Abstract
BioImage Suite is an NIH-supported medical image analysis software suite developed at Yale. It leverages both the Visualization Toolkit (VTK) and the Insight Toolkit (ITK) and it includes many additional algorithms for image analysis especially in the areas of segmentation, registration, diffusion weighted image processing and fMRI analysis. BioImage Suite has a user-friendly user interface developed in the Tcl scripting language. A final beta version is freely available for download 1
1 Introduction
Biomedical imaging and image analysis technology have made substantial progress over the last 20 years. However, a major bottleneck that blocks the full use of these techniques is still the relative lack of easy-to-use, readily-available software. The key reason for this problem is that while sophisticated image analysis methods are constantly being developed, these methods are not being translated into user-friendly well documented software packages. Part of the reason, in our experience, is that software development obeys the 80/20 rule which suggests that 80% of the work is done in 20% of the time. While 80% of the package makes the methods usable in the hands of its developers and perhaps closely supported collaborators, it is insufficient for making the software truly available to a wider audience.
Over the last ten years, we have developed a body of software collectively known as BioImage Suite as a result of the needs of different projects in structural [3] and functional neuroimaging, cardiac image analysis[13], image guided epilepsy neurosurgery [18], abdominal fat quantification [23], targeted imaging of angiogenesis and diffusion tensor imaging [3]. BioImage Suite is in wide use in the Yale Image Analysis and Processing Group, the Yale Magnetic Resonance Research Center, the laboratories of local close collaborators, and is freely available for download. While the software development has focused on the analysis of human images, it has also been applied to mouse CT and MRI and to a lesser extent optical microscopy data.
There are a number of software packages available for medical image analysis that provide some overlapping capabilities; some are commercial but most are research-based. The neuroimaging packages such as Slicer [19], Brainsuite [17] and SPM [20] are the best developed. AIR [1] is limited to image registration. The general purpose commercial software package Analyze [2] has strengths in visualization, segmentation, registration and measurement.
2 A Brief Description
BioImage Suite is developed using a combination of C++ and Tcl in the same fashion as that pioneered by VTK [15]. In practice, most of the underlying computationally expensive algorithms are implemented in C++ (as classes deriving from related VTK or ITK [5] classes) and the user interface is for the most part developed in the Tcl/Tk scripting environment (using the [Incr Tcl/Tk ] and Iwidgets extensions. Further, a custom written C++ wrapper around the Tk graphical user interface library enables the creation of complex graphical components from C++. BioImage Suite is a collection of programs which utilize the same underlying code infrastructure (a rough count using the UNIX ‘wc’ command gives an estimate of about 300,000 lines of code – naturally this is an overestimate as it includes blank lines, comments etc.) and provide the same look and feel but are tuned to specific applications. This modular design encourages the creation of re-usable components (e.g. viewers). The current version of BioImage Suite consists of (a) a number of a graphical applications (GUI) and (b) a set of command-line utilities thus providing support for and batch-mode processing. All software has been compiled and tested primarily on Linux and MS-Windows, and to a lesser extend on Mac OS X and a variety of other UNIX platforms.
Algorithms currently in BioImage Suite
In addition to the extensive set of methods available within the above libraries, we have implemented and customized the following algorithms: Pre-processing: A custom reimplementation of the bias field correction method of Styner et al. [22] has been performed which incorporates automated histogram fitting for determining the appropriate numbers of classes, and additional spatial constraints. Voxel Classification: Methods for voxel classification are available using simple histogram, single channel MRF and exponential-fit methods [12]. Deformable Surface Segmentation: BioImage Suite has a strong and unique interactive deformable surface editor tool [13] which allows for easy semi-interactive segmentation of different anatomical structures and embeds common snake-like deformable models. Registration: BioImage suite includes a clean reimplementation of the work of Studholme et al. [21] for rigid/affine registration using a highly efficient conjugate gradient optimization scheme. These methods have been successfully used to align serial MRI data as well as multimodal data (e.g. CT/PET/SPECT to MRI). It also includes a full complement of non-rigid point-based registration methods [3], intensity-only and integrated feature intensity methods [10]. Diffusion Weighted MR Imaging: BioImage suite has methods for computation and visualization of basic voxel-wise measures from diffusion tensor images (e.g. fractional anisotropy) as well as fiber tracking methods using traditional (streamlining) and novel (anisotropic front propagation) methods [3]. Cardiac Image Analysis: The shape-based cardiac deformation method of Papademetris et al. [13] is included in BioImage Suite (but requires in addition the presence of the Abaqus finite element package [6]). fMRI Activation Detection: BioImage Suite has a clean and fast reimplementation of the standard General Linear Model (GLM) [4] method for fMRI activation detection, in addition to tools for performing region of interest analysis (ROI), multisubject composite maps, etc. The registration tools (described above) can be used for motion correction, distortion correction and intra-subject registration.
Key Components
An illustration of several key components of BioImage Suite are shown in Figure 1. These include the two core viewers (the mosaic multiple slice viewer and the orthogonal viewer); the landmark tool which includes common functionality for drawing and processing curves on any given slice (or on volume renderings and surfaces) as well as simple ROI analysis; the surface processing tool which includes functions for surface decimation and smoothing, marching cubes iso-contour extraction etc. and the colormap editor for manipulating how images are displayed. In addition there are common controls grouping related algorithms such as the histogram control, the image processing control and the segmentation/bias field correction control also shown in Figure 1. These components are building blocks for creating application specific programs.
Figure 1. Example Views of BioImage Suite.
A Initial menu. B The mosaic viewer capable of displaying multiple parallel slices in any orthogonal orientation. C1–C5 The multi-functional viewer tool displaying G orthogonal sections with linked cursors (C1), volume rendering (C2), single slice (C3) combination of 3D and slice views (C4), oblique slice (C5). D Histogram Tool. E Image Processing Tool. F Colormap editor. G Landmark Tool. H Surface Processing Tool. I Segmentation/Bias Field Correction Tool. J Basic DTI Tool.
Graphical Applications
The components are linked together to yield several individual applications including the Brainsegment and Brainregister tools for brain segmentation and registration. The latter also includes the multisubject compositing tool for creating composite functional maps, and the distortion correction algorithms for fMRI. The Surfaceeditor tool is for manual/interactive segmentation of both brain, cardiac and abdominal image data. In surfaceeditor, each surface consists of a stack of curves (one per slice) and each curve is parameterized using a non-uniform b-spline. The user is able to edit the b-spline in a curve editor window and have the surface updated interactively. These techniques enable the generation of surfaces which are smooth in all three orthogonal directions, unlike the more common slice-by-slice editing packages which typically display the result in only one orientation. The ElectrodeEditor tool is used for intra-cranial electrode localization in epilepsy neurosurgery planning. The fMRITool includes both individual and group fMRI activation analysis methods. The DTITool is a diffusion weighted MR image analysis package and Vessel is a tool for vessel extraction from angiography images. A recent addition is the DataTree tool which provides a database-like interface to large datasets, and is especially useful in a surgical planning context. All of these programs have the same look and feel as they use much of the same underlying components.
Command Line Tools
All of the registration tools in BioImage Suite, plus some support utilities, can be accessed through command line programs, in addition to being available in the Brainregister graphical tool. We provide these because: (a) they enable access to the registration algorithms from custom software packages already in use in other laboratories and (b) given that nonlinear registration is a computationally intensive task, the command line tools are more easily used for batch mode processing. A custom batch mode registration generation script was also developed and takes as input a well defined setup file consisting of reference and target images/surfaces. The batch file generates as output a standard Unix makefile consisting of all the commands that need to be executed for registration with appropriately defined dependencies. Using the make utility enables us to leverage its flexibility for partitioning jobs into multiple processors (using the -j flag) and recovery after system crashes (the dependency mechanism of the makefile enables the process to restart exactly where it was stopped as a result of a system crash, etc.).
Surgical Environment Integration
BioImage Suite can integrate with the BrainLAB VectorVision Cranial image-guided neurosurgery system via VectorVison Link (VVLink), a custom designed client/server architecture [14, 9], which extends functionality from the Visualization Toolkit. VVLink enables bi-directional data transfer of image data sets and visualizations and tool positions in real time. VV Link was designed explicitly with BioImage Suite as it’s target client, however the interface used is generic and other applications can take advantage of it.
3 Availability, Ongoing work and Future Plans
Availability
BioImage Suite is freely available for download in a final beta version from the webpage www.bioimagesuite.org. Binary versions are available for:
MS-Windows
Linux (3 versions gcc2.96 – for Redhat 7.x and gcc3.2/gcc3.3 for more modern distributions).
Mac OS X 10.4 (powerpc) (both aqua and X11 versions).
Mac OS X 10.4 (intel) (x11 version only – compiled in OpenDarwin under VMware!)
Other Unix versions: Solaris 10.0 (both x86 and sparc), IRIX 6.5 and Free BSD 6.0, which are updated less frequently.
The binary distributions include precompiled binary versions of VTK 4.4, ITK 2.4.1 and Tcl/Tk 8.4.11, and additional software – see the webpage for more details. We utilize extensively virtual machines running under VMware Server [16] for development and testing on different operating systems. In addition, the BioImage Suite subversion repository is hosted in a virtual machine to make it more easily migratable to a different server, should the need arise.
The next release candidate version will be made available in early July 2006. We anticipate a first official release under the GNU General Public License a few weeks later, at which point source code will also be made publicly available. Support for users is available at the BioImage Suite forum (accessible from the main webpage). An early version of the BioImage Suite manual is also available; completion of the manual (aside from final testing) is the major task remaining prior to the first ull official relase.
Open Source Licensing Details
Most of the code in BioImage Suite will be made available under the GPL. A single core library will remain close source for now as it contains (i) propertiary code needed for Vector Vision Link integration (ii) code derived from Numerical Recipes which may not be released as open source and (iii) some code derived from the work of researchers who have not yet released their work under an open source license. This library consists of less than 5% of all code in BioImage Suite and practically all the interesting/useful pieces of code in the software will be made freely available. Eventually we hope to eliminate the closed source library requirement, however it is currently the best compromise solution that enables us to freely distribute the software.
Future Plans
We are currently working on the implementation of a database front-end to BioImage Suite – the Data Tree Manager [8] is the first version of the user interface for this. Ongoing work also aims to include additional algorithms in the BioImage Suite core as was discussed in [11].
Figure 2. Example Visualizations from Applications of BioImage Suite.
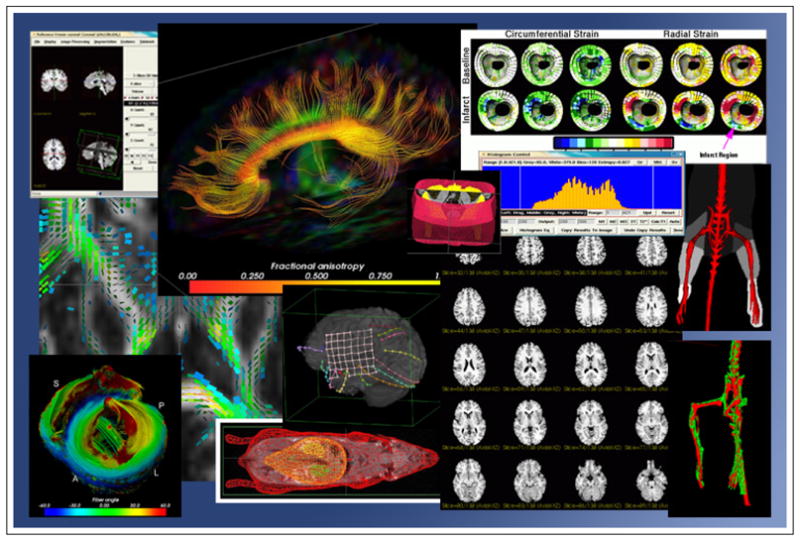
Acknowledgments
BioImage Suite is supported by the National Institutes of Health (NIH)/National Institute of Biomedical Imaging and Bioengineering (NIBIB) under grant 1 R01 EB006494-01 (Papademetris X. PI). Initial work on the Datatreee tool and its application to neurosurgery was also supported by NIH/NIBIB under grant R01 ED00473-01 (BRP) (Duncan J.S. PI).
Footnotes
This paper represents an updated version of [11].
References
- 1.Automated Image Registration (AIR) http://bishopw.loni.ucla.edu/air5/. 1.
- 2.Analyze. http://www.analyzedirect.com/. 1.
- 3.Duncan J, Papademetris X, Yang J, Jackowski M, et al. Geometric strategies for neuroanatomical analysis from MRI. NeuroImage. 2004;23:S34–S45. doi: 10.1016/j.neuroimage.2004.07.027. 1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friston KJ, Jezzard P, Turner R. The analysis of functional MRI time-series. Human Brain Mapping. 1994;1:153–171. 2. [Google Scholar]
- 5.Ibanez L, Schroeder W. The ITK Software Guide: The Insight Segmentation and Registration Toolkit. Kitware, Inc; Albany, NY: 2003. www.itk.org. 2. [Google Scholar]
- 6.Hibbit, Karlsson, Sorencen . Abaqus Version 6. 3. Rhode Island, USA: 2002. 2. [Google Scholar]
- 7.Kass M, Witkin A, Terzopoulos D. Proc Int Conf on Computer Vision. 1987. Snakes: Active contour models; pp. 259–268. [Google Scholar]
- 8.The Data Tree Manager Manual. http://bioimagesuite.org/public/datatree.html. 3.
- 9.Neff Markus. Master’s thesis. Technical University of Munich; 2003. Design and implementation of an interface facilitating data exchange between an igs system and external image processing software. This project was jointly performed at BrainLAB AG (Munich, Germany) and Yale University (New Haven, CT U.S.A). 2. [Google Scholar]
- 10.Papademetris X, Jackowski A, Schultz RT, Staib LH, Duncan JS. MICCAI. 2004. Integrated intensity and point-feature nonrigid registration. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papademetris X, Jackowski M, Rajeevan N, Constable RT, Staib LH. Bioimage suite: An integrated medical image analysis suite. The Insight Journal. 2005. http://hdl.handle.net/1926/37. 1, 3. [PMC free article] [PubMed]
- 12.Papademetris X, Shkarin P, Staib LH, Behar KL. MRI-based whole body fat quantification in mice. IPMI. 2005 doi: 10.1007/11505730_31. 2. [DOI] [PubMed] [Google Scholar]
- 13.Papademetris X, Sinusas AJ, Dione DP, Constable RT, Duncan JS. Estimation of 3D left ventricular deformation from medical images using biomechanical models. IEEE Trans Med Imag. 2002 Jul;21(7) doi: 10.1109/TMI.2002.801163. 1, 2. [DOI] [PubMed] [Google Scholar]
- 14.Papademetris X, Vives KP, DiStasio M, Staib LH, Neff M, Flossman S, Frielinghaus N, Zaveri H, Novotny EJ, Blumenfeld H, Constable RT, Hetherington HP, Duckrow RB, Spencer SS, Spencer DD, Duncan JS. Development of a research interface for image guided intervention: Initial application to epilepsy neurosurgery. International Symposium on Biomedical Imaging ISBI. 2006:490–493. 2. [Google Scholar]
- 15.Schroeder W, Martin K, Lorensen B. The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics. Kitware, Inc; Albany, NY: 2003. www.vtk.org. 2. [Google Scholar]
- 16.VWware Server. http://www.vmware.com/products/server/. 3.
- 17.Shattuck DW, Leahy RM. Brainsuite: An automated cortical surface identification tool. MICCAI. 2000:50–61. doi: 10.1016/s1361-8415(02)00054-3. 1. [DOI] [PubMed] [Google Scholar]
- 18.Skrinjar O, Nabavi A, Duncan JS. Model-driven brain shift compensation. Medical Image Analysis. 2002 Dec;6(4):361–373. doi: 10.1016/s1361-8415(02)00062-2. 1. [DOI] [PubMed] [Google Scholar]
- 19.3D Slicer. http://www.slicer.org. 1.
- 20.Statistical Parametric Mapping (SPM) http://www.fil.ion.ucl.ac.uk/spm/. 1.
- 21.Studholme C, Hill D, Hawkes D. Automated 3D registration of magnetic resonance and positron emission tomography brain images by multiresolution optimisation of voxel similarity measures. Med Phys. 1997;24(1):25–35. doi: 10.1118/1.598130. 2. [DOI] [PubMed] [Google Scholar]
- 22.Styner M, Brechbuhler C, et al. Parametric estimate of intensity inhomogeneities applied to MRI. IEEE Trans Med Imag. 2000:153–165. doi: 10.1109/42.845174. 2. [DOI] [PubMed] [Google Scholar]
- 23.Weiss R, Taksali SE, Dufour S, Yeckel CW, Papademetris X, Kline G, et al. The “Obese Insulin Sensitive adolescent” – importance of adiponectin and lipid partitioning. J Clin Endocrinol Metab. 2005 Mar; doi: 10.1210/jc.2004-2305. 1. [DOI] [PubMed] [Google Scholar]



