Abstract
Registration plays an important role in group analysis of diffusion-weighted imaging (DWI) data. It can be used to build a reference anatomy for investigating structural variation or tracking changes in white matter. Unlike traditional scalar image registration where spatial alignment is the only focus, registration of DWI data requires both spatial alignment of structures and reorientation of local signal profiles. As such, DWI registration is much more complex and challenging than scalar image registration. Although a variety of algorithms has been proposed to tackle the problem, most of them are restricted by the zdiffusion model used for registration, making it difficult to fit to the registered data a different model. In this paper we describe a method that allows any diffusion model to be fitted after registration for subsequent multifaceted analysis. This is achieved by directly aligning DWI data using a large deformation diffeomorphic registration framework. Our algorithm seeks the optimal coordinate mapping by simultaneously considering structural alignment, local signal profile reorientation, and deformation regularization. Our algorithm also incorporates a multi-kernel strategy to concurrently register anatomical structures at different scales. We demonstrate the efficacy of our approach using in vivo data and report detailed qualitative and quantitative results in comparison with several different registration strategies.
Keywords: Diffeomorphism, Diffusion-weighted imaging, Image registration, Signal profile reorientation, Explicit orientation optimization
1. Introduction
Diffusion-weighted imaging (DWI) is widely used to non-invasively study white matter microstructure and fiber tracts in the human brain. The information provided by DWI is helpful for identifying pathological damages associated with brain diseases (e.g., stroke Schaefer et al., 2000, Alzheimer’s disease Hanyu et al., 1998; Wee et al., 2011, 2012; Zhang et al., 2013, and schizophrenia Shi et al., 2012) and brain changes associated with normal development (Yap et al., 2011).
To quantify white matter changes, a common space is required where images of patients and healthy controls can be spatially normalized and compared. Image registration is used to build such space and to spatially normalize the images by warping them to the space.
Traditional scalar image registration techniques are not directly applicable to diffusion-weighted images. When diffusion-weighted images corresponding to different diffusion gradient directions are put together, each voxel location encodes a vector-valued signal profile that provides information on the segment of the fiber bundle that traverses the voxel. As such, registration of diffusion-weighted images requires not only the spatial alignment of anatomical structures, as in scalar image registration, but also the reorientation of signal profiles with respect to the surrounding anatomical structures, which is not considered in scalar image registration. DWI registration is thus much more complicated and challenging than scalar image registration.
A common approach to registering diffusion-weighted images is to fit some diffusion model to the images to estimate angular quantities, such as orientation distribution functions (ODFs), and then incorporate such information into a registration algorithm for structural alignment. There are a number of choices of diffusion models as well as registration algorithms, leading to a variety of DWI registration methods.
Early work uses the relatively simple diffusion tensor model (Alexander et al., 2001; Cao et al., 2006; Yeo et al., 2009, 2010; Zhang et al., 2006). Alexander et al. (2001) introduced the preservation of principal direction (PPD) algorithm for the reorientation of diffusion tensors during image alignment. Instead of PPD, Yeo et al. (2009) used a finite strain reorientation strategy (Alexander et al., 2001) together with a diffeomorphic demons algorithm (Vercauteren et al., 2009) for registration. Zhang et al. (2006) broke down the image into uniform regions and estimated an affine transformation for each region by explicitly optimizing tensor orientation within that region. Cao et al. (2006) proposed a large deformation diffeomorphic metric mapping (LDDMM) algorithm (Beg et al., 2005) to tackle large-deformation non-linear registration of directional vector fields.
However, the diffusion tensor model can only characterize one principal fiber direction at each voxel and thus is unable to handle complex fiber configurations such as crossings. It has been found that at least one third of voxels in white matter have complex fiber configurations (Behrens et al., 2007). Obviously, failure to reorient the signal profiles in those voxels will lead to misalignments of microstructure.
To deal with crossing fibers, a number of researchers (Geng et al., 2011; Du et al., 2012; Hong et al., 2009; Raffelt et al., 2011; Yap et al., 2011; Dhollander et al., 2011; Zhang et al., 2012) attempted to use more complicated diffusion models. Geng et al. (2011) aligned ODFs represented by spherical harmonics (SHs) using an elastic registration algorithm. Du et al. (2012) integrated a similarity metric for the ODFs, which is defined in a Riemannian manifold, into a variant of LDDMM algorithm (Glaunès et al., 2008). Yap et al. (2011) extracted coarse-to-fine features from the ODFs for hierarchically refined alignment. Instead of using ODFs, Hong et al. (2009) performed registration with the help of T2-weighted images and subsequently reoriented the fiber orientation distribution (FOD). Raffelt et al. (2011) registered DWI data by mapping the FODs through a subject-template-symmetric diffeomorphic framework.
However, the aligned data generated by the above approaches are not in the form of diffusion-weighted images. The ability to produce diffusion-weighted images as final registration outcome is important for common-space analysis using diffusion models without well-defined warping and reorientation methods.
To overcome this problem recent studies propose to register DWI data directly in the Q-space (Dhollander et al., 2011; Zhang et al., 2012). Dhollander et al. (2011) tackled the problem by virtue of an SH-based reorientation algorithm together with a diffeomorphic demons algorithm (Vercauteren et al., 2009). We (Zhang et al., 2012) achieved a similar goal by using a set of diffusion basis functions (DBFs) (Yap and Shen, 2012) and a geodesic shooting algorithm simplified proposed by Ashburner and Friston (2011). Both methods regard spatial alignment and local signal profile reorientation as two separate components, and perform optimization by alternating between (i) computing the spatial mapping without considering reorientation, and (ii) reorienting the data using the resulting mapping. Although this strategy is simple, it ignores the crucial role reorientation plays in correspondence establishment.
As shown by Yeo et al. (2009), a better but more complicated strategy is to integrate the two components into a single cost function and explicitly take into account reorientation during registration. In this paper we describe a method that is able to register DWI data in the Q-space in a single framework where image matching, data reorientation, and deformation regularization are considered simultaneously. Part of this work has been reported in our recently published conference paper (Zhang et al., 2013). Herein, we provide additional derivations, implementation details, and experimental results that are not available in the conference version. Compared with the conference paper, this paper uses a more general symmetric tensor model, instead of Watson distribution, as the DBFs (Yap and Shen, 2012). In addition, the cost function is reformulated such that the solution satisfies the Euler–Lagrange equation.
2. Outline of the approach
Our method consists of two components: (1) DWI data reorientation (Section 3) and (2) an LDDMM-based registration algorithm (Section 4). The first component achieves reorientation in the Q-space while the second one provides a registration framework where alignment and reorientation are considered simultaneously.
The first component is realized based on the work of Yap and Shen (2012), where reorientation is achieved by three steps: (i) decomposing the diffusion signal profile into a set of weighted DBFs; (ii) reorienting each DBF independently using a local transformation; (iii) recomposing the reoriented DBFs to obtain the desired profile. Compared with the SH-based reorientation scheme as used by Dhollander et al. (2011), this strategy avoids the computational complexity of SHs as well as the loss of sharp directional information when the maximum order of the SH basis functions is insufficient (see Yap and Shen (2012) for detailed discussion).
The second component involves the LDDMM algorithm (Beg et al., 2005). Based on the spatial mapping estimated by the LDDMM algorithm, a Jacobian matrix can be computed at each voxel location and used for DBF reorientation. The interaction between the two components is mathematically expressed as a single cost function (Section 4) and, during optimization, spatial alignment and local reorientation are considered simultaneously.
To simultaneously register anatomical structures at different scales we use a multi-kernel strategy (Risser et al., 2011). This is to introduce a natural multi-resolution property to our registration algorithm and to provide an intuitive way of parameter tuning based on the desired scales that should be captured by the registration. Details are given in Section 5.1.
This work has three major contributions. First, we propose a non-rigid registration algorithm for direct registration of DWI data. This allows any diffusion model to be fitted to the aligned data for subsequent multifaceted analysis. Second, we incorporated spatial alignment and local reorientation into a single cost function. In contrast to the works of Dhollander et al. (2011) and Hsu et al. (2012), our method does not rely on multi-shell data, which require long acquisition time. Last but not least, we derive the gradient of the cost function and describe in detail the numerical implementation.
3. Reorientation of DWI data
We now briefly review the major concepts involved in reorientation using DBFs (Yap and Shen, 2012).
3.1. Decomposition of signal profile
Let S(qi) be the diffusion signal measured in direction qi (i = 1, …, M). It can be represented by a set of N DBFs:
where f(qi∣λ1, λ2, μj) is the j-th DBF, wj is the associated weight, and f0 is a constant component representing isotropic diffusion. Specifically, the j-th DBF is defined by
| (1) |
where b is the diffusion weighting and is a symmetric diffusion tensor. λ1 and λ2 control the shape of the tensor, {μj} is a pre-defined set of tensor principal directions and I is an identity matrix representing an isotropic tensor. We generated {μj} via spherical tessellation by subdividing the faces of an icosahedron.
If λ1 ≫ λ2, Dj can be approximated by . Then, we have
which is essentially the p.d.f. of the Watson distribution (Jupp and Mardia, 1989). This simplified model has been used in our previous work (Zhang et al., 2012; Zhang et al., 2013). In this work we use the more general model (1).
Let S be the signal vector, then we have S = Fw, where S = [S(q1), S(q2), …, S(qM)]T, w = [w0, w1, …, wN]T, and
Since typically, M < N + 1, this is a set of under-determined linear equations, which can be solved by an L1-regularized least-squares solver with a non-negative constraint (see Yap and Shen (2012) for details).
3.2. Reorientation of signal profile
At each voxel, a local affine transformation A is used to reorient the directions of the DBFs, i.e. . A is estimated locally from a typically non-linear mapping and hence varies spatially. The reoriented DBF matrix, F′, is calculated based on as follows
The orientation-rectified signal profile is then computed as S′ = F′ w. Note that the isotropic component is not reoriented.
4. LDDMM-based DWI registration
LDDMM techniques (Beg et al., 2005; Miller et al., 2006; Vialard et al., 2012; Ashburner and Friston, 2011) are designed for diffeomorphic registration when the object of interest undergoes large shape variation. Initially formulated for scalar image registration (Beg et al., 2005), it was later extended for registering vector fields (Cao et al., 2005). In this work, we further extend it for DWI registration.
Let I0 be the source image and I1 be the target image. The goal of the LDDMM algorithm is to minimize the following cost function
| (2) |
where υt is a time-dependent velocity field that needs to be estimated, σ > 0 is a regularization constant, and ϕs,t is a mapping induced by υt, transforming a voxel from its position at time s to its position at time t (e.g., ϕ1,0(x) transforms a voxel at position x at time 1 to its position y = ϕ1,0(x) at time 0). , where V is a Hilbert space in which the velocity field resides, i.e. υ ∈ L2([0, 1], V) and L is a proper differential operator which, when appropriately chosen, guarantees a diffeomorphic solution (Beg et al., 2005). Instead of realizing L directly, diffeomorphism can be achieved by defining a smoothing kernel K = (L†L)−1. Here we use a multi-Gaussian kernel scheme (Risser et al., 2011) for simultaneous multi-scale registration.
Since our focus is on DWI registration, we further assume that I is a vector-valued image representing diffusion signal vector S at each position x, i.e. I(x) ≡ S(x). We denote the i-th element of I(x) as Ii(x). We define the action of a mapping ϕ on I as
| (3) |
where W is a vector-valued weight image associated with I, containing a sparse weight vector w at each x, i.e. W(x) ≡ w(x), and Fϕ(x) is a matrix-valued image containing the reoriented DBFs, with the voxel at x given by
where D is the Jacobian operator. From (3), we can see that ϕ spatially transforms the sparse weights and reorients the DBFs via Fϕ(x). W is computed by fitting the DBFs to the DWI data as described in Section 3.
To reflect reorientation, we rewrite the cost function (2) as
| (4) |
where W0 is the weight image associated with I0. By computing the variation of E(υ) w.r.t. υ, the gradient of (4) can be obtained as
| (5) |
where
Ω is a bounded domain in ℝd and x ∈ Ω. The detailed derivation is given in Appendix A.
Apart from υt, there are three other terms in (5). If no reorientation is involved, the first two terms will vanish and it is easy to show that (5) is equivalent to the gradient for scalar image registration as given in Beg et al. (2005). When reorientation is in effect, the first two terms contribute to updating {υt} by using mappings1 estimated in the preceding iterations to reorient the directions of the DBFs. This reveals how spatial alignment and reorientation interact with each other. In contrast, spatial alignment and reorientation are regarded as two separate components by Dhollander et al. (2011) and Zhang et al. (2012).
5. Numerical implementation details
We now describe the numerical implementation details of our algorithm, including multi-Gaussian kernel, mapping computation, and gradient descent optimization.
5.1. Multi-kernel scheme
Instead of defining L, we follow the approach proposed by Risser et al. (2011) to define a multi-Gaussian kernel K to achieve the desired smoothness. Specifically, K is realized by a set of weighted Gaussian kernels
where al is the weight of the l-th Gaussian kernel and Σl is a diagnal covariance matrix defined by a scale factor σl, i.e. Σl = σlI, where I is an identity matrix.
To estimate al we need to perform a pre-registration step to estimate the maximum update of υ at each position x, i.e. τl = max({∥δυ∥∣ δυ at ∀x ∈ Ω}), where δυ can be computed via (5) by setting ϕt,0 = ϕt,1 = id, where id is an identity mapping, υt = 0, and K = N(0,Σ1). Then al is calculated as the reciprocal of τl and normalized such that Σal = 1.
5.2. Mapping computation
Suppose the time interval is [0, 1], there are n time points {tp∣p = 1, 2, …, n} evenly distributed in this interval, i.e. tp = p/n and the time point 0 is denoted by t0. To update the gradient using (5), we have to compute the forward mappings {ϕtp,1} and backward mappings {ϕtp,0} at each time point tp. This can be obtained via a backward integration by assuming that ϕtn,1 = ϕt0,0 = id. Specifically, the forward mapping ϕt,1 and backward mapping ϕt,0 can be computed by concatenating a set of small mappings (Ashburner and Friston, 2011), i.e.
where {υtp} is a series of velocity fields.
5.3. Gradient descent
Once we have computed the gradient using (5), we can use any gradient descent algorithm to solve (4). In this work we use line search for gradient descent. A summary of the algorithm is given in Algorithm 1.
Algorithm 1.
LDDMM-based DWI registration
| Input: I0, I1, {μj}, {qi}, λ1, λ2, b, n, {σl} and {α} (step size) |
| Initialization {υtp ∣ υtp ← 0, ∀p} |
| 1: estimate kernel weights {al} |
| 2: repeat |
| 3: for each p do |
| 4: |
| 5: |
| 6: end for |
| 7: compute current energy E using (4) |
| 8: for each p do |
| 9: compute gradient (∇υ E)tp using (5) |
| 10: end for |
| 11: (E′, α) ← linesearch (E, {υtp}, {α}, {(∇υ E)tp}) |
| 12: if E′ < E then |
| 13: for each p do |
| 14: υtp ← υtp − α(∇υ E)tp |
| 15: End For |
| 16: end if |
| 17: until E′ ≥ E ∨ α = inf {α} ∨ maximum number of iterations |
| 18: function linesearch (E, {υtp}, {α}, {(∇υ E)tp}) |
| 19: for each α do |
| 20: for each p do |
| 21: |
| 22: end for |
| 23: for each p do |
| 24: |
| 25: |
| 26: end for |
| 27: compute current energy E′ using (4) |
| 28: if E′ < E then |
| 29: return E′, α |
| 30: end if |
| 31: end for |
| 32: return E, inf {α} |
| 33: end function |
6. Experiments
DWI data were acquired from 11 adults using a Siemens 3T TIM Trio MR Scanner with an EPI sequence. Diffusion gradients were applied in 120 non-collinear directions with diffusion weighting b = 2000s/mm2. The imaging matrix was 128 × 128 with a field of view of 256 × 256mm2. 80 contiguous slices with thickness of 2mm covered the whole brain. A diffusion tensor model was fitted to the signal vector at each voxel, leading to a field of diffusion tensors. The eigenvalues corresponding to the principal eigenvectors were then computed for each tensor. By using a region of interest (ROI) defined at the corpus callosum, which is known to contain coherent single-orientation fiber bundles, we computed λ1 by averaging the eigenvalues corresponding to the first principal directions. We then computed λ2 as the mean of the eigenvalues corresponding to the second and third principal eigenvectors.
We randomly chose an image as the target image and used the rest as source images. For each image, we fit the DBFs, with 321 directions uniformly distributed on a unit sphere, to estimate the associated weight image, which was then used to obtain a reconstructed version of the image. A set of affine transformations was estimated between the target image and each source image using their anisotropy images computed from the reconstructed data. We then reconstructed each source image again using the associated weight image by taking into account the affine transformation (see (3)). The resulting reconstructed source images were used for both DWI and diffusion tensor imaging (DTI) registration. We compared our method (Section 4), which we will refer to as LDDMM-DWI, with the following registration strategies:
Naïve LDDMM-DWI (Zhang et al., 2012): This is a registration scheme that iteratively (1) warps and reorients the source and target images based on an estimated mapping, (2) estimates a new mapping that further aligns the two resulting images via a geodesic shooting algorithm, (3) concatenates the estimated mapping with the one estimated in the previous iteration. The source image is reconstructed using the composite mapping together with an affine transformation, whereas the target is reconstructed without any transformation. At each stage the reconstruction is done by using an increasing number of diffusion directions.
DTI registration: For each image we computed a DT image by fitting the diffusion tensor model to the reconstructed data. We then registered each source DT image to the target DT image using DTI-TK (Zhang et al., 2006).
For LDDMM-DWI, we ran line search for 30 iterations. For naïve LDDMM-DWI (Zhang et al., 2012), we performed the registration in 6 stages: 30 iterations in the first two stages, 20 in the middle stages, and 10 and 5 in the last two stages. In each stage, the number of diffusion directions was 1, 6, 21, 81, 120, and 321, respectively. The default parameters provided on the DTI-TK website2 were used for DTI registration.
To quantify the comparison, we computed the voxel-wise root mean square (RMS) error between the target image and each of source image, warped and reoriented using the estimated affine and non-linear mapping. Averaging the resulting RMS error images across subjects for each method leads to the mean images shown in Fig. 1. For reference, we also show the mean RMS error image of source images warped and reoriented using affine transformations alone. For each mean RMS error image we computed its statistics (i.e. mean, standard deviation, median, and the 90-th percentile) over all voxels and report the results in Table 1. For each method, we also computed the mean intensity value of the RMS error image associated with each subject and used these means to perform two-tailed student’s t-tests using LDDMM-DWI as the baseline. The results, given in the same table, indicate that the performance differences are statistically significant.
Fig. 1.
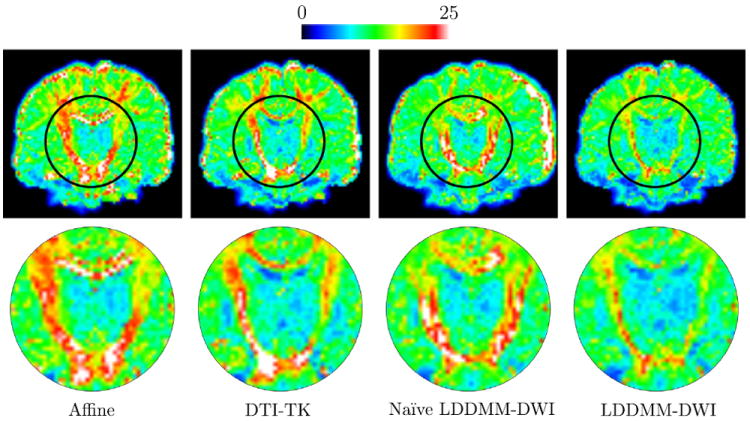
The mean RMS error images (top) and the close-ups of regions marked by black circles (bottom). Note that the images were not sliced for left–right symmetry. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Statistics of the mean RMS error images given by various registration strategies.
| Method | Mean ± s.d. | Median | 90%-ile | p-value |
|---|---|---|---|---|
| Affine | 14.7 ± 6.3 | 13.4 | 21.4 | p < 10−6 |
| DTI-TK | 13.3 ± 6.1 | 12.2 | 19.6 | p < 0.01 |
| Naïve LDDMM-DWI | 12.9 ± 5.4 | 11.9 | 18.3 | p < 0.05 |
| LDDMM-DWI | 11.7 ± 4.5 | 11.3 | 16.5 | – |
From Fig. 1, it is clear that LDDMM-DWI significantly improves the alignment of white matter structures, such as the internal capsule. Affine registration and naïve LDDMM-DWI do not take into account reorientation in the optimization process and are hence less effective in white matter alignment. Although DTI-TK explicitly considers reorientation during optimization, it can only handle one principal fiber direction per voxel and thus ambiguity will occur where fiber configuration is complex. The quantitative results given in Table 1 show that LDDMM-DWI outperforms the other methods and reduces overall mismatching error (e.g., smaller median).
To further demonstrate the efficacy of LDDMM-DWI, we compared the similarity of ODFs (for high-anisotropy voxels) computed from the target image and each source image using the symmetrized Kullback–Leibler (sKL) divergence (Chiang et al., 2008)
where p1 and p2 are the ODFs and s is a vector defined on the unit sphere S2. The mean sKL divergence images resulting from each method are given in Fig. 2 and the statistics of the mean images are given in Table 2. Similarly, we can see that LDDMM-DWI works much better than other registration methods and reduces white matter misalignment.
Fig. 2.
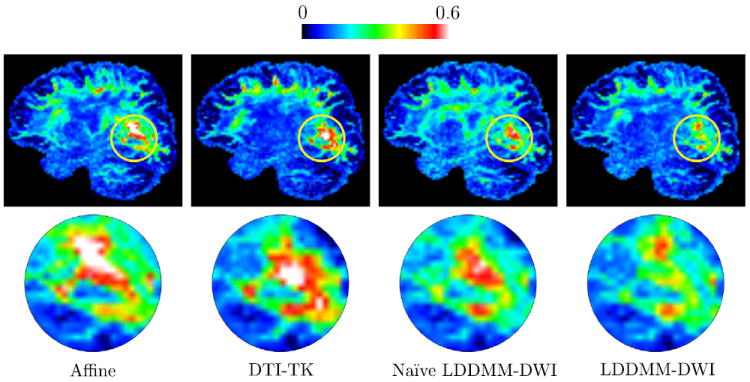
The mean sKL divergence images (top) and the close-ups of regions marked by yellow circles (bottom). Note that the low-anisotropy regions were not masked out for display purpose. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Statistics of the mean sKL divergence images given by different registration strategies.
| Method | Mean ± s.d. | Median | 90%-ile | p-value |
|---|---|---|---|---|
| Affine | 0.22 ± 0.10 | 0.21 | 0.35 | p < 0.01 |
| DTI-TK | 0.21 ± 0.10 | 0.20 | 0.36 | p ≈ 0.01 |
| Naïve LDDMM-DWI | 0.21 ± 0.08 | 0.21 | 0.32 | p < 0.05 |
| LDDMM-DWI | 0.19 ± 0.09 | 0.19 | 0.31 | – |
Figure 3 shows the anisotropy images of the target image and the mean of the aligned source images, from which we can see that LDDMM-DWI registers all main structures reasonably well. ODFs computed from the target image and the LDDMM-DWI mean image are shown in Fig. 4. It can be observed that the results produced by LDDMM-DWI are in close agreement at voxel level with the target image. This is important for applications such as white matter tractography (Yap et al., 2011; Yap et al., 2011), which is sensitive to errors in local fiber orientations.
Fig. 3.
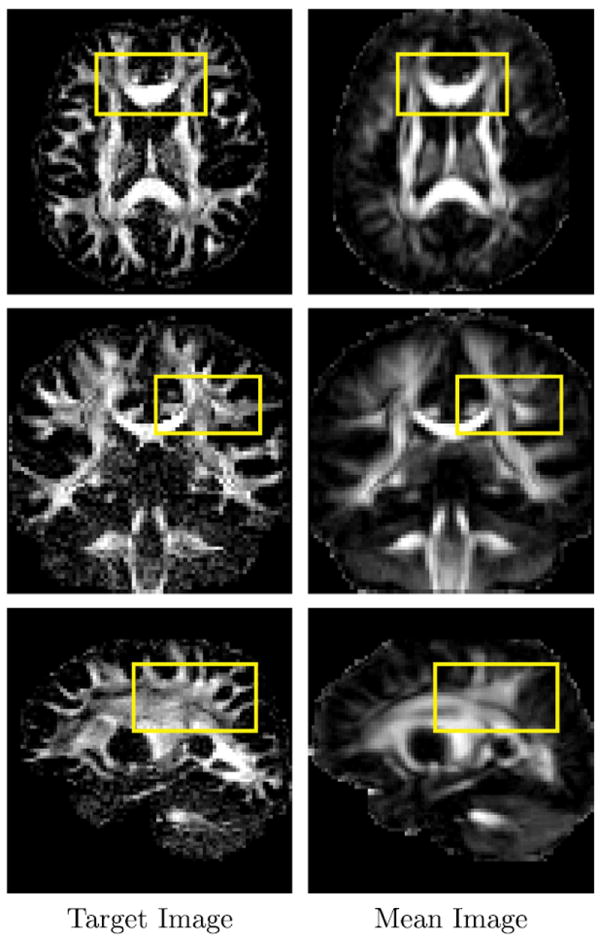
Anisotropy images of the target image and the mean of the align source images. The ODFs in regions marked by yellow rectangles are shown in Fig. 4. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
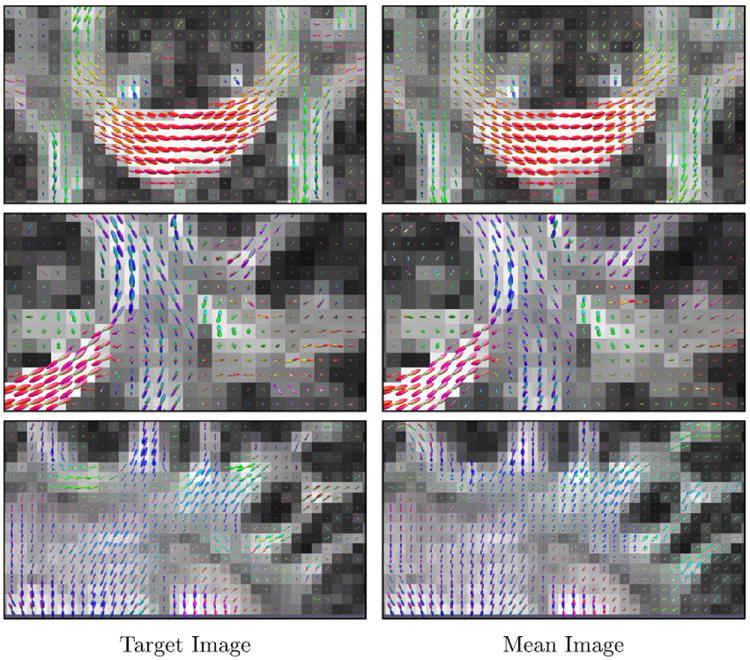
ODFs in the regions marked by yellow rectangles in Fig. 3. The target anisotropy image is used as the background. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
7. Conclusion and discussions
We have described a method for directly registering the DWI data under large deformation. This is achieved by incorporating a DWI data reorientation technique (Yap and Shen, 2012) into an LDDMM algorithm (Beg et al., 2005), optimizing spatial alignment and local reorientation simultaneously. Unlike most of the existing methods, our approach eventually results in a set of diffusion-weighted images, thus allowing the fitting of any diffusion model for subsequent analysis. Experimental results indicate that our method significantly outperforms several other registration strategies as well as the method proposed in our earlier work (Zhang et al., 2012), which does not explicitly consider reorientation during optimization. In the future, to overcome the large computation cost of our method, general-purpose computing on graphics processing units (GPGPU) can be employed to significantly speed up the computation of the parallelizable portion of the algorithm.
Acknowledgments
This work was supported in part by a UNC BRIC-Radiology startup fund, NSF Grants (EECS-1148870 and EECS-0925875) and NIH Grants (EB006733, EB008374, EB009634, MH088520, AG041721, MH100217, and MH091645).
Appendix A. Derivation of the gradient of the cost function
To minimize the cost function (4), we consider a small perturbation of υ ∈ L2 ([0, 1], V) along direction h ∈ L2 ([0, 1], V). The variation of energy functional E(υ) w.r.t υ can be obtained as
where ∂h E(υ) is the Gâteaux derivative of E and ∇υ E is the Fréchet derivative of E.
The variation of can be obtained easily as follows
| (A.1) |
Now focusing on the variation of , we have
where Ω is a bounded domain in ℝd and y ∈ Ω. at y is given by
Note that we drop y for simplicity, i.e. .
The differentiation of each non-zero element in the above matrix yields
Note that now . According to the proof given by Beg et al. (2005),
Hence, can be obtained by simply taking differentiation w.r.t y on both sides of the above equation, i.e.
where Dy· is the value of D· at y, D2· is the Hessian operator, is the value of D2· at y, giving a vector-valued matrix. For instance, represents the Jacobian at position . Then we have
We now deal with the two terms in the above equation one by one:
and
where we have used and for simplification. Hence, we have
Also, we have
Now we are ready to compute , where G is a vector-valued image and the i-th element of its voxel at y is given by Gi (y). We have
where
and [w0, …, wN]T is the sparse weights of W0 at position y. Let bi = [f0, bi1, ⋯, biN]T, we can then compute Gi(y) as
Now we can rewrite ∂h E2 (v) as
Let , then and . By changing the variable, we have
where now . As ν, h ∈ V, we have the following two equations (see (Cao et al., 2005) for details on the property of the reproducing kernel Hilbert space)
Hence, the variation of E2 (v) is given by
| (A.2) |
Since h is arbitrary in L2 ([0, 1], V), we obtain the gradient as given in (5) by combining results (A.1) and (A.2).
Footnotes
The mappings at each iteration are generated by integrating {υt} over time. See Section 5.2 for details.
References
- Schaefer PW, Grant PE, Gonzalez RG. Diffusion-weighted MR imaging of the brain. Radiology. 2000;217(2):331–345. doi: 10.1148/radiology.217.2.r00nv24331. [DOI] [PubMed] [Google Scholar]
- Hanyu H, Sakurai H, Iwamoto T, Takasaki M, Shindo H, Abe K. Diffusion-weighted MR imaging of the hippocampus and temporal white matter in Alzheimer’s disease. J Neurol Sci. 1998;156(2):195–200. doi: 10.1016/s0022-510x(98)00043-4. [DOI] [PubMed] [Google Scholar]
- Wee C-Y, Yap P-T, Li W, Denny K, Brownyke J, Potter G, Welsh-Bohmer K, Wang L, Shen D. Enriched white-matter connectivity networks for accurate identification of MCI patients. NeuroImage. 2011;54(3):1812–1822. doi: 10.1016/j.neuroimage.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee C-Y, Yap P-T, Zhang D, Denny K, Browndyke JN, Potter GG, Welsh-Bohmer KA, Wang L, Shen D. Identification of MCI individuals using structural and functional connectivity networks. NeuroImage. 2012;59(3):2045–2056. doi: 10.1016/j.neuroimage.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wee C-Y, Niethammer M, Shen D, Yap P-T. Large deformation image classification using generalized locality-constrained linear coding. In: Mori, Sakuma I, Sato Y, Barillot C, Navab N, editors. MICCAI 2013. Vol. 8149. Springer; Heidelberg: 2013. pp. 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Yap P-T, Gao W, Lin W, Gilmore JH, Shen D. Altered structural connectivity in neonates at genetic risk for schizophrenia: a combined study using morphological and white matter networks. NeuroImage. 2012;62:1622–1633. doi: 10.1016/j.neuroimage.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap P-T, Fan Y, Chen Y, Gilmore J, Lin W, Shen D. Development trends of white matter connectivity in the first years of life. PLoS ONE. 2011;6(9):e24678. doi: 10.1371/journal.pone.0024678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D, Pierpaoli C, Basser P, Gee J. Spatial transformations of diffusion tensor magnetic resonance images. IEEE Trans Med Imag. 2001;20(11):1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- Cao Y, Miller MI, Mori S, Winslow RL, Younes L. Diffeomorphic matching of diffusion tensor images. Proceedings of the 2006 Conference on Computer Vision and Pattern Recognition Workshop; 2006. pp. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Vercauteren T, Fillard P, Peyrat J-M, Pennec X, Golland P, Ayache N, Clatz O. DT-REFinD: diffusion tensor registration with exact finitestrain differential. IEEE Trans Med Imag. 2009;28(12):1914–1928. doi: 10.1109/TMI.2009.2025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap P-T, Wu G, Zhu H, Lin W, Shen D. TIMER: tensor image morphing for elastic registration. NeuroImage. 2009;47(2):549–563. doi: 10.1016/j.neuroimage.2009.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap P-T, Wu G, Zhu H, Lin W, Shen D. F-TIMER: fast tensor image morphing for elastic registration. IEEE Trans Med Imag. 2010;29:1192–1203. doi: 10.1109/TMI.2010.2043680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yushkevich PA, Alexander DC, Gee JC. Deformable registration of diffusion tensor MR images with explicit orientation optimization. Med Image Anal. 2006;10(5):764–785. doi: 10.1016/j.media.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. NeuroImage. 2009;45(1):S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Beg MF, Miller MI, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis. 2005;61(2):139–157. [Google Scholar]
- Behrens T, Berg HJ, Jbabdi S, Rushworth M, Woolrich M. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? NeuroImage. 2007;34(1):144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Ross TJ, Gu H, Shin W, Zhan W, Chao Y-P, Lin C-P, Schuff N, Yang Y. Diffeomorphic image registration of diffusion MRI using spherical harmonics. IEEE Trans Med Imag. 2011;30(3):747–758. doi: 10.1109/TMI.2010.2095027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Goh A, Qiu A. Diffeomorphic metric mapping of high angular resolution diffusion imaging based on Riemannian structure of orientation distribution functions. IEEE Trans Med Imag. 2012;31(5):1021–1033. doi: 10.1109/TMI.2011.2178253. [DOI] [PubMed] [Google Scholar]
- Hong X, Arlinghaus L, Anderson A. Spatial normalization of the fiber orientation distribution based on high angular resolution diffusion imaging data. Mag Reson Med. 2009;61(6):1520–1527. doi: 10.1002/mrm.21916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffelt D, Tournier J-D, Fripp J, Crozier S, Connelly A, Salvado O. Symmetric diffeomorphic registration of fibre orientation distributions. NeuroImage. 2011;56(3):1171–1180. doi: 10.1016/j.neuroimage.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Yap P-T, Chen Y, An H, Yang Y, Gilmore JH, Lin W, Shen D. SPHERE: spherical harmonic elastic registration of HARDI data. NeuroImage. 2011;55(2):545–556. doi: 10.1016/j.neuroimage.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhollander T, Veraart J, Van Hecke W, Maes F, Sunaert S, Sijbers J, Suetens P. Feasibility and advantages of diffusion weighted imaging atlas construction in Q-space. In: Fichtinger G, Martel A, Peters T, editors. MICCAI 2011. Vol. 6892. Springer; Heidelberg: 2011. pp. 166–173. [DOI] [PubMed] [Google Scholar]
- Zhang P, Niethammer M, Shen D, Yap P-T. Large deformation diffeomorphic registration of diffusion-weighted images. In: Ayache N, Delingette H, Golland P, Mori K, editors. MICCAI 2012. Vol. 7511. Springer; Heidelberg: 2012. pp. 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaunès J, Qiu A, Miller M, Younes L. Large deformation diffeomorphic metric curve mapping. Int J Comput Vis. 2008;80(3):317–336. doi: 10.1007/s11263-008-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap P-T, Shen D. Spatial transformation of DWI data using non-negative sparse representation. IEEE Trans Med Imag. 2012;31(11):2035–2049. doi: 10.1109/TMI.2012.2204766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Diffeomorphic registration using geodesic shooting and Gauss-Newton optimisation. NeuroImage. 2011;55(3):954–967. doi: 10.1016/j.neuroimage.2010.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Niethammer M, Shen D, Yap P-T. Large deformation diffeomorphic registration of diffusion-weighted images with explicit orientation optimization. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. MICCAI 2013. Vol. 8150. Springer; Heidelberg: 2013. pp. 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risser L, Vialard F-X, Wolz R, Murgasova M, Holm DD, Rueckert D. Simultaneous multi-scale registration using large deformation diffeomorphic metric mapping. IEEE Trans Med Imag. 2011;30(10):1746–1759. doi: 10.1109/TMI.2011.2146787. [DOI] [PubMed] [Google Scholar]
- Hsu Y-C, Hsu C-H, Tseng W-YI. A large deformation diffeomorphic metric mapping solution for diffusion spectrum imaging datasets. NeuroImage. 2012;63(2):818–834. doi: 10.1016/j.neuroimage.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Jupp P, Mardia K. A unified view of the theory of directional statistics. Int Stat Rev. 1989;57:261–294. [Google Scholar]
- Miller MI, Trouvé A, Younes L. Geodesic shooting for computational anatomy. J Math Imag Vis. 2006;24(2):209–228. doi: 10.1007/s10851-005-3624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialard F-X, Risser L, Rueckert D, Cotter C. Diffeomorphic 3D image registration via geodesic shooting using an efficient adjoint calculation. Int J Comput Vis. 2012;97(2):229–241. [Google Scholar]
- Cao Y, Miller MI, Winslow RL, Younes L. Large deformation diffeomorphic metric mapping of vector fields. IEEE Trans Med Imag. 2005;24(9):1216–1230. doi: 10.1109/tmi.2005.853923. [DOI] [PubMed] [Google Scholar]
- Chiang M-C, Barysheva M, Lee A, Madsen S, Klunder A, Toga A, McMahon K, Zubicaray G, Meredith M, Wright M, Srivastava A, Balov N, Thompson P. Brain fiber architecture, genetics, and intelligence: a high angular resolution diffusion imaging (HARDI) study. In: Metaxas D, Axel L, Fichtinger G, Székely G, editors. MICCAI 2008. Vol. 5241. Springer; Heidelberg: 2008. pp. 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap P-T, Gilmore J, Lin W, Shen D. PopTract: population-based tractography. IEEE Trans Med Imag. 2011;30(10):1829–1840. doi: 10.1109/TMI.2011.2154385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap P-T, Gilmore JH, Lin W, Shen D. Longitudinal tractography with application to neuronal fiber trajectory reconstruction in neonates. In: Fichtinger G, Martel A, Peters T, editors. MICCAI 2011. Vol. 6892. 2011. pp. 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]


