Abstract
Early diagnosis of oral squamous cell carcinoma (OSCC) is an attractive strategy to increase survival rate of patient. An integrated separation approach of reversed phase liquid chromatography and hydrophilic interaction chromatography combining with time of flight mass spectrometer has been firstly developed for performing global saliva metabonomics analysis for early diagnosis of OSCC. This approach was designed to overcome the limitations of a single chromatographic method due to different polarity of endogenous metabolites. As a result, 14 potential salivary metabolites were identified. Eight biomarkers up-regulated in OSCC patients are compared with control and six down-regulated groups. Receiver operating characteristic analysis was exploited to evaluate the diagnostic power of the candidate biomarkers, and related metabolic pathways have also been studied. Five salivary biomarkers (propionylcholine, N-Acetyl-L-phenylalanine, sphinganine, phytosphingosine, and S-carboxymethyl-L-cysteine) in combination yielded satisfactory accuracy (AUC = 0.997), sensitivity (100%), and specificity (96.7%) in distinguishing early stage of OSCC from the control. In this study, a comprehensive saliva metabonomics analysis for identifying potential biomarkers to early diagnose OSCC is successfully demonstrated, which has the advantages of non-invasive, simple, reliable, and low-cost. These novel metabolic biomarkers have obvious clinical utility that will help to diagnose OSCC at its early stage.
Oral cancer, a subtype of head and neck cancer, is an cancerous tissue growth located in the oral cavity1,2. More than 90% of oral cancers are squamous cell carcinoma (OSCC) originating in the tissues those line the lips, the oral cavity, and pharynx. Approximately 42,000 people in the US will be newly diagnosed with oral cancer in 2013. The World Health Organization has reported oral cancer with a death rate of 45% at five years from diagnosis(for all stages combined at time of diagnosis)3. If the oral cancer was found at early stages of development, it has an 80 to 90% survival rate. Unfortunately, most of these cancers are hard to discover in the early stages because of a lack of public awareness and screening method, which would generally result in a poor prognosis and a low survival rate4.
Currently, the most definitive method for oral cancer diagnosis and screening is a scalpel biopsy. It is time-consuming and needs extensive experience. CT, or CAT (co-axial tomography) scan technology and magnetic resonance imaging technology have developed rapidly over the last few decades. However, CT is only able to detect the actual presence of masses and only a biopsy can verify that the mass is malignant. Therefore, novel diagnostic technologies are urgently needed to diagnose OSCC at its early stage. Now, molecular-based biomarkers have been used to diagnose OSCC drawing more and more attention. Saliva as diagnose medium offers an easy, inexpensive, safe, and noninvasive approach5.
Saliva, non-invasive and stress-free alternative to blood, is widely accepted as a potential medium for clinical diagnostics. It comprises the secretions of three major glands namely parotid gland, submandibular gland and sublingual gland and hundreds of minor salivary glands6,7. It is one of the most complex, versatile, and important body fluids, which reflects a large range of physiological needs and information8. Saliva biomarkers such as proteins and DNA have been used to detect OSCC over the last few decades9,10,11. Metabonomics as modern high-throughput technology has developed rapidly, and opens a door to biomarker discovery12. It is the systematic study of small-molecular-weight substances in cells, tissues or whole organisms as influenced by multiple factors13,14,15. Recent metabonomics analysis demonstrates its applicability for the diagnosis and prognosis of OSCC. High performance liquid chromatography-mass spectrometry (HPLC-MS)16 and nuclear magnetic resonance (NMR)17 spectroscopy were performed to discriminate OSCC patients from the healthy controls. Metabonomics analysis by considering the low sensitivity and limited dynamic range of NMR, HPLC-MS based metabonomics approach has been successfully used as a tool in the diagnosis of diseases with excellent reproducibility and sensitivity18,19,20. Recently, a faster metabonomics analysis technique, ultrahigh performance liquid chromatography-mass spectrometry (UPLC-MS) has been used as to diagnose oral cancer5,21, diabetes22, colorectal cancer23, hepatocellular carcinoma24, and chronic renal failure. However, an integrated UPLC-MS method including reversed phase liquid chromatography (RPLC), and hydrophilic interaction chromatography (HILIC) based saliva metabonomics analysis of OSCC has not been reported hitherto as far as we know.
HILIC allows different selectivity and the better retention of polar analytes which generally have poor retention on RPLC. It was first introduced by Alpert in 199025. In HILIC, the sensitivity of the mass spectrometer is enhanced due to the high organic content in the mobile phase and improving the efficiency of ionization. Saliva contains approximately 99% water26, therefore, many endogenous metabolites are expected to be highly polar. It was very necessary to combine RPLC and HILIC techniques for a comprehensive saliva metabonomics analysis to discover more potential biomarkers for the early diagnosis of OSCC.
In this work, RRLC-MS and HILIC-MS analysis in positive and negative ion modes were examined to investigate saliva samples from OSCC patients and healthy volunteers. Multivariate data analysis was performed to highlight discriminated variables (Metabonomics research strategy was provided in Supplemental Figure 1). After refining the model, the fourteen potential biomarkers for the early diagnosis of OSCC have been identified. Eight biomarkers up-regulated in OSCC patients compare with controls and six down-regulated. The purpose of this study is to develop a comprehensive saliva metabonomics analysis for identifying potential biomarkers to early diagnosis of OSCC.
Results
Salivary metabolomics analysis
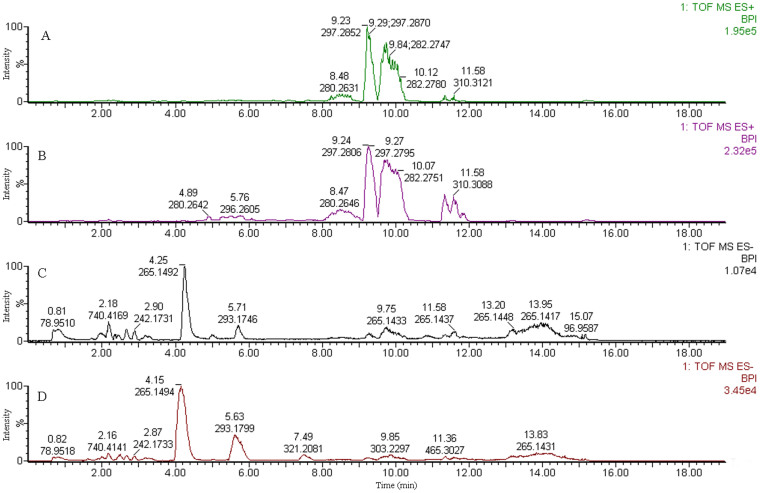
The separation conditions of saliva samples on both columns were optimized. Taking the RPLC as example, typical UPLC-TOF/MS base peak intensity (BPI) chromatograms of saliva samples from the control group and the OSCC group in both positive- and negative-ion modes was shown in Figure 1. From the BPI chromatograms, more marked variations can be seen in the patient group than in the control group. RPLC has better retention for weakly polar component. In addition, HILIC allows varied selectivity and the better retention for polar analytes. The integrated utilization of two separation modes could enlarge metabolite identification.
Figure 1. Typical base peak intensity (BPI) chromatograms by RPLC-MS.
(A) the control group in positive-ion mode (ESI+); (B) the patient group in ESI+; (C) the control group in negative-ion mode (ESI−); (D) the patient group in ESI−; The mass range m/z 50–1000.
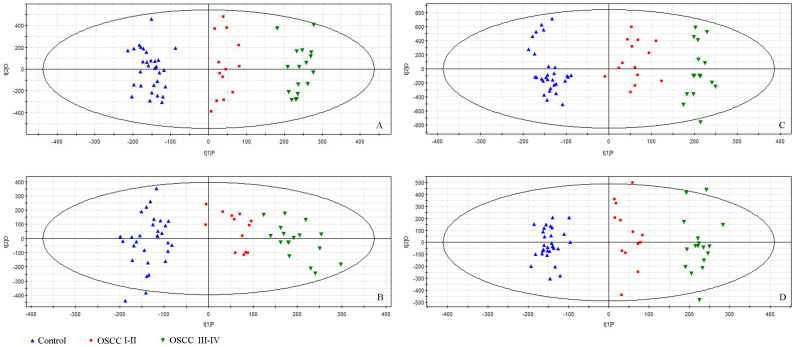
Using MarkerLynx software for peak detection, 8319 peaks of positive ions and 6298 negative ions in RPLC, 2379 peaks of positive ions and 2269 negative ions in HILIC were obtained. Although fewer peaks were extracted from the HILIC column, these peaks can also be used as a comprehensive saliva metabonomics profiling as well as RPLC. The variables were exported into SIMCA-P 11.0 for multivariate data analysis to detect any inherent trend within the date. All saliva samples were divided into three groups: healthy controls as group 1 (HC); early stage of OSCC including stages I and II as group 2 (OSCC I–II); and advanced stage of OSCC including stages III and IV as group 3 (OSCC III–IV). PCA analysis was tested first in our study, however, there are no obvious separation trends observed for groups of OSCC I–II and OSCC III–IV. For this reason, a supervised method, OPLS-DA27, was applied in the data analysis. As can be seen from the Figure 2, satisfactory clustering trends among HC, OSCC I–II, and OSCC III–IV are observed in the scores plot, indicating that the possibility of using saliva metabonomics for staging OSCC.
Figure 2. The scores plot of the OPLS-DA model of the UPLC-MS date from control group, OSCC I–II group, and OSCC III–IV group.
(A) RPLC in positive mode; (B) RPLC in negative mode; (C) HILIC in positive mode; (D) HILIC in negative mode.
In OPLS-DA, R2Y (cum) and Q2 (cum) parameters were used for the evaluation of the models, indicating the fitness and prediction ability, respectively1. Q2 (cum) > 50% shows that the mode is useful; if the Q2 (cum) > 90%, the mode is excellent. In Figure 2A, the classification resulted in one predictive component and two orthogonal components, with excellent modeling and predictive abilities (R2(X) = 52.7%, R2(Y) = 97.6%, Q2(Y) = 95.1%) for positive-ion mode in RPLC separation. Other parameters including the R2X(cum), R2Y (cum) and Q2 (cum) values obtained from RPLC and HILIC separation in both positive- and negative-ion modes were summarized in Table 1. From these results, we can find that the OPLS-DA models were valid for all four modes.
Table 1. Parameters of OPLS-DA models based on RPLC and HILIC.
| R2X (cum) | R2Y (cum) | Q2 (cum) | Imported variables no a. | VIP value > 1 variables no. | |
|---|---|---|---|---|---|
| RPLC | |||||
| Positive-ion mode | 0.527 | 0.976 | 0.951 | 8319 | 102 |
| Negative-ion mode | 0.314 | 0.911 | 0.771 | 6298 | 227 |
| HILIC | |||||
| Positive-ion mode | 0.625 | 0.966 | 0.908 | 2379 | 163 |
| Negative-ion mode | 0.534 | 0.979 | 0.934 | 2269 | 131 |
aNumber of the variables imported to build the model.
Early OSCC biomarker discovery
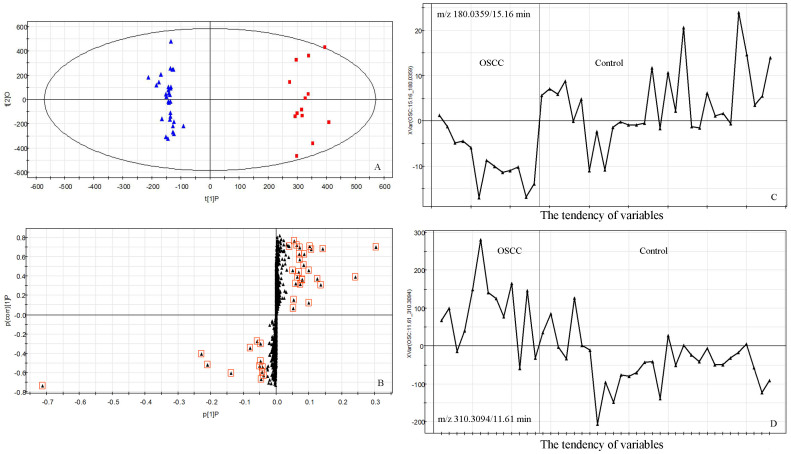
In order to identify discriminating variables used in the early stage detection of OSCC, an S-plot model was used. VIP was used as an important parameter to select the biomarkers. Variables with VIP value greater than one were considered as great value28,29. A nonparametric Mann-Whitney U test was performed in succession, and variables with significant differences between OSCC patients and control individuals (P < 0.05) were retained. Figure 3 is the strategy for selection of interesting variables for HC vs OSCC I–II in RP column with positive-ion mode. S-plot of HC vs OSCC I–II was shown in Figure 3B, which is a scatter plot that combines the covariance and correlation for the model variables with respect to model component scores. Eighty-seven variables were highlighted in S-plot (VIP > 1 and P < 0.05). Then the variables were confirmed by the raw data plots. Two examples of the tendency of variables in OSCC and control were shown in Figure 3C and 3D. Finally, fifty-five variables were selected in RPLC with positive-ion mode. The data acquired from other modes were correspondingly analyzed by the same method. Therefore, a total of fifty-five discriminate variables as interesting biomarker candidates were found in OSCC relative to the control group of RPLC in positive-ion mode, thirty-two discriminate variables of RPLC in negative-ion mode, twenty-eight discriminate variables of HILIC in positive-ion mode, and thirty-seven discriminate variables of HILIC in negative-ion mode.
Figure 3. The strategy for selection of interesting variables for HC vs OSCC I–II in RP column with positive-ion mode.
(A) Score plots for OPLS-DA models; (B) S-plot; The tendency of variables (C) m/z 180.0359/15.16 min and (D) m/z 310.3094/11.61 min in OSCC and control.
Identification of potential biomarkers
The elemental composition was calculated from the acquired high resolution MS data using the Masslynx 4.1 analysis software. The spectra routinely collected with 7000 mass resolution, ~0.6 mDa precision and ~10 ppm tolerances is adequate to generate an initial list of possible chemical compositions. The element limits were set to C, H, N, O, and S. We take the ion of m/z 89.0226/2.59 min in HILIC with negative-ion mode as an example to illustrate the detailed process of biomarker identification. Based on the elemental composition analysis software (Supplemental Figure 2), the maximum number of chemical formulas for m/z 89.0226 was 12. Among these formulas, the selection of the best match was strongly dependent on its i-FIT value, which is an index of the deviations observed from the predicted masses and intensities of monoisotopic peaks corresponding to a given chemical formula. The elemental composition was determined to be C3H5O3 (cal. 89.0239) based on the lowest i-FIT value. After searching various metabolomic databases with the molecular formula C3H5O3 and m/z 89.0226, lactic acid was considered as possible compound. Then, it was finally confirmed by comparison with standard sample. Eventually, fourteen metabolites were tentatively identified as potential biomarkers for early diagnosis of OSCC and were listed in Table 2. In fourteen potential biomarkers, seven salivary biomarkers can only be separated by RPLC technique; Four salivary biomarkers can only be separated by HILIC technique, while three other salivary biomarkers can be separated both by RPLC and HILIC techniques. These results suggested that combining RPLC and HILIC techniques for a comprehensive saliva metabonomics analysis expanded the scope of screening biomarkers for the early diagnosis of OSCC. Among these metabolites, four potential biomarkers (lactic acid, succinic acid, ornithine and carnitine) were confirmed using standard samples. However, other structural assignments were incomplete and these structures could be confirmed in future studies.
Table 2. Potential biomarkers and their identification results.
| m/z | Adduct | VIP (Trendb) | Column | P value | Identified results | Related pathway |
|---|---|---|---|---|---|---|
| 89.0226/2.59 | [M − H] | 1.29 (↑) | HILIC | 0.032 | Lactic acid c | Glycolysis |
| 181.0635/3.64 | [M − H] | 1.80 (↑) | HILIC | 0.030 | Hydroxyphenyllactic acid | Tyrosine metabolism |
| 214.1450/1.66 | [M − H] | 1.35 (↑) | HILIC | 0.022 | N-nonanoylglycine | Fatty acids oxidation |
| 141.0138/4.53 | [M − H] | 1.82 (↑) | HILIC | 0.024 | 5-hydroxymethyluracil | Oxidative stress |
| 116.9278/14.81 | [M − H] | 1.98 (↑) | RP | 0.003 | Succinic acid c | TCA cycle |
| 131.0701/2.07 | [M − H] | 1.45 (↑) | RP | 0.030 | Ornithine c | Urea cycle |
| 258.1698/2.43 | [M − H] | 1.17 (↑) | RP | 0.015 | Hexanoylcarnitine | Energy metabolism |
| 321.2885/8.67 | [M + H] | 1.35 (↑) | RP | <0.0001 | Propionylcholine | - |
| 142.0973/2.34 | [M − H2O − H] | 1.49 (↓) | RP | 0.039 | Carnitine c | Beta oxidation of long chain fatty acids |
| 142.0028/2.34 | [M − H] | 2.42 (↓) | RP | 0.045 | 4-hydroxy-L-glutamic acid | Arginine and proline metabolism |
| 415.2137/4.20 a | [2M + H] | 4.42 (↓) | RP/HILIC | <0.0001 | Acetylphenylalanine | Phenylalanine metabolism |
| 302.3048/4.87 a | [M + H] | 3.95 (↓) | RP/HILIC | 0.001 | Sphinganine | Sphingolipid metabolism |
| 318.2967/3.72 a | [M + H] | 4.02 (↓) | RP/HILIC | <0.0001 | Phytosphingosine | Sphingolipid metabolism |
| 180.0359/15.16 | [M + H] | 2.39 (↓) | RP | <0.0001 | S-carboxymethyl-L-cysteine | - |
aRetention time of RPLC.
bChange trend compared with controls. (↑): up-regulated. (↓): down-regulated.
cvalidated with standard sample.
Explanation of change trend and characterization of potential biomarkers
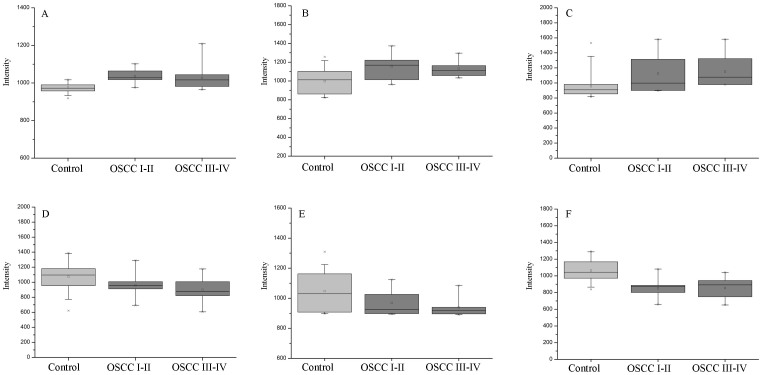
In all biomarkers, eight potential biomarkers were up-regulated in saliva of OSCC patients and six potential biomarkers were down-regulated. To provide an intuitive comparison, the change trends (up- or down-regulated) of six representative biomarkers in OSCC I–II and OSCC III–IV compared to controls are also provide in box plots in Figure 4.
Figure 4. The box plots of six potential biomarkers in distinguishing OSCC patients at stage I–II and III–IV from healthy controls.
(A) propionylcholine, (B) succinic acid, (C) lactic acid, (D) acetylphenylalanine, (E) carnitine, (F) phytosphingosine. Horizontal lines represent from bottom to top: the minimum, 25th, 50th, and 75th percentiles and the maximum.
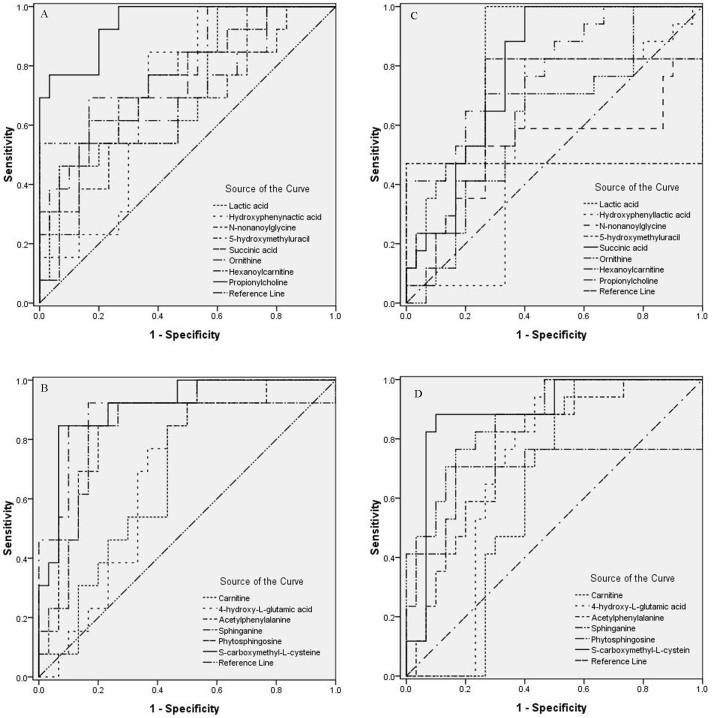
In order to characterize these potential biomarkers in early stage of OSCC, receiver operating characteristic (ROC) analysis was performed. The fourteen identified potential biomarkers were divided into two groups, eight up-regulated in OSCC patients (Figure 5A) and six down-regulated (Figure 5B). Table 3 shows the detailed sensitivity, specificity levels and 95% confidence interval of the fourteen identified potential salivary biomarkers for OSCC early prediction. As a single biomarker in saliva, S-carboxymethyl-L-cysteine had a sensitivity of 84.6% and a specificity of 93.3% for early predicting OSCC. The eight up-regulated metabolites, lactic acid, hydroxyphenyllactic acid, N-nonanoylglycine, 5-hydroxymethyluracil, succinic acid, ornithine, hexanoylcarnitine and propionylcholine provided the areas under curve (AUC) values of 0.708, 0.710, 0.721, 0.718, 0.785, 0.710, 0.733 and 0.946, respectively in HC vs OSCC I–II mode. The six down-regulated metabolites, i.e., carnitine, 4-hydroxy-L-glutamic acid, acetylphenylalanine, sphinganine, phytosphingosine, and S-carboxymethyl-L-cysteine, provided AUC values of 0.700, 0.695, 0.838, 0.818, 0.910, and 0.913, respectively in HC vs OSCC I–II mode.
Figure 5. ROC analysis for potential biomarkers in diagnose OSCC.
(A) Eight up-regulated metabolites of OSCC in OSCC I–II mode; (B) Six down-regulated metabolites of OSCC in OSCC I–II mode; (C) Eight up-regulated metabolites in OSCC of OSCC III–IV mode; (D) Six down-regulated metabolites of OSCC in OSCC III–IV mode.
Table 3. ROC curve analysis of identified potential salivary biomarkers in OSCC I–II.
| HC vs OSCC I–II | |||||
|---|---|---|---|---|---|
| Metabolite | AUCs | 95% confidence interval | Sensitivity (%) | Specificity (%) | Std. Errora |
| Lactic acid | 0.708 | 0.539–0.876 | 46.2 | 93.3 | 0.086 |
| Hydroxyphenyllactic acid | 0.710 | 0.557–0.863 | 92.3 | 46.7 | 0.078 |
| N-nonanoylglycine | 0.721 | 0.549–0.892 | 69.2 | 73.3 | 0.088 |
| 5-hydroxymethyluracil | 0.718 | 0.532–0.904 | 53.8 | 96.7 | 0.095 |
| Succinic acid | 0.785 | 0.636–0.933 | 69.2 | 83.3 | 0.076 |
| Ornithine | 0.710 | 0.530–0.891 | 53.8 | 86.7 | 0.092 |
| Hexanoylcarnitine | 0.733 | 0.574–0.893 | 61.5 | 83.3 | 0.082 |
| Propionylcholine | 0.946 | 0.882–1.011 | 76.9 | 96.7 | 0.033 |
| Carnitine | 0.700 | 0.546–0.854 | 92.3 | 50.0 | 0.079 |
| 4-hydroxy-L-glutamic acid | 0.695 | 0.542–0.848 | 92.3 | 50.0 | 0.078 |
| Acetylphenylalanine | 0.838 | 0.705–0.972 | 92.3 | 76.7 | 0.068 |
| Sphinganine | 0.818 | 0.660–0.976 | 84.6 | 83.3 | 0.081 |
| Phytosphingosine | 0.910 | 0.816–1.005 | 92.3 | 83.3 | 0.048 |
| S-carboxymethyl-L-cysteine | 0.913 | 0.822–1.004 | 84.6 | 93.3 | 0.047 |
aUnder the nonparametric assumption.
The ROC curve was also plotted for the model of HC vs OSCC III–IV (Figure 5C and 5D). The detailed parameters of potential salivary biomarkers for OSCC III–IV prediction were provided in Table 4. Compared with Table 3, the AUC value of each potential biomarker of HC vs OSCC I–II is larger than HC vs OSCC III–IV, except lactic acid and 4-hydroxy-L-glutamic acid, indicated that other twelve biomarkers have better ability in distinguishing OSCC I–II from the controls.
Table 4. ROC curve analysis of identified potential salivary biomarkers in OSCC III–IV.
| HC vs OSCC III–IV | |||||
|---|---|---|---|---|---|
| Metabolite | AUCs | 95% confidence interval | Sensitivity (%) | Specificity (%) | Std. Errora |
| Lactic acid | 0.837 | 0.723–0.951 | 100.0 | 73.3 | 0.058 |
| Hydroxyphenyllactic acid | 0.569 | 0.395–0.742 | 82.4 | 60.0 | 0.089 |
| N-nonanoylglycine | 0.527 | 0.328–0.727 | 52.9 | 73.3 | 0.102 |
| 5-hydroxymethyluracil | 0.471 | 0.233–0.708 | 47.1 | 96.7 | 0.121 |
| Succinic acid | 0.786 | 0.658–0.914 | 88.2 | 66.7 | 0.065 |
| Ornithine | 0.671 | 0.490–0.851 | 82.4 | 73.3 | 0.092 |
| Hexanoylcarnitine | 0.612 | 0.446–0.777 | 70.6 | 60.0 | 0.084 |
| Propionylcholine | 0.788 | 0.655–0.921 | 64.7 | 80.0 | 0.068 |
| Carnitine | 0.629 | 0.470–0.789 | 94.1 | 46.7 | 0.081 |
| 4-hydroxy-L-glutamic acid | 0.708 | 0.556–0.860 | 94.1 | 56.7 | 0.077 |
| Acetylphenylalanine | 0.765 | 0.628–0.902 | 82.4 | 70.0 | 0.070 |
| Sphinganine | 0.590 | 0.495–0.885 | 70.6 | 83.3 | 0.099 |
| Phytosphingosine | 0.875 | 0.776–0.973 | 76.5 | 83.3 | 0.050 |
| S-carboxymethyl-L-cysteine | 0.888 | 0.784–0.992 | 88.2 | 90.0 | 0.053 |
aUnder the nonparametric assumption.
To demonstrate the utility of salivary biomarkers in combination for the early diagnosis of OSCC, five metabolites (AUC > 0.8) comprising propionylcholine, acetylphenylalanine, sphinganine, phytosphingosine, and S-carboxymethyl-L-cysteine were selected to form a biomarker group. These biomarkers were first combined by a conventional binary logistic regression (LR) prediction model and then subjected to ROC analysis. The results showed that the potential biomarker group provided an AUC value of 0.997, with a sensitivity of 100% and a specificity of 96.7% in distinguishing OSCC I–II from control. The ROC curve was also plotted for the LR model of HC vs OSCC III–IV. As a result, the AUC value was 0.971 (sensitivity 86.7%; specificity 94.1%; 95% confidence interval = 0.989–1.006) for the LR model of OSCC III–IV. The AUC value of LR mode in HC vs OSCC I–II is larger than that of HC vs OSCC III–IV, which indicated that these five biomarkers in combination have superior performance in diagnosis of OSCC at stage I and II from control.
These results demonstrated that five salivary biomarkers (propionylcholine, acetylphenylalanine, sphinganine, phytosphingosine, and S-carboxymethyl-L-cysteine) in combination will improve the sensitivity and specificity for the OSCC early detection (stage I and II). Therefore, these salivary biomarkers might have important clinical value for the diagnosis of OSCC in its early stage.
Discussion
Saliva testing, a non-invasive alternative to serum testing, is rapidly advancing in recent years. Additionally, it is inexpensive, and easy to use. The collection of saliva can reduce the discomfort for patients, particularly if repeated sampling is necessary.
With our experiment, the metabolites obtained included sphingolipids (sphinganine and phytosphingosine), carnitines (carnitine, hexanoylcarnitine), choline derivative (propionylcholine), carboxylic acid (lactic acid), carboxylic acid (succinic acid), pyrimidine (5-hydroxymethyluracil), benzyl alcohol derivative (hydroxyphenyllactic acid), and amino acids and derivatives (ornithine, N-nonanoylglycine, acetylphenylalanine, 4-hydroxy-L-glutamic acid, and S-carboxymethyl-L-cysteine), in accordance with the defined chemical class category in the Human Metabolome Database. Description for the selected biomarkers were grouped as up-regulated and down-regulated metabolites.
For up-regulated metabolites, lactic acid plays a role in several biochemical processes and is an end product of glycolysis. Most cancer cells depend on aerobic glycolysis rather than oxidative phophorylation for energy production30. To fulfill tumor cell needs, the glycolytic switch is associated with elevated glucose uptake and lactic acid release31. Compared with control, a higher level of lactic acid was observed in the early stage of OSCC patients. The excessive proliferation of cancer cells requires more energy, and as a result, tumors often produce large amounts of lactic acid by carrying out glycolysis even under aerobic conditions. This phenomenon is known as the ‘Warburg effect’32. Lactic acid also associated with pyruvate metabolism. Increased lactic acid may have the relationship with the decreased pyruvate entering into tricarboxylic acidcycle (TCA)21, which is a series of enzyme-catalyzed chemical reactions of key importance in all living cells. Succinic acid is a dicarboxylic acid, which is the intermediate metabolite in TCA cycle33. In the early stage of OSCC, succinic acid content in saliva increased as compared to the healthy people, probably because the increased metabolic utilization by the TCA cycle in oral cancer cells. We found that the levels of ornithine were markedly higher in patients with OSCC than in healthy controls. Over expression of ornithine has also been reported in other researches34. Ornithine is an amino acid produced in the urea cycle by the splitting off urea from arginine35. It is a central part of the urea cycle and is also a precursor of citrulline and arginine36. Another amino acid and derivative, N-nonanoylglycine is normally minor metabolites of fatty acids. Elevated level of acylglycines appears in the saliva of OSCC patients probably due to oxidation disorders of various fatty acids. Hexanoylcarnitine is an acylcarnitine. Unusual acylcarnitines can be observed in disturbances in energy production and in intermediary metabolism in the organism. Detection of the qualitative pattern of acylcarnitines can be of diagnostic and therapeutic importance. The hexanoylcarnitine content is elevated in OSCC patients probably because energy metabolism is up-regulated in OSCC. Propionylcholine is a choline derivative, precursor of acetylcholine, an important neurotransmitter and synthesis. Acetylcholine is critical substance in phospholipid biosynthesis. The salivary propionylcholine content is higher compared with healthy subjects probably due to the increased expression of the process of choline phosphorylation. Hydroxyphenyllactic acid is a tyrosine metabolite, in which the level is elevated with a deficiency of the enzyme p-hydroxyphenylpyruvate oxidase. In the OSCC patients, the content of salivary hydroxyphenyllactic acid is higher compared with healthy subjects. This may be related to the deficiency of the enzyme p-hydroxyphenylpyruvate oxidase. In our study, a similar tyrosine metabolism disorder was found in OSCC as in phenylketonuria and tyrosinemia37. 5-Hydroxymethyluracil, an oxidation damage product, is formed when cells are under oxidative stress. In the OSCC patients, the content of 5-hydroxymethyluracil was elevated probably because the redox state of the body was changed resulting in oxidative stress in cancer patients. There are various reasons, such as nutritional intake disorders and immune system activation in OSCC patients. Similar oxidative stress disorder in OSCC was found in other researches38.
For down-regulated metabolites, the obvious decrease of acetylphenylalanine reveals an abnormal phenylalanine metabolism in OSCC patients. Acetylphenylalanine is a product of enzyme phenylalanine N-acetyltransferase in the pathway phenylalanine metabolism18 (Supplemental Figure 3) which indicates a disturbance of glycine N-acyltransferase in OSCC. Sphingolipid metabolism is thus seen to be abnormal in OSCC patients compared to controls. Sphinganine and phytosphingosine are down-regulated in saliva of OSCC patients, which are all involved in ceramide (N-Acylsphingosine) synthesis and metabolism (Supplemental Figure 4). Ceramide is regarded as important cellular signals for inducing apoptosis. Therefore, sphinganine and phytosphingosine have attracted considerable interest. Sphinganine is a blocker postlysosomal cholesterol transport by inhibition of low-density lipoprotein-induced esterification of cholesterol and cause unesterified cholesterol to accumulate in perinuclear vesicles39. Phytosphingosine is structurally similar to sphingosine. In OSCC, sphinganine and phytosphingosine are both down-regulated with P value < 0.001. These bioactive sphingolipid metabolites may have the potential to serve as biomarkers for OSCC. Carnitineis an essential factor in beta-oxidation of long chain fatty acids and its most important known metabolic function is the transport of fat into the mitochondria of muscle cells40. Carnitine is synthesized from lysine and methionine41. The acetyl-CoA generated in beta-oxidation enters the TCA cycle, where it is further oxidized to CO2, producing more reduced energy carriers. The carnitine content is lower in OSCC patients probably because fatty acid metabolism is down-regulated in OSCC.
Separation technology plays a critical role in metabolomics study. In our work, an integrated separation approach by combining RPLC and HILIC with TOF-MS has been developed for performing global metabolomics analysis in human saliva and identified more potential biomarkers for the early diagnosis of OSCC. The results demonstrated that different separation approaches enlarged metabolite coverage. A total of fourteen potential biomarkers have a close relationship with early stage of OSCC. Eight potential biomarkers were up-regulated in saliva of OSCC patients and six potential biomarkers were down-regulated. Five salivary biomarkers (propionylcholine, acetylphenylalanine, sphinganine, phytosphingosine, and S-carboxymethyl-L-cysteine) in combination yielded satisfactory accuracy (AUC = 0.997), sensitivity (100%), and specificity (96.7%) in distinguishing OSCC patients at stage I–II from the control. In the future, subsequent research should be carried out to clinically validate these potential biomarkers in large patient cohorts before they can be used in real clinical diagnostics.
Methods
Chemicals
Acetonitrile (HPLC grade) and methanol (HPLC grade) were purchased from Fisher (USA). Distilled water (18.2 MΩ) was purified “in-house” using an ULUPURE system (Chengdu Ultrapure Technology Co., Ltd, Chengdu, China). Ammonium acetate and ammonium formate (Ke long Chemical Reagent Factory, Chengdu, China) were used in this work. Formic acid (HPLC grade), lactic acid and carnitine were purchased from J&K Chemical Ltd (Beijing, China). Succinic acid and ornithine were purchased from Damas-beta (Shanghai, China).
Study participants
Thirty Chinese diagnosed with OSCC were recruited from the West China Hospital of Stomatology, West China School of Stomatology, Sichuan University (25 males and 5 females, clinical stage: 4 of stage I, 9 of stage II, 3 of stage III and 14 of stage IV). The mean age was 55 years (range: 29–72). All OSCC patients were diagnosed based on clinical and histopathologic criteria. The detailed clinical characteristics of saliva samples used in this study were provided in the Supplemental Table 1. OSCC stage was established according to the Tumor Nodes Metastasis (TNM) staging system (based on a combination of tumour size or depth (T), lymph node spread (N), and presence or absence of metastases (M)), promulgated by the American Joint Committee on Cancer (AJCC). The disease status and staging of OSCC patients were obtained from clinical records. The OSCC patients were all recruited with no history of receiving medication and surgical operation and none had been treated with chemotherapy and radiotherapy before sample collection. The control group contained thirty age-gender matched cancer-free healthy Chinese volunteers with 25 males and 5 females, whose mean age was 47 years (range: 25–69).
Ethics statement
The Ethical Committee of the West China Hospital of Stomatology, Sichuan University, approved the protocol. All of the volunteers and patients signed an Ethical Committee consent form agreeing to serve as saliva donors for the experiments. The methods were carried out in “accordance” with the approved guidelines.
Saliva collection and preparation
All the donors were asked to refrain from smoking, eating, drinking, or oral hygiene procedures for at least 1 hour prior to samples collection, and then rinse their mouth thoroughly with water. Saliva samples were collected between 9:00 and 11:00 a.m. in a private room using standard techniques. Roughly three milliliters of unstimulated whole saliva was obtained. The samples, once collected, were centrifuged at 12000 rpm for 20 min at 4°C to remove insoluble materials, cell debris and food remnants. Equal amounts of supernatant (400 μL) were transferred to fresh tubes and frozen at −40°C until the laboratory analysis.
Before the analysis, frozen saliva was thawed and dissolved at room temperature. A mixture of acetonitrile/methanol (75:25 v/v, 800 μL) was added to saliva (400 μL) in a 1.5 mL Eppendorf tube to precipitate proteins. The mixture was allowed to stand for 10 min after vortexing for 60 s, and then the samples were centrifuged at 12000 rpm for 20 min at 4°C. The supernatant were filtered through syringe filters (0.22 μm, Jinteng) before UPLC-MS analysis.
UPLC-MS analysis
RPLC separation was performed on an ACQUITY UPLC™ BEH C18 column (50 mm × 2.1 mm i.d., 1.7 μm, Waters, Milford, USA). HILIC separation was performed on ACQUITY UPLC™ BEH Amide column (100 mm × 2.1 mm i.d., 1.7 μm, Waters, Milford, USA). The column was maintained at 45°C. The injected sample volume was 10 μL for each run in the full loop injection mode. The flow rate of the mobile phase was 0.2 mL/min. In RPLC mode, gradient elution was performed with the following solvent system: (A) 0.1% formic acid-water with 1 mM ammonium formate, (B) acetonitrile (ACN). The gradient started with 95% A and decreased to 50% A in 2 min, 50% ~ 5% A in 13 min, holding at 5% A for 1 min, then turned to 95% A immediately, and holding at 95% A for 5 min. Isocratic elution was performed in HILIC mode with the following solvent system: (A) 95:5 ACN-10 mM aqueous ammonium acetate, (B) 50:50 ACN-10 mM aqueous ammonium acetate; 65% A and 35% B over 19 min.
Mass spectrometry experiments were performed on an orthogonal accelerated time of flight mass spectrometer (Waters, Milford, USA) equipped with an electrospray ion source. Data were acquired in both positive- and negative-ion V-geometry mode for each chromatography separation technique which generated four separate UPLC-MS analysis. The capillary and cone voltages were set to 2000 and 100 V, cone gas 30 L/h, desolvation gas 750 L/h, source temperature 110°C, desolvation temperature 300°C. The scan range was from m/z 50 to 1000 in the full scan mode and data were collected in centroid mode. Independent reference Lock-mass ions via the LockSpray™ interface was used to ensure mass accuracy during data acquisition. Leucine-Enkephalin (Sigma-Aldrich, [LE + H]+, m/z, 556.2771) was used as the reference compound. The solution of LE was infused through the reference probe at the flow rate of 0.04 mL/min with the help of a second LC pump (Waters).
Data handling and statistical analysis
The UPLC-TOF/MS data from the saliva samples of both separation modes were analyzed to identify potential discriminant biomarkers. Data acquisition and handling were performed by Masslynx 4.1 (Waters). MarkerLynx application manager (Waters, Manchester, UK) has been used for peak finding, filtering, and alignment. The parameters used were retention time range 0–19 min; retention time window was set to 0.2 min; mass range 50–1000 Da. After a suitable processing, the resulting three dimensional data matrix containing m/z-retention time pairs, sample names and their normalized chromatographic peak areas (variables) was exported into SIMCA-P 11.0 (Umetrics, Sweden) for subsequent processing by multivariate data analysis including principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA). These statistical methods were carried out on data to identify discriminant metabolites in saliva between OSCC patients and healthy individuals. The discriminating variables were selected according to Variable Importance in the Projection (VIP) values and VIP > 1.0 is considered relevant for group discrimination28. The nonparametric Mann-Whitney U test was applied to assess the differences in these potential biomarkers between the OSCC and healthy control group. Results were considered significant at a 2-tailed P value of <0.05. The areas under curve (AUC) of receiver operating characteristic (ROC) curves were constructed to evaluate the diagnostic effectiveness of potential biomarkers using SPSS 16.0. The ROC curve procedure is a useful method for evaluating the performance of classification schemes that categorize cases into one of the two groups. AUC value is known to be a useful measure of overall predictor quality, with a value of 1.0 for a perfect predictor and 0.5 for a random predictor.
Metabolites were identified by searches of databases: Human Metabolome Database (http://www.hmdb.ca/), MASS Bank (http://www.massbank.jp/), KEGG (http://www.genome.jp/kegg/ligand.html), and PubChem compound database (http://www.ncbi.nlm.nih.gov) using exact molecular weights. Commercial standard reagents were used to support identification of metabolites.
Author Contributions
Q.W. designed and performed experiments, analyzed data, and wrote the main manuscript text. P.G. and X.W. provided saliva samples of OSCC. Y.D. designed and supervised experiments, and reviewed and submitted the manuscript as the corresponding author.
Supplementary Material
Supporting Information
Acknowledgments
The authors are grateful to the financial support from National Recruitment Program of Global Experts (NRPGE), the Hundred Talents Program of Sichuan Province (HTPSP), and the Startup Funding of Sichuan University for setting up the Research Center of Analytical Instrumentation.
References
- Lozano R. et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende T. M. B., Freire M. d. S. & Franco O. L. Head and neck cancer. Cancer 116, 4914–4925 (2010). [DOI] [PubMed] [Google Scholar]
- Liu S. A. et al. Nutritional factors and survival of patients with oral cancer. Head & neck 28, 998–1007 (2006). [DOI] [PubMed] [Google Scholar]
- Ehsan S. et al. Disease Specific Biomarkers of Abdominal Aortic Aneurysms Detected by Surface Enhanced Laser Desorption Ionization Time of Flight Mass Spectrometry. Eur J Vasc Endovasc 44, 52–54 (2012). [DOI] [PubMed] [Google Scholar]
- Wang Q., Gao P., Wang X. & Duan Y. Investigation and identification of potential biomarkers in human saliva for the early diagnosis of oral squamous cell carcinoma. Clin Chim Acta 427, 79–85 (2013). [DOI] [PubMed] [Google Scholar]
- Pfaffe T., Cooper-White J., Beyerlein P., Kostner K. & Punyadeera C. Diagnostic Potential of Saliva: Current State and Future Applications. Clin Chem 57, 675–687 (2011). [DOI] [PubMed] [Google Scholar]
- Cubbon S., Antonio C., Wilson J. & Thomas-Oates J. Metabolomic applications of HILIC–LC–MS. Mass Spectrom Rev 29, 671–684 (2010). [DOI] [PubMed] [Google Scholar]
- Pereira H., Martin J. F., Joly C., Sébédio J. L. & Pujos-Guillot E. Development and validation of a UPLC/MS method for a nutritional metabolomic study of human plasma. Metabolomics 6, 207–218 (2010). [Google Scholar]
- Shintani S., Hamakawa H., Ueyama Y., Hatori M. & Toyoshima T. Identification of a truncated cystatin SA-I as a saliva biomarker for oral squamous cell carcinoma using the SELDI ProteinChip platform. Int J Oral Maxillofac Surg 39, 68–74 (2010). [DOI] [PubMed] [Google Scholar]
- Zimmermann B. G. & Wong D. T. Salivary mRNA targets for cancer diagnostics. Oral Oncol 44, 425–429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci 69, 3341–3350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratlin J. L., Serkova N. J. & Eckhardt S. G. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res 15, 431–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimetti M., Cacciatore S., Graziano A. & Tenori L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics 8, 465–474 (2012). [Google Scholar]
- Nicholson J. K. & Lindon J. C. Systems biology: metabonomics. Nature 455, 1054–1056 (2008). [DOI] [PubMed] [Google Scholar]
- Mishur R. J. & Rea S. L. Applications of mass spectrometry to metabolomics and metabonomics: Detection of biomarkers of aging and of age-related diseases. Mass Spectrom Rev 31, 70–95 (2012). [DOI] [PubMed] [Google Scholar]
- Yan S. K. et al. Metabonomic approach to the diagnosis of oral squamous cell carcinoma, oral lichen planus and oral leukoplakia. Oral Oncol 44, 477–483 (2008). [DOI] [PubMed] [Google Scholar]
- Tiziani S., Lopes V. & Günther U. L. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia 11, 269–276 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. et al. LC-MS based serum metabonomic analysis for renal cell carcinoma diagnosis, staging, and biomarker discovery. J Proteome Res 10, 1396–1405 (2011). [DOI] [PubMed] [Google Scholar]
- An Z. et al. Integrated Ionization Approach for RRLC− MS/MS-based Metabonomics: Finding Potential Biomarkers for Lung Cancer. J Proteome Res 9, 4071–4081 (2010). [DOI] [PubMed] [Google Scholar]
- Theodoridis G., Gika H. G. & Wilson I. D. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trends Ana. Chem 27, 251–260 (2008). [Google Scholar]
- Wei J. et al. Salivary metabolite signatures of oral cancer and leukoplakia. Int J Cancer 129, 2207–2217 (2011). [DOI] [PubMed] [Google Scholar]
- Tsutsui H. et al. Practical analytical approach for the identification of biomarker candidates in prediabetic state based upon metabonomic study by ultraperformance liquid chromatography coupled to electrospray ionization time-of-flight mass spectrometry. J Proteome Res 9, 3912–3922 (2010). [DOI] [PubMed] [Google Scholar]
- Qiu Y. et al. Serum metabolite profiling of human colorectal cancer using GC− TOFMS and UPLC− QTOFMS. J Proteome Res 8, 4844–4850 (2009). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. Pseudotargeted Metabolomics Method and Its Application in Serum Biomarker Discovery for Hepatocellular Carcinoma Based on Ultra High-Performance Liquid Chromatography/Triple Quadrupole Mass Spectrometry. Anal Chem 85, 8326–8333 (2013). [DOI] [PubMed] [Google Scholar]
- Alpert A. J. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J Chromatogr A 499, 177–196 (1990). [DOI] [PubMed] [Google Scholar]
- Liu J. & Duan Y. Saliva: A potential media for disease diagnostics and monitoring. Oral Oncol 48, 569–577 (2012). [DOI] [PubMed] [Google Scholar]
- Eriksson L. Multi-and megavariate data analysis; MKS Umetrics AB, 2006. [Google Scholar]
- Jia L., Chen J., Yin P., Lu X. & Xu G. Serum metabonomics study of chronic renal failure by ultra performance liquid chromatography coupled with Q-TOF mass spectrometry. Metabolomics 4, 183–189 (2008). [Google Scholar]
- Huo T. et al. Metabonomic study of biochemical changes in the serum of type 2 diabetes mellitus patients after the treatment of metformin hydrochloride. J Pharm Biomed Anal 49, 976–982 (2009). [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C. & Thompson C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhup S., Kumar Dadhich R., Ettore Porporato P. & Sonveaux P. Multiple biological activities of lactic acid in cancer: influences on tumor growth, angiogenesis and metastasis. Curr Pharm Des 18, 1319–1330 (2012). [DOI] [PubMed] [Google Scholar]
- Kim J. W. & Dang C. V. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res 66, 8927–8930 (2006). [DOI] [PubMed] [Google Scholar]
- Hori S. et al. A metabolomic approach to lung cancer. Lung Cancer 74, 284–292 (2011). [DOI] [PubMed] [Google Scholar]
- Sugimoto M., Wong D. T., Hirayama A., Soga T. & Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6, 78–95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavri-Damelin D. et al. Ornithine transcarbamylase and arginase I deficiency are responsible for diminished urea cycle function in the human hepatoblastoma cell line HepG2. Int J biochem Cell B 39, 555–564 (2007). [DOI] [PubMed] [Google Scholar]
- Deignan J. L. et al. Ornithine deficiency in the arginase double knockout mouse. Mol Genet Metab 89, 87–96 (2006). [DOI] [PubMed] [Google Scholar]
- Gerlo E. et al. Gas chromatographic–mass spectrometric analysis of N-acetylated amino acids: The first case of aminoacylase I deficiency. Anal Chim Acta 571, 191–199 (2006). [DOI] [PubMed] [Google Scholar]
- Bahar G., Feinmesser R., Shpitzer T., Popovtzer A. & Nagler R. M. Salivary analysis in oral cancer patients. Cancer 109, 54–59 (2007). [DOI] [PubMed] [Google Scholar]
- Ryland L. K., Fox T. E., Liu X., Loughran T. P. & Kester M. Dysregulation of sphingolipid metabolism in cancer. Cancer Biol Ther 11, 138–149 (2011). [DOI] [PubMed] [Google Scholar]
- Rebouche C. J. & Paulson D. J. Carnitine metabolism and function in humans. Annu Rev Nutr 6, 41–66 (1986). [DOI] [PubMed] [Google Scholar]
- Busquets S. et al. L-Carnitine: An adequate supplement for a multi-targeted anti-wasting therapy in cancer. Clin Nutr 31, 889–895 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information