryonic stem cells (Zhao et al., 2012). EpCAM has been used as a marker of human embryonic stem cells, mouse embryonic stem cells, embryonic carcinoma cells, and hepatocellular carcinoma. In this study, J1ES, a mouse embryonic stem cell line, was used, and we examined whether EP166 can specifically bind or not. As shown in Fig. 4C, EP166 binds with undifferentiated J1ES cells. On the other hand, EP166 cannot bind with J1ES cells anymore when the cells differentiate into embryonic bodies (Fig. 4D). These results strongly indicate that EP166, an aptamer against EpCAM, could be used as a specific marker for embryonic stem cells.
As described in the introduction section, EpCAM is overexpressed in most cancers and is an ideal marker for clinical uses in cancer diagnosis/treatment, imaging, and prognosis. Therefore, besides application as a stem cell marker, the aptamer against EpCAM could be applied in a variety of clinical fields.
Acknowledgments
This work was supported by grants from KRIBB and the Research Program (2011-0020507, 2013M3A9A7046301 and 2011-0030028) of the Korea National Research Foundation.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Breaker R.R. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432:838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]
- Dalerba P., Dylla S.J., Park I.K., Liu R., Wang X., Cho R.W., Hoey T., Gurney A., Huang E.H., Simeone D.M., et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan Y.Y., Riehle K.J., Lazaro C., Teoh N., Haque J., Campbell J.S., Fausto N. Isolation of multipotent progenitor cells from human fetal liver capable of differentiating into liver and mesenchymal lineages. Proc. Natl. Acad. Sci. USA. 2006;103:9912–9917. doi: 10.1073/pnas.0603824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fang X., Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc. Chem. Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez B., Denzel S., Mack B., Conrad M., Gires O. EpCAM is involved in maintenance of the murine embryonic stem cell phenotype. Stem Cells. 2009;27:1782–1791. doi: 10.1002/stem.97. [DOI] [PubMed] [Google Scholar]
- Hao P.P., Lee M.J., Yu G.-R., Kim I.-H., Cho Y.-G., Kim D.-G. Isolation of EpCM+/CD133− hepatic progenitor cells. Mol. Cells. 2013;36:424–431. doi: 10.1007/s10059-013-0190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.K., Jung H., Kim D.H., Kim E.Y., Chung J.W., Cho Y.S., Park S.G., Park B.C., Bae K.-H., Lee S.C. Regulation of adipocyte differentiation by LAR tyrosine phosphatase in human mesenchymal stem cells and 3T3-L1 preadipocytes. J. Cell Sci. 2009;122:4160–4167. doi: 10.1242/jcs.053009. [DOI] [PubMed] [Google Scholar]
- Kim W.K., Jung H., Kim E.Y., Kim D.H., Cho Y.S., Park B.C., Park S.G., Ko Y., Bae K.-H., Lee S.C. RPTPμ tyrosine phosphatase promotes adipogenic differentiation via modulation of p120 catenin phosphorylation. Mol. Biol. Cell. 2011;22:4883–4891. doi: 10.1091/mbc.E11-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.Y., Kim W.K., Kang H.J., Kim J.-H., Chung S.J., Seo Y.S., Park S.G., Lee S.C., Bae K.-H. Acetylation of malate dehydrogenase 1 promotes adipogenic differentiation via activating its enzymatic activity. J. Lipid Res. 2012;53:1864–1876. doi: 10.1194/jlr.M026567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.Y., Han B.S., Kim W.K., Lee S.C., Bae K.-H. Acceleration of adipogenic differentiation via acetylation of malate dehydrogenase 2. Biochem. Biophys. Res. Comm. 2013;441:77–82. doi: 10.1016/j.bbrc.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Kim E.Y., Kim J.W., Kim W.K., Han B.S., Park S.G., Chung B.H., Lee S.C., Bae K.-H. Selection of aptamers for mature white adipocytes by Cell SELEX using flow cytometry. PLoS One. 2014;9:e97747. doi: 10.1371/journal.pone.0097747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.F., Stovall G.M., Ellington A.D. Aptamer therapeutics advance. Curr. Opin. Chem. Biol. 2006;10:282–289. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Litvinov S.V., Balzar M., Winter M.J., Bakker H.A., Briaire-de Bruijn I.H., Prins F., Fleuren G.J., Warnaar S.O. Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J. Cell Biol. 1997;139:1337–1348. doi: 10.1083/jcb.139.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.-Y., Lu R.-M., Liao M.-Y., Yu J., Chung C.-H., Kao C.-F., Wu H.-C. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J. Biol. Chem. 2010;285:8719–8732. doi: 10.1074/jbc.M109.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Kieu C., Papior P., Baeuerle P.A., Munz M., et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- Mi J., Liu Y., Rabbani Z.N., Yang Z., Urban J.H., Sullenger B.A., Clary B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010;6:22–24. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng V.Y., Ang S.N., Chan J.X., Choo A.B.H. Characterization of epithelial cell adhesion molecule as a surface marker on undifferentiated human embryonic stem cells. Stem Cells. 2010;28:29–35. doi: 10.1002/stem.221. [DOI] [PubMed] [Google Scholar]
- Patriarca C., Macchi R.M., Marschner A.K., Mellstedt H. Epithelial cell adhesion molecule expression (CD326) in cancer: a short review. Cancer Treat. Rev. 2012;38:68–75. doi: 10.1016/j.ctrv.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Sefah K., Shangguan D., Xiong X., O’Donoghue M.B., Tan W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- Shamah S.M., Healy J.M., Cload S.T. Complex target SELEX. Acc. Chem. Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- Shangguan D., Li Y., Tang Z., Cao Z.C., Chen H.W., Mallikaratchy P., Sefah K., Yang C.J., Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Zhu Z., An Y., Zhang W., Zhang H., Liu D., Yu C., Duan W., Yang C.J. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- Stingl J., Raouf A., Emerman J.T., Eaves C.J. Epithelial progenitors in the normal human mammary gland. J. Mammary Gland Biol. Neoplasia. 2005;10:49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- Sundberg M., Jansson L., Ketolainen J., Pihlajamaki H., Suuronen R., Skottman H., Inzunza J., Hovatta O., Narkilahti S. CD marker expression profiles of human embryonic stem cells and their neural derivatives, determined using flow-cytometric analysis, reveal a novel CD marker for exclusion of pluripotent stem cells. Stem Cell Res. 2009;2:113–124. doi: 10.1016/j.scr.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Tan W., Wang H., Chen Y., Zhang X., Zhu H., Yang C., Yang R., Liu C. Molecular aptamers for drug delivery. Trends Biotechnol. 2011;29:634–640. doi: 10.1016/j.tibtech.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Zhao W., Ji X., Zhang F., Li L., Ma L. Embryonic stem cell markers. Molecules. 2012;17:6196–6136. doi: 10.3390/molecules17066196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.