Abstract
Virus detection and/or identification traditionally rely on methods based on cell culture, electron microscopy and antigen or nucleic acid detection. These techniques are good, but often expensive and/or time-consuming; furthermore, they not always lead to virus identification at the species and/or type level. In this study, Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) was tested as an innovative tool to identify human polioviruses and to identify specific viral protein biomarkers in infected cells. The results revealed MALDI-TOF MS to be an effective and inexpensive tool for the identification of the three poliovirus serotypes. The method was firstly applied to Sabin reference strains, and then to isolates from different clinical samples, highlighting its value as a time-saving, sensitive and specific technique when compared to the gold standard neutralization assay and casting new light on its possible application to virus detection and/or identification.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) is a high throughput technology based on the comparison of the protein fingerprint obtained by microbial cells with a database of reference spectra by means of the use of various algorithms integrated in systems recently made commercially available. In the last few years this tool has been increasingly studied and applied for the identification and typing of microorganisms1,2,3,4. MALDI-TOF MS is referred to as a “soft” ionization technique, because it causes minimal or no fragmentation and allows the molecular ions of analytes to be identified, even in complex mixtures of biopolymers3,4,5,6,7,8.
Many studies investigating the application of MALDI-TOF MS in clinical microbiology laboratories or addressing its use in experimental approaches related to bacteria and fungi have been published; however, very few studies have been conducted about its application to virus research and diagnostics8,9,10,11,12,13. This could be a consequence of the high molecular weight of the viral proteins (>20 KDa): in microbiology diagnosis the identification is based on a low molecular weight range (2–20 KDa), where the majority of the peaks detected corresponds to microorganism ribosomal proteins8. Moreover, while for bacteria there is a simple and standardized protocol of protein extraction8, for viruses, requiring a cell substrate to be in vitro cultivated, the identification of specific viral proteins can be affected by cellular debris14.
Traditionally, the gold standard method for the direct detection of viruses is cell culture, although the use of this method often requires several days or weeks before the results can be obtained. Electron microscopy and direct antigen detection methods based on enzyme immunoassay or immunofluorescence are widely used for virus detection and/or identification, although some of these techniques are less sensitive than cell culture14. Finally, genic amplification techniques based on the polymerase chain reaction (PCR) provide a rapid and sensitive alternative for virus detection and/or identification15.
Considering the well-founded ability of MALDI-TOF MS to determine the molecular weight of individual specific polypeptides, the aim of the present study was to develop a simple method exploiting the MALDI-TOF MS platform application for the identification of viruses and the detection of specific viral biomarkers in order to obtain a profile of identification useful to differentiate between virus-infected and uninfected cells.
We focused on three members of the Picornaviridae family, Enterovirus genus, Enterovirus C species (www.ictvdb.org), that includes poliovirus serotypes 1, 2 and 316.
Poliovirus serotype presents slightly different capsid proteins, conferring cellular receptor specificity and virus antigenicity17. Poliovirus type 1 is the most common serotype encountered in nature; however, all the three serotypes are extremely infectious and dangerous, being the most frequent causative agents of poliomyelitis.
The choice of poliovirus as an experimental model was firstly based on the relevance of this viral agent in causing one of the most severe human pathologies of the central nervous system and on the importance in the globalization context: thus, their efficient identification and, most importantly, their eradication represent one of the main goals for the World Health Organization18,19.
Furthermore, poliovirus provides an ideal experimental viral model, considering its very simple structure: the virion is composed by a non-enveloped icosahedral protein coat built up of 60 copies of 4 structural proteins (VP4, VP2, VP3, VP1, listed as mapped in the viral genome)17,20,21,22,23 and a viral genome-linked protein, VPg, covalently attached to the 5′end of viral RNA, which acts as a primer during RNA synthesis.
Results
MALDI-TOF MS analysis of type 1, 2 and 3 Sabin poliovirus strains
In order to develop a simple method exploiting the MALDI-TOF MS platform application in the virologic field we used poliovirus type 1, 2 and 3 Sabin reference strains and LLC-MK2 confluent cell monolayers (Rhesus monkey kidney epithelial cell line LLC-MK2; ATCC CCL-7) for their cultivation and titration, using the limit dilution method (“tissue culture infectious dose50”, “TCID50”); the experimental infections were performed using a multiplicity of infection of 0.01 TCID50/cell.
LLC-MK2 infected cells and the respective culture media were harvested when a robust cytopathic effect was clearly observable and the recovered viral particles were subjected to a linear sucrose gradient.
The gradient profile showed a peak that spans through fractions 2 to 6, at the expected gradient region (Supplementary Fig. 1a). These fractions were pooled and subjected to ultracentrifugation. Poliovirus concentration, morphology and purity were confirmed by electron microscopy analysis after negative staining, that also enabled us to exclude any evident cellular contamination (Supplementary Fig. 1b). The highly purified poliovirus particles were subjected to protein extraction for subsequent MALDI-TOF MS analysis of low (2–20 KDa range) and high molecular weight (17–90 KDa) proteins.
The purified virion spectra analysis, performed in the low molecular weight range (2–20 KDa) (Fig. 1a), showed only two significant peaks of about 3,800 and 7,600 m/z (common to the three purified poliovirus strains, corresponding to the mass of VPg (genome-associated viral protein) and VP4 poliovirus proteins (Table 1). The analysis of the spectra obtained from purified virions performed in the high molecular weight range (17–90 KDa) (Fig. 2b) showed three peaks around 26,000, 30,000 and 33,000 m/z corresponding to VP3, VP2 and VP1 poliovirus capsid proteins (Table 1).
Figure 1. Representative MALDI-TOF mass spectra of purified poliovirus Sabin reference strains and ClinPro Tools statistical analysis.
(a,b) Spectra of human poliovirus type 1, 2, 3 purified particles, shown in the m/z range of (a) 2 to 20 KDa (low molecular weight) and (b) 17 to 90 KDa (high molecular weight). Molecular weight values of poliovirus significant peaks are indicated on peak tops. (c,d) Analysis of discriminating peaks within the three poliovirus serotypes by using ClinPro Tools software (“Poliovirus type 1”, blue; “Poliovirus type 2”, red; “Poliovirus type 3”, green): “2D Peak distribution view” with the details at low (c) and high (d) molecular weight of the two best separating peaks; “PCA-3D dot plots window” of the spectra showing in 3 separate clusters the spectra of the 3 different poliovirus serotypes in the low (c) and high (d) molecular weight range; “Spectra view” in the low (c) and high (d) molecular weight range of the average spectra of the replicates of each poliovirus serotype. The red rectangles highlight the three discriminating peaks revealed in the mass range 2–20 KDa (7,522; 7,579; 7,635 m/z), and the four discriminating peaks in the mass range 17–90 KDa (30,197; 33,257; 33,554; 38,601 m/z). The blue rectangles are not discriminating peaks according to the applied Supervised Neural Network (SNN) statistical method.
Table 1. Proteic peaks detected for poliovirus 1, 2 and 3 and their statistical significance.
| Protein | Sabin Strains | Theoretical Da* | Observed** m/z | SD | p-value |
|---|---|---|---|---|---|
| VPg | Poliovirus1 | 3,890 | 3,807 Da | +/−2 Da | 0.000008 |
| Poliovirus2 | 3,720 | 3,788 Da | +/−1 Da | 0.000004 | |
| Poliovirus3 | 3,530 | 3,760 Da | +/−2 Da | 0.000002 | |
| VP4 | Poliovirus1 | 7,530 | 7,613 Da | +/−3 Da | <0.000001 |
| Poliovirus2 | 7,370 | 7,578 Da | +/−4 Da | <0.000001 | |
| Poliovirus3 | 7,450 | 7,522 Da | +/−2 Da | <0.000001 | |
| VP3 | Poliovirus1 | 26,570 | 26,622 Da | +/−25 Da | 0.98 |
| Poliovirus2 | 26,430 | 26,759 Da | +/−30 Da | 0.85 | |
| Poliovirus3 | 26,270 | 26,320 Da | +/−25 Da | 0.43 | |
| VP2 | Poliovirus1 | 30,090 | 30,240 Da | +/−35 Da | 0.87 |
| Poliovirus2 | 30,050 | 30,130 Da | +/−40 Da | 0.94 | |
| Poliovirus3 | 30,150 | 30,197 Da | +/−30 Da | 0.000012 | |
| VP1 | Poliovirus1 | 33,460 | 33,530 Da | +/−45 Da | 0.00008 |
| Poliovirus2 | 33,150 | 33,257 Da | +/−45 Da | <0.000001 | |
| Poliovirus3 | 33,480 | 33,555 Da | +/−50 Da | 0.00008 |
Da: Dalton.
*: theoretical mass available by GenBank accession numbers AAN85442.1, AAN85443.1, AAN85444.1 for poliovirus type 1, 2 and 3, respectively.
**: average value of the multiple measurements.
m/z: mass/charge expressed as Dalton (Da).
SD: Standard Deviation of the multiple measurements (see Methods section).
p-value: significance of the mass differences of each protein evaluated with ANOVA test (see Methods section).
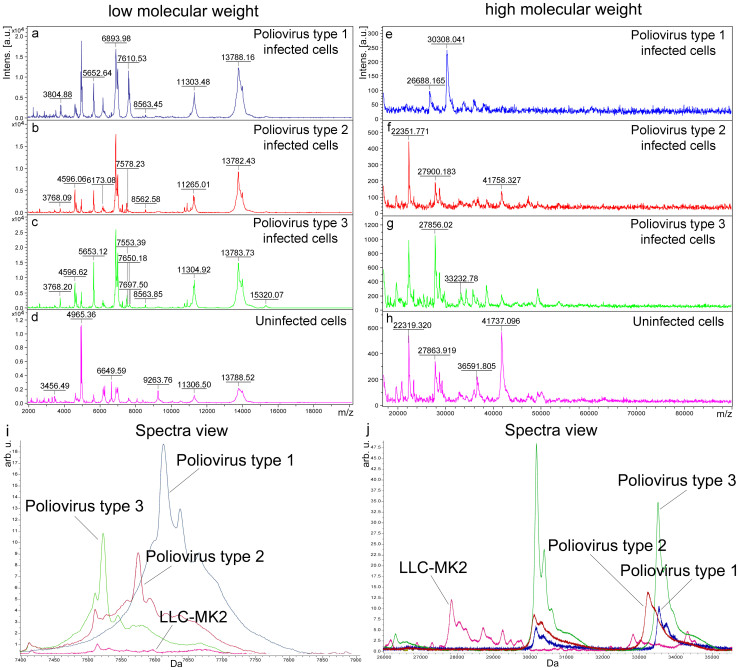
Figure 2. Representative MALDI-TOF mass spectra of Sabin poliovirus-infected and uninfected LLC-MK2 cells and comparative analysis by ClinPro Tools software.
The spectra of LLC-MK2 cells infected with human poliovirus types 1, 2, 3 (“Poliovirus type 1-infected cells”, “Poliovirus type 2-infected cells”, “Poliovirus type 3-infected cells”), with pointed average masses, are compared in the m/z range 2 to 20 KDa (a–c) and 17 to 90 KDa (e–g) with the spectra of LLC-MK2 uninfected cells (d,h). (i,j) Spectra view of the Average spectra profiles in the mass range 7,400–7,900 Da (i) and in the mass range 26,000 to 35,000 Da (j) of LLC-MK2 cells infected with the 3 different poliovirus serotypes with LLC-MK2 uninfected cells.
In order to exclude any non-specific peaks the non-inoculated matrix solutions (4-HCCA and SA) and the culture medium alone (E-MEM) were also subjected to MALDI-TOF MS analysis and no peaks were observed (data not shown).
Differential identification of the three purified Sabin poliovirus serotypes by MALDI-TOF MS and validation of the results by statistical analysis
In order to corroborate the evidence of the differences observed among the purified poliovirus strains protein spectra obtained by MALDI-TOF MS both in the low and in the high molecular weight range (see Fig. 1a,b), these spectra were imported into ClinPro Tools Software: data analysis began with raw data pre-treatment, normalization, baseline subtraction, peak defining, and recalibration; then, the automatic comparison of multiple spectra was performed. First the multivariate unsupervised statistical tool Principal Component Analysis (PCA) was applied to the data sets.
In Figure 1c (“low molecular weight”) and Figure 1d (“high molecular weight”) the statistical analysis of the spectra of the three different Sabin poliovirus serotypes is shown. In particular, in the “2D Peak distribution view” the two best separating peaks of the current statistic sort order are shown. In the “PCA-3D dot plot window” the plots of the spectra in a three-dimensional space are reported showing the spectra replicates of the three different poliovirus serotypes analyzed in the mass range 2–20 KDa (Fig. 1c) and 17–90 KDa (Fig. 1d) as three separate clusters.
Subsequently to reveal the discriminating peaks able to differentiate the 3 serotypes, 3 supervised statistical algorithms were applied (Supervised Neural Network, Genetic Algorithm and QuickClassifier).
Notably, the results obtained using the Supervised Neural Network (SNN) algorithm, that shows the highest values of Recognition Cabability (RC) and Cross Validation (CV), revealed three main discriminating peaks around 7,500–7,600 Da, in the low mass range (7,522, 7,578 and 7,613 m/z; RC: 100%; CV: 96,48%) (Fig. 1c, “Spectra view” and Supplementary Fig. 2a), and four discriminating peaks around 30,000–38,000 Da in the high mass range (30,197, 33,257, 33,555 and 38,601 m/z) (Fig. 1d, “Spectra view” and Supplementary Fig. 2b) with a RC and CV values of 100% and 96.3%, respectively.
MALDI-TOF MS analysis of Sabin poliovirus protein bands extracted from polyacrylamide gel
In order to assess whether the peaks observed in the spectra derived from MALDI-TOF MS analysis of purified virions corresponded to viral proteins, the poliovirus protein pattern obtained after sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) was analyzed. Three major bands, corresponding to the viral capsid proteins VP1, VP2, VP3, were clearly observable: two of them (VP1 and VP2) migrated quite closely and were cut together, while the VP4 protein band was very faint and the protein VPg band was not detectable (data not shown).
The spectra obtained by MALDI-TOF MS analysis of VP1, VP2 and VP3 bands directly excised from Coomassie stained gel revealed a mass/charge ratio matching the corresponding molecular weights (Supplementary Fig. 3a and Table 1). For the VP4 band a very weak signal on the gel after SDS-PAGE was observed, so that after the excision of the band the concentration of the protein was lower than the detection limit of MALDI-TOF MS and than not detectable. The expected correspondence between the molecular weights obtained by SDS-PAGE and MALDI-TOF MS analysis observed for the viral proteins was corroborated by the correspondence of the molecular weight marker bands in the range of interest (6,5 to 36,0 KDa) observed when excised and subjected to the same protein extraction protocol followed by mass spectrometry analysis (Supplementary Fig. 3b and Table 1).
MALDI-TOF MS analysis of Sabin poliovirus-infected cells: detection of differentiating viral peaks
In order to verify whether substantial differences exist between viral and cellular protein peak patterns, concentrated stocks of the three poliovirus Sabin reference strains were also prepared, not subjected to sucrose gradient purification, in order to obtain a pellet of poliovirus particles together with residual infected cell debris. Similarly, we collected uninfected LLC-MK2 cells by applying the same procedure.
To identify the protein mass profiles of the three poliovirus types, the corresponding spectra were automatically acquired. The macroscopic comparison of the different spectra in the “stack view mode” indicates that significant differences do exist between viral particles (poliovirus types 1, 2 and 3) combined with the residual infected cell debris and uninfected LLC-MK2 spectra (Fig. 2).
The mass spectra of poliovirus strains with residual infected cell debris compared at both low (Fig. 2a,b,c) and high (Fig. 2e,f,g) molecular weight with those of uninfected cells (Fig. 2d,h) revealed the presence of specific viral proteins.
In order to verify the discriminant viral peaks also in cells infected with poliovirus Sabin reference strains, the “Spectra view” in the range 7,400 to 7,900 Da (Fig. 2i) and in the range 26,000 to 35,000 Da (Fig. 2j) was created with ClinPro Tools software. From this analysis, performed directly on the infected and uninfected cells, only the VP4 protein (with a mass included in the low molecular weight range) corresponding peak, resulted a discriminant peak among the three different poliovirus types.
MALDI-TOF analysis of poliovirus isolates from different clinical samples
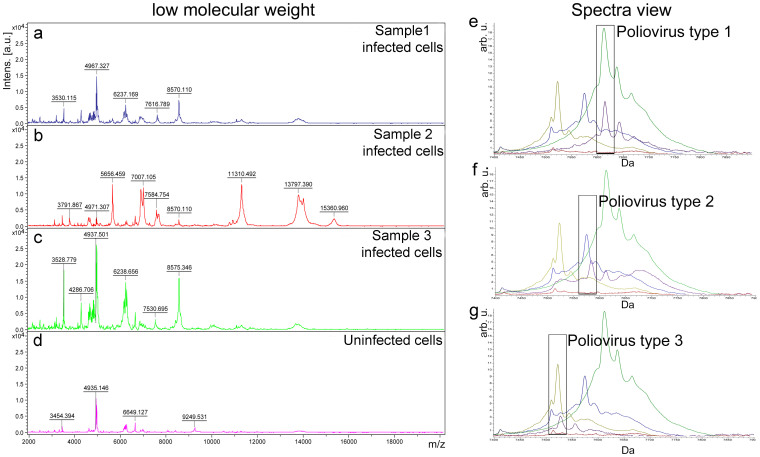
In order to further explore the applicability of this technology an in-house viral database was created. A Main Spectrum Profile (MSP) was generated, at low molecular weight, for each of the three poliovirus serotypes and for the uninfected LLC-MK2 cells. The four MSP spectra obtained by this procedure were deposited in the database for further blind identification of poliovirus clinical isolates. We performed the MALDI-TOF MS analysis on poliovirus types 1, 2 and 3 representative isolates from our collection, derived from different clinical samples (cerebrospinal fluid, throat swab and stools), identified by neutralization assay and stored at −80°C. The protein spectra (Fig. 3a,b,c) of the five poliovirus serotypes isolated from these samples were correctly identified as poliovirus without differentiation among serotypes with a score value between 1.9 and 2.0 for all the replicates tested when compared with the newly created MSP virus database by using the MALDI Biotyper software.
Figure 3. Representative MALDI-TOF mass spectra of LLC-MK2 cells infected with poliovirus clinical isolates and of uninfected cells, and ClinPro Tools comparative analysis between clinical isolates and Sabin poliovirus type 1, 2, and 3 reference strains.
The spectra of LLC-MK2 cells infected with poliovirus isolated from different human samples (“sample 1 infected cells”, “sample 2 infected cells”, “sample 3 infected cells”), identified by neutralization assay as poliovirus type 1, 2 and 3, respectively, are compared in the m/z range of 2 to 20 KDa (a–c) with the protein profile of LLC-MK2 uninfected cells (d). Molecular weight values of poliovirus clinical isolates and cellular peaks are indicated on peak tops. (e–g) Spectra view of the average spectra profiles in the mass range 7,400–7,900 Da of the three Sabin reference poliovirus strains (green, blue and yellow for poliovirus type 1, 2 and 3, respectively) compared with the poliovirus strains isolated from clinical samples (violet; poliovirus type 1, 2 and 3, in panels e, f and g, respectively) and with LLC-MK2 uninfected cells (red). Rectangles highlight the match between the VP4 protein peaks of clinical isolates and those of the corresponding Sabin reference strain.
Poliovirus clinical isolates serotypes differentiation
In order to detect and visualize the specific viral biomarker VP4 of the three different poliovirus serotypes isolated from the five clinical samples, all the spectra used to create the MSP spectra (the same that were previously used for the creation of the in-house database) of Sabin reference strains-infected cells, and those of uninfected LLC-MK2 cells, were imported into ClinPro Tools software. The average spectra of the cells infected with the three Sabin reference strains, as well as those of uninfected LLC-MK2 cells, were compared with the average spectra of each clinical isolate (Fig. 3e,f,g), previously identified by neutralization assay. As observed for purified Sabin reference strains and for Sabin reference strains-infected cells, also for the polioviruses identified from clinical samples the VP4 protein showed mass differences among the three serotypes. The molecular weight of VP4 protein of the five poliovirus clinical isolates overlapped the mass value of the corresponding Sabin reference serotype. In order to evaluate the reliability of VP4 protein as a discriminating peak at the serotype level, the Receiver Operating Characteristic (ROC) curve was created and the Area Under Curve (AUC) value was calculated. The VP4 protein of each poliovirus clinical isolates was compared to the VP4 of each Sabin reference strain-infected cells so that 3 ROC curves for each poliovirus clinical isolates were created; when correspondence between the serotype of poliovirus clinical isolate and the serotype of Sabin reference strain-infected cells was found the ROC curve gave an AUC values of 0. Otherwise the AUC values were >0.9.
VP4 protein as a biomarker for Enterovirus
In order to verify the absence of misidentification with members of the Picornaviridae family other than polioviruses, human coxsackievirus B1 and human echovirus 9 (both members of Enterovirus B species) were analyzed by MALDI-TOF MS. The protein spectra obtained in low molecular weight range (2–20 KDa) of human coxsackievirus B1 and echovirus 9 infected- and uninfected-LLC-MK2 cells are shown in Figure 4a. As expected for Enterovirus genus, the spectra showed also the specific peak of about 7,500 Da related to VP4 protein.
Figure 4. Representative MALDI-TOF mass spectra of LLC-MK2 cells infected with the Picornaviridae family members human coxsackievirus B1 and human echovirus 9 and uninfected cells, and ClinPro Tools comparative analysis with the Sabin poliovirus type 1, 2, and 3 strains.
(a) Spectra of LLC-MK2 cells infected with human coxsackievirus B1 and human echovirus 9 (“Coxsackie B1-infected cells” and “Echovirus 9-infected cells”) compared with the protein profile of LLC-MK2 uninfected cells in the m/z range of 2 to 20 KDa. Molecular weight values are indicated on peak tops. (b,c) Spectra view of the average spectra of human coxsackievirus B1 (b) and human echovirus 9 (c) compared by ClinPro Tools in the mass range 7,400–7,900 Da with the average spectra of the 3 Sabin poliovirus strains (poliovirus type 1; poliovirus type 2; poliovirus type 3) and uninfected LLC-MK2 cells.
The VP4 proteins of human coxsackievirus B1 and human echovirus 9 were compared with those of the three Sabin poliovirus serotypes in the molecular range 7,400 Da to 7,900 Da (Fig. 4b,c) giving AUC values > 0.9 in all the cases.
Discussion
This study aimed to evaluate the application of MALDI-TOF MS to virus identification, and to the detection of specific viral biomarkers, differentiating between virus-infected and uninfected cells, thus extending the use of this technology to a novel application.
The mass spectrometry analysis applied to the study of viral structure and identification, although far from conclusive, has received increasing amounts of consensus, due to its high level of accuracy, rapidity and low operational costs7. Nevertheless, applications of MALDI-TOF MS for virus identification, in particular those with diagnostic purposes, couple this technique with PCR24, often resulting in very expensive procedures. On the other hand, other studies that have used the spectra obtained from infected cells with the aim of detecting specific viral peaks have neither compared them with those obtained with purified virions, nor made an accurate analysis of the uninfected cells protein peaks, thus rendering the results often unclear and inconclusive11.
In this study, the very efficient technique adopted to obtain highly purified poliovirus preparations combined with SDS-PAGE analysis allowed us to confirm the specificity of the peaks detected by MALDI-TOF MS analysis, confirming its ability to discriminate viral protein peaks from uninfected cells peaks and to detect specific poliovirus protein biomarkers.
Moreover, MALDI-TOF MS analysis applied to the three Sabin poliovirus serotypes revealed characteristic peak profiles for each of them, as demonstrated by statistical analysis, showing three independent clusters for the three serotypes, as expected considering that the genes coding for poliovirus capsid proteins widely diverged25.
Promising results were obtained by applying MALDI-TOF MS analysis to the identification of poliovirus strains isolated from different clinical samples by conventional techniques: the method has proven to give a correct identification of poliovirus even if not at the serotype level by using MALDI Biotyper software. This is probably due to the fact that in the three poliovirus Sabin reference serotypes spectra, in the mass range 2–20 KDa (the only exploitable for identification purposes), differences were revealed only for the VP4 capsid protein.
The results showed a significant match between the molecular weight of the VP4 protein of each of the reference poliovirus strains and the corresponding clinical isolates at the serotype level. Moreover, no significant match between the molecular weight of the VP4 protein of each of the reference poliovirus strains and human coxsackievirus B1 and human echovirus 9 was found. For this reason the VP4 protein may be considered a good viral biomarker to identify poliovirus strains at the serotype level.
One of the main advantages of this approach is that the identification of poliovirus strains by MALDI-TOF MS analysis can be obtained after a 5-days-procedure (starting from the cytopathic effect observation upon sample cultivation), significantly shortening the time needed to perform the gold standard neutralization test (about 20 days). This confirms the ability of MALDI-TOF MS platform of shortening significantly the times for conventional identification methods in some specific cases (in the case of polioviruses, the neutralization assay) and, moreover, the significant reduction of reagent costs and the need of experienced personnel.
Enzyme immunoassays and immunofluorescence assays for poliovirus identification have been described26,27, but not yet recommended for laboratory diagnosis, because the WHO gold standard is the neutralization assay.
Furthermore, as compared to the molecular methods used for poliovirus identification at the species and at the serotype level (such as reverse transcription and PCR [RT-PCR], Real Time RT-PCR and oligonucleotide microarray hybridization)28,29,30, MALDI-TOF MS is less cumbersome and cheaper, not requiring separate areas of manipulation of the samples in the different phases, preventing from risk of genome contamination not requiring expertise by technicians, and could be considered the best method for protein identification.
These results, though they need to be further refined, support the potential of MALDI-TOF MS technology for virus identification and provide a promising basis to extend its application to many other RNA and DNA viral agents of medical interest.
Methods
Cell cultivation
Confluent cultures of Rhesus monkey kidney epithelial cell line (LLC-MK2; American Type Cell Culture, ATCC CCL-7) were propagated in different lots of Earle's modified Minimum Essential Medium (E-MEM), supplemented with 2 mM L-glutamine, antibiotics (100 U/ml penicillin, 100 mg/ml streptomycin), and 10% (growth medium) or 1% (maintenance medium) fetal bovine serum (FBS). Culture media and supplements were from PAA (The cell culture company, Velizy-Villacoublay, France).
Reference viral strains
Sabin poliovirus type 1, 2 and 3 reference strains (PHLS, Central Public Health Laboratory Service, London, UK), human coxsackievirus B1 (ATCC VR28) and human echovirus 9 (ATCC VR1050) were cultivated and titrated in LLC-MK2 cells.
Poliovirus clinical isolates
Human poliovirus strains representative of the three serotypes isolated from five different biological samples, identified by neutralization assay according to standard procedures31 and stored at −80°C in our laboratory collection were included in the study: one poliovirus type 1 isolate (from cerebrospinal fluid), two poliovirus type 2 isolates (from stools and throat swab) and two poliovirus type 3 isolates (from stools and throat swab) strains, derived from patients with different clinical signs and symptoms referred to viral meningitis, gastroenteritis and pharyngitis.
Virus titration
For poliovirus titration, the 50% tissue culture infectious dose (TCID50), was calculated by using the Reed and Muench formula on the basis of microscopic observations (DFC 320, Leica, Milan, Italy) of the cytopathic effect in LLC-MK2 cell monolayers infected with the poliovirus strains (10-fold viral dilutions, from 10−2 to 10−9; 6 replicas/dilution using a 96 - well microtiter plate for each viral serotype31).
Virus cultivation
Sabin reference polioviruses, human coxsackievirus B1 and human echovirus 9 were cultivated in LLC-MK2 cells: viruses were inoculated at a multiplicity of infection (MOI) of 0.01 TCID50/cell in flasks with confluent cell monolayers. After 1 hour-absorption, the viral inoculum was removed and the cells re-fed with maintenance medium (E-MEM) without serum. Infected cells were incubated at 37°C in a CO2 humidified incubator for 48 h post infection (p.i.). Then the maintenance medium was collected in 50 ml conical sterile centrifuge tubes and stored at −80°C. The cells monolayers were re-fed with fresh E-MEM without serum and this step was repeated till the complete cytopathic effect had developed (usually at 96 h p.i.). Finally, the cells were scraped off the flasks, collected with the medium in 50 ml conical sterile centrifuge tubes and added to the previously frozen culture medium.
Poliovirus strains isolated from clinical samples and identified by neutralization assay were cultivated in LLC-MK2 cells. Specifically, each isolate was inoculated at a MOI of 5 TCID50/cell in cell monolayers grown in flasks and incubated at 37°C for 1 h in a CO2 humidified incubator. After 1 hour-absorption, the viral inoculum was removed, the flasks re-fed with maintenance E-MEM without serum and daily checked till a complete cytopathic effect was observed. Finally, the cells were scraped off the flasks and collected in 50 ml conical sterile centrifuge tubes with the medium.
Sabin poliovirus reference strains, poliovirus clinical isolates, and human coxsackievirus B1, human echovirus 9 strains concentration
After freezing and thawing and cell removing by centrifugation at 2,000 × g for 15 minutes (3,153 rpm, Centrifuge 5804 R, Eppendorf, Milan, Italy), the virus particles and residual cell debris obtained as described above were immediately ultracentrifuged at 154,300 × g for 70 minutes at 4°C (30,000 rpm; SW41 Ti Rotor Optima™ XPN, Beckman Coulter, Milan, Italy) and then subjected to protein extraction for the subsequent analysis by MALDI-TOF MS.
Sabin poliovirus reference strains purification by sucrose gradient
Poliovirus particles released from infected cells were obtained by a freezing-thawing cycle and cell removing by centrifugation at 2,000 × g for 15 minutes (3,153 rpm, Centrifuge 5804 R, Eppendorf, Milan, Italy). Ammonium sulfate (Sigma-Aldrich, Milan, Italy) was then added to the supernatant at a concentration of 40 g/100 ml, and the viral particles and the residual cell debris were pelleted by centrifugation at 3,000 × g (3,861 rpm, Centrifuge 5804 R, Eppendorf, Italy) for 2 hours at 4°C. The pellet was dissolved in 2 ml phosphate buffer saline (PBS solution) with 1% Nonidet P-40 detergent (Sigma-Aldrich, Milan, Italy) to “clean” the viral particles32.
Sucrose gradient solutions were prepared in 13.2 ml-Ultraclear tubes (Beckman Coulter, Milan, Italy) by using ultrapure analytical grade sucrose (Sigma-Aldrich, Milan, Italy) and sterile Tris-HCl 10 mM (pH 7.4), NaCl 0.05 M32.
Sterile solutions were prepared at a final concentration of a) 15% (w/v): sucrose 15 g per 100 ml Tris-HCl 10 mM (pH 7.4), NaCl 0.05 M, and b) 45% (w/v): sucrose 45 g per 100 ml Tris-HCl 10 mM (pH 7.4), NaCl 0.05 M, respectively. Gradients were prepared by using a peristaltic pump (Minipuls™ 3, Gilson, Middleton, USA) and a gradient maker (PBI, Milan, Italy).
The viral pellet, suspended in PBS as described above, was carefully layered onto the gradient tubes (1 ml of virus suspension/tube) and centrifuged at 103,700 × g using a SW41 Ti Rotor (24,600 rpm, Optima™ XPN, Beckman Coulter, Milan, Italy) for 4 hours at 4°C.
Sucrose gradient fractions were collected and evaluated for their protein content by spectrophotometric analysis (Beckman, DU® -65, Milan, Italy) at 750 nm, following Lowry assay33 performed by using a commercial kit (DC Protein Assay, Bio-Rad, Milan, Italy) according to the manufacturer's instructions. Finally, the fractions corresponding to the purified virions were pooled and centrifuged at 154,300 × g for 70 minutes at 4°C. The pellet was resuspended in 50 μl of E-MEM without serum and further analyzed by electron microscopy, as previously described34, before protein extraction.
Protein extraction and target plate preparation for MALDI-TOF MS analysis
A short protein extraction protocol was used. Each of the poliovirus preparations (purified Sabin reference strains; Sabin strains combined with residual infected cell debris; polioviruses from clinical samples), human coxsackievirus B1, and echovirus 9 strains together with residual infected cell debris were additioned with 30 μl 70% formic acid (HCOOH) (Sigma-Aldrich, Milan, Italy) by vigorous mixing, than with 30 μl 100% acetonitrile (CAN) (Sigma-Aldrich, Milan, Italy) and further vigorous mixing. This mixture was subjected to centrifugation at 16,100 × g for 2 minutes (13,200 rpm; Centrifuge 5415R, Eppendorf, Italy). MALDI-TOF MS analysis was performed both in a low (2–20 KDa) and high (17–90 KDa) molecular weight range by using a MicroFlex LT mass spectrometer (Bruker Daltonics, Germany supplied by Becton Dickinson, Italy) instrument. For the analysis in the low molecular weight range, 1 μl of the supernatant was spotted (20 spots for each strain) onto a MSP 96 polished steel target plate (Bruker Daltonics, Germany), air-dried at room temperature, and overlaid with 1 μl of matrix solution (alpha-cyano-4-hydroxycinnamic acid [4HCCA], diluted in 50% CAN and 2.5% trifluoroacetic acid [TFA], Sigma-Aldrich, Milan, Italy) followed by air-drying. For the analysis in the high molecular weight range, a saturated solution of synapinic acid (SA) was used as the matrix (50 mg/ml SA in 30:70 CAN:TFA acid 0.1%). Equal volumes (2 μl) of supernatant and matrix solution were mixed. One microliter of this solution was spotted onto the target plate (20 spots for each sample) and air-dried at room temperature.
SDS-PAGE and protein extraction from polyacrylamide gel
The methods used for SDS-PAGE and protein extraction were modified from those originally described by Susnea et al.5 and by Mirza et al.35, respectively.
Briefly, viral peptides from purified virions, treated with Laemmli buffer (50 mM Tris-HCl pH 6.8; 100 mM dithiothreitol; 25% (w/v) glycerol; 2% SDS; 0.1% bromophenol blue) and heated at 95°C for 5 minutes, were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with the following concentration conditions: 4% acrylamide/N,N bisacrylamide for the stacking gel and 20% for the running gel. A low range protein standard (Sigma-Aldrich, Milan, Italy) (MW 6,500–66,000) was used as the molecular weight marker.
Coomassie brilliant blue (Bio-Rad, Milan, Italy) gel staining was used according to Mirza et al.35.
For protein extraction, the individual bands were excised and put into 1.5 ml-colorless tubes. After washing with 300 μl of high purity liquid chromatography (HPLC) grade water, each excised gel band was vortexed for 10 minutes in 100 μl 10% acetic acid solution. The solution was removed and the gel piece washed thoroughly with: 300 μl HPLC grade water, then with 100 μl CAN, then again with HPLC grade water, and finally with methanol for 20 minutes. To destain the gel completely, each gel piece was dipped into a 100 μl solution of formic acid:HPLC grade water:isopropanol (1:3:2, v/v/v) and vortexed for 30 minutes (until the gel piece turned colourless). Finally, each gel piece was partially dried, crushed into small pieces, and the proteins were extracted by adding 10 μl 4HCCA, when used for MALDI-TOF MS analysis in the low molecular weight range and 10 μl of 50 mg/ml saturated SA solution when used for MALDI-TOF MS analysis in the high molecular weight range. Molecular weight marker bands in the range of interest (6,500 to 36,000 Da) were also excised and subjected to the same procedure.
MALDI-TOF MS instrumental settings
Spectra were created by performing MALDI-TOF MS analysis on protein extracts obtained from: purified Sabin poliovirus reference strains; virus-infected cells both with reference strains and poliovirus clinical isolates; uninfected cells; SDS-PAGE protein bands corresponding to viral peptides from purified virions and molecular weight marker bands. All the spectra were manually acquired by using a Microflex LT mass spectrometer and the FlexControl software (version 3.3.108.0) (Bruker Daltonics, Germany). In the 2–20 KDa range, the instrument parameters were set on the MBT-FC method (positive linear mode; laser frequency 60 Hz). To improve the quality of the spectra, each mass spectrum was generated from the data deriving from several single laser shots in 200-shot steps from different positions of the sample spot, acquiring only the spectra with an intensity ≥ 104 arbitrary units as suggested by the manufacturer. To optimize the experimental conditions, a solution of Bruker Bacterial Test Standard (BTS, Bruker Daltonics, Germany) (2–20 KDa range) was used to calibrate the system, according to manufacturer's instructions.
For the analysis of larger proteins, in the range of 17–90 KDa, the parameters used in the MALDI Biotyper Software package were set on the LP-44 kDa method. In this case, several single laser shots in 100-shot steps from different positions of the sample were used to generate the spectra with an intensity ≥ 104 arbitrary units. To optimize the experimental conditions, a solution of Protein Standard II (Bruker Daltonics, Germany) (20–90 KDa range) was used to calibrate the system, according to the manufacturer's instructions. Calibration was successful when the protein peaks of the standard were in the range of +/−300 ppm (parts per million) as compared to the respective expected molecular weight.
Statistical analysis by using ClinPro Tools software
The acquired spectra were processed with FlexAnalysis Software (Version 3.3, Bruker Daltonics, Germany). Data analysis began with raw data pre-treatment, including baseline substraction, normalization of a set of spectra, internal peak alignment using prominent peaks, and a peak picking procedure. Subsequently, the data derived from purified polioviruses (Sabin reference strains) were imported into ClinPro Tools Software version 2.2 (Bruker Daltonics, Germany) for statistical analysis. This software was used for the visual comparison of the loaded spectra, as well as for identifying specific peaks to discriminate among the three analyzed poliovirus serotypes.
To compare individual serotypes, the same number of spectra for each strain was needed to be analyzed by using ClinPro Tools. Two different analysis were performed for the low (2 to 20 KDa) and the high (17 to 90 KDa) molecular weight range. In order to validate each statistical analysis, a total of 30 spectra (specifically ten replicates for each of the investigated viral serotypes), obtained by the MBT-FC method (low molecular weight), and a total of 30 spectra (specifically ten replicates for each of the investigated viral serotypes) obtained by the LP-44 KDa method (high molecular weight), were uploaded into the software and automatically recalibrated.
As reported in the manufacturer's procedure, the task of recalibration is to reduce mass shifts occurred during the multiple measurement of the same sample obtained in a single or in different experiments. The recalibration was carried out with a specific algorithm already present in ClinPro Tools software. In this study the most important parameter (Maximum Peak Shift) is set on 1,000 ppm.
Moreover, as part of recalibration step, a list of masses which occur very frequently within the entire data set is generated. The number of these masses is called the Maximum Quality Value. After recalibration it is checked how many of these masses can be found in each spectrum using the Maximum Peak Shift parameter as maximum shift. The number of found masses is called the Spectrum Quality Value. The Spectrum Quality Threshold is computed as the product:
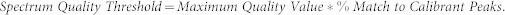 |
All spectra with a Spectrum Quality Value < Spectrum Quality Threshold are marked as “Not Recalibratable” and excluded. The value of the % Match to Calibrant Peaks parameter is set to 30%.
After the recalibration process, the software automatically created an average spectrum from all the replicates of each poliovirus serotype. The obtained average spectra were compared with each other and a report of peaks was available.
The data included in this peak report were used to assess statistical information. To give a significance value for differences in each m/z peaks ANOVA test was performed and a p-value was calculated. A p-value < 0.05 was considered significant (discriminating peak).
Statistical testing of the datasets was performed on the basis of multivariate unsupervised principal component analysis (PCA) and the results were displayed in a three-dimensional score plot, which was generated automatically by the software. PCA reduces the variables of the complex dataset, generating a set of new variables called the Principal Component (PC)36. The same spectra were analyzed by statistical supervised algorithm to find the peaks with the probably highest serotypes separation capability under a uni/multivariate view of the data. In this study 3 statistical algorithms included in the software are used: QuickClassifier (univariate supervised algorithm), Supervised Neural Network and Genetic Algorithm (multivariate supervised algorithms).
These algorithms automatically determined the best number of peaks to be integrated in the statistical model, in order to differentiate the different serotypes, on the basis of the Recognition Capability (RC) parameter. Moreover the algorithms evaluated the reliability of the created statistical model by the Cross Validation (CV) parameter; this parameter can be used also to predict how this model will behave in the future.
The algorithm with the highest score of RC and the highest value of CV, also taking into account the peaks number used to obtain the model, was chosen as a reference model in order to analyze the spectra of the clinical isolates. The presence/absence of each discriminating peak was evaluated by comparing the average spectra automatically created from all the replicates of each poliovirus serotype. Moreover, the discriminatory power for each putative biomarker in the model was further described via analysis of area under the receiver operating characteristic (ROC) curve (AUC) using ClinPro Tools software.
The ROC curve gives a graphical overview about the specificity and the sensitivity of a test, and in this case an evaluation of the discrimination quality of a peak. The ROC curve view takes into account only one peak as a test criterion and indicates this peak by an Area Under Curve (AUC) value. An AUC value of 0 indicates that the considered peak is not discriminating, while an AUC of 1 indicates that the considered peak is discriminating.
For each clinical isolate the ROC curves were generated for all the possible combinations of the peak representative for the VP4 protein (VP4-clinical isolates vs VP4-Sabin poliovirus type 1; VP4-clinical isolates vs VP4-Sabin poliovirus type 2; VP4-clinical isolates vs VP4-Sabin poliovirus type 3). The same analysis was performed also for the VP4 protein of coxsackievirus B1 and echovirus 9.
MALDI-TOF MS in-house viral database
For each of the three Sabin reference strains, the raw spectra acquired by MALDI-TOF MS (obtained from sucrose gradient-purified poliovirus, as well as from the poliovirus strains with residual infected cell debris), were analyzed by FlexAnalysis software to carry out “Smoothing” and “Baseline” and to select spectra with an intensity ≥ 104 arbitrary units. In order to create a viral database to be used for further application to clinical isolates, a minimum of ten spectra for each reference strain, was uploaded in MALDI BioTyper software (Version 3.1, Bruker Daltonics, Germany) to calculate in the range 2–20 KDa a reference Main Spectrum Profile (MSP) for each serotype by the automated function of BioTyper software (BioTyper MSP Creation Standard Methods), following the manufacturer's suggestions, as previously described6,10. MSPs were created by extracting information on peak position, intensity and frequency on the basis of an unbiased algorithm. Uninfected LLC-MK2 cells MSP spectra were included in the generated database, to verify the specificity of the tool.
Poliovirus identification by MALDI BioTyper software
The spectra of the poliovirus isolates from different clinical samples, manually acquired by using the MBT-FC method, were imported into MALDI BioTyper for the identification by the created in-house viral database. The MALDI BioTyper software, in the range of 2–20 KDa, compares each sample mass spectrum to the reference mass spectra present in the database using a pattern matching approach that is based on statistical multivariant analysis and takes into account peak position and intensity. The software calculates an arbitrary unit score value between 0 and 3, indicating the similarity between sample and reference spectra and finally displays the top ten matching database records. The results were displayed into the “Detected species” window as specified by the manufacturer with an identification score value ≥ 1.7 for a reliable identification. Scores < 1.7 were considered as unreliable.
Author Contributions
A.C., M.C.A. and C.C.: conceived and designed the experiments I.R., M.B., F.M. and D.G.: performed the experiments I.R. and M.B.: statistical analysis A.C., M.C.A., C.G., M.C.M., C.C. and F.D.C.: revised the data A.C., M.C.A., I.R., M.B., C.G. and C.C.: wrote the paper A.C. and C.C.: supervised the project A.C.: provided grants, samples and reagents.
Supplementary Material
Supplementary Information
Acknowledgments
This study was supported by the Ministry of University and Scientific Research Grant FIL, Parma, Italy, and by the grant “The Biobank of microorganisms and viruses pathogenic to humans as a starting point for the study of the infectious diseases and zoonoses” financed by the Bureau of the Council of Ministers - Italian National Committee for Biosafety, Biotechnology and Life Sciences.
References
- Welker M. & Moore E. R. B. Application of whole-cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst. Appl. Microbiol. 34, 2–11 (2011). [DOI] [PubMed] [Google Scholar]
- Fenselau C. & Demirev P. A. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20, 157–171 (2001). [DOI] [PubMed] [Google Scholar]
- Lavigne J. P. et al. Mass spectrometry: a revolution in clinical microbiology? Clin. Chem. Lab. Med. 51, 257–270 (2013). [DOI] [PubMed] [Google Scholar]
- Wieser A., Schneider L., Jung J. & Schubert S. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review). Appl. Microbiol. Biotechnol. 93, 965–974 (2012). [DOI] [PubMed] [Google Scholar]
- Susnea I. et al. Application of MALDI-TOF-Mass Spectrometry to Proteome Analysis Using Stain-Free Gel Electrophoresis. Top. Curr. Chem. 331, 37–54 (2013). [DOI] [PubMed] [Google Scholar]
- Calderaro A. et al. Identification of Borrelia species after creation of an in-house MALDI TOF database. PLoS One 9, e88895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzini A. & Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infec. 16, 1614–1619 (2010). [DOI] [PubMed] [Google Scholar]
- Croxatto A., Prod'hom G. & Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 36, 380–407 (2012). [DOI] [PubMed] [Google Scholar]
- Cobo F. Application of MALDI-TOF Mass Spectrometry in Clinical Virology: A Review. Open Virol. J. 7, 84–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. et al. Proteomic profiling of hepatitis B virus-related hepatocellular carcinoma with magnetic bead-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Acta Biochim. Biophys. Sin. 43, 542–550 (2011). [DOI] [PubMed] [Google Scholar]
- Erukhimovitch V., Karpasasa M. & Huleihel M. Spectroscopic detection and identification of infected cells with herpes viruses. Biopolymers 91, 61–67 (2009). [DOI] [PubMed] [Google Scholar]
- Chou T. C., Hsu W., Wang C. H., Chen Y. J. & Fang J. M. Rapid and specific influenza virus detection by functionalized magnetic nanoparticles and mass spectrometry. J. Nanotechnol. 9, 52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderaro A. et al. MALDI-TOF MS analysis of human and animal Brachyspira species and benefits of database extension. J. Proteom. 78, 273–280 (2013). [DOI] [PubMed] [Google Scholar]
- Storch G. A. Diagnostic Virology. Clin. Inf. Dis. 31, 739–751 (2000). [DOI] [PubMed] [Google Scholar]
- Piqueur M. A., Verstrepen W. A., Bruynseels P. & Mertens An. H. Improvement of a real time RT-PCR assay for the detection of enterovirus RNA. J. Virol. 6, 1–3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles N. J. et al. Picornaviridae. In: Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committe on Taxonomy of Viruses. Ed: King, A. M. Q.,Adams,M. J., Carsten, E. B. & Lefkowitz, E. J. 855–880 (Elsevier, San Diego, USA, 2012). [Google Scholar]
- Paul A. V., Schultzt A., Pincus S. E., Oroszlant S. & Wimmer E. Capsid protein VP4 of poliovirus is N-myristoylated (protein modification/fatty acylation of proteins/picornaviruses). Proc. Natl. Acad. Sci. USA 84, 7827–7831 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassly N. C. The final stages of the global eradication of poliomyelitis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 368 (1623), 20120140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew O. Reaching the last one percent: progress and challenges in global polio eradication. Curr. Opin. Virol. 2, 188–198 (2012). [DOI] [PubMed] [Google Scholar]
- Filman D. J. et al. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. Embo J. 8, 1567–1579 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Tomas C. B. & Baltimore D. Morphogenesis of poliovirus II. Demonstration of a new intermediate, the proviron. J. Virol. 12, 1122–1130 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscufo N., Yafal A. G., Rogove A., Hogle J. & Chow M. A mutation in VP4 defines a new step in the late stages of cell entry by poliovirus. J. Virol. 67, 5075–5078 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegers K. J. & Dernick R. Poliovirus-specific polypeptides in infected Hela cells analysed by isoelectric focusing and 2D-analysis. J. Gen. Virol. 52, 61–69 (1981). [DOI] [PubMed] [Google Scholar]
- Sjöholm M. I. L., Dillner J. & Carlson J. Multiplex Detection of Human Herpesviruses from Archival Specimens by Using Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol. 46, 540–545 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L. I., Tolskaya E. A. & Agol V. I. Antigenic and Structural Relatedness among non-Capsid and Capsid Polypeptides of Polioviruses belonging to Different Serotypes. J. Gen. Virol. 52, 279–289 (1981). [DOI] [PubMed] [Google Scholar]
- Payment P., Tremblay C. & Trudel M. Rapid identification and serotyping of poliovirus isolates by an immunoassay. J. Virol. Methods 5, 301–308 (1982). [DOI] [PubMed] [Google Scholar]
- Rigonan A. S., Mann L. & Chonmaitree T. Use of monoclonal antibodies to identify serotypes of enterovirus isolates. J. Clin. Microbiol. 36, 1877–1881 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham R. et al. Rapid detection of poliovirus by reverse transcription and polymerase chain amplification: application for differentiation between poliovirus and non poliovirus enteroviruses. J. Clin. Microbiol. 31, 395–399 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laassri M. et al. Genomic analysis of vaccine-derived poliovirus strains in stool specimens by combination of full-length PCR and oligonucleotide microarray hybridization. J. Clin. Microbiol. 43, 2886–2894 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick D. R. et al. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by Real-Time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J. Clin. Microbiol. 47, 1939–1941 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. D., Dulbecco R., Eisen H. N. & Ginsberg H. Microbiology. 4th Ed.Lippincott Williams and Wilkins (Philadelphia, USA, 1990). [Google Scholar]
- Minor P. D. Comparative Biochemical Studies of Type 3 Poliovirus. J. Virol. 34, 73–84 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L. & Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951). [PubMed] [Google Scholar]
- Arcangeletti M. C. et al. Electron microscopy as a reliable tool for rapid and conventional detection of enteric viral agents:a five-year experience report. Acta Biomed. 76, 165–170. (2005). [PubMed] [Google Scholar]
- Mirza U. A. et al. Extraction and characterization of adenovirus proteins from sodium dodecylsulfate polyacrylamide gel electrophoresis by matrix-assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 11, 356–361 (2000). [DOI] [PubMed] [Google Scholar]
- Joliffe I. T. Principal Component Analysis. 2nd edition (Springer-Verlag New York) (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information