Abstract
Objective. This review is to evaluate the diagnostic value of serum GPC3 for hepatocellular carcinoma (HCC) due to conflicting results reported. Methods. NCBI PubMed and Embase were comprehensively searched for studies that have used serum GPC3 level as a diagnostic index for HCC. The quality of the included studies was assessed. Subgroup analyses were conducted to evaluate the sensitivity and specificity of GPC3 as a HCC marker. Statistical analysis was performed with the software STATA version 12.0. Results. A total of 22 studies were included. The qualities of included studies were relatively poor. Among them, 18 studies have shown that serum GPC3 is a specific biomarker for HCC, and the pooled sensitivity and specificity of these studies were 69 and 93%, respectively. The other 4 studies have reported conflicting results, which were not caused by races, infection status of HBV and HCV, or assay reagents but due to one common experimental design of enrolling liver cirrhosis patients as control subjects. Conclusions. This meta-analysis indicates that serum GPC3 is elevated in HCC patients compared with healthy individuals, but more studies are needed to evaluate its effectiveness to differentially diagnose HCC and liver cirrhosis.
1. Introduction
Primary hepatocellular carcinoma (HCC) is one of the most malignant tumors and is the most common cause of cancer-related death worldwide [1]. Nowadays, laparoscopic surgery and drug intervention with Sorafenib are widely utilized to treat HCC patients. But their 5-year survival rate remains low [2, 3] because many patients are diagnosed at the late stage of HCC and lose the opportunity of effective medical interventions. Thus, diagnosis of HCC at an early stage is of utmost importance for reducing HCC-associated mortality.
European Association for the Study of the Liver (EASL) recommends patients with liver diseases to do liver ultrasound and examine serum α-fetoprotein (AFP) level every six months [4]. However, ultrasound is an indirect diagnostic method with accuracy greatly depending on the skill of operators and has limited ability to distinguish HCC from nonneoplastic nodules [4]. Similarly, serum AFP level is not an accurate biomarker for HCC because its sensitivity only ranges from 40 to 65% [5]. Therefore, a novel biomarker with superior diagnostic accuracy is greatly desired.
Glypican-3 (GPC3) belongs to the glypican family of heparan-sulfate proteoglycans [6]. GPC3 is normally expressed in fetal liver and placenta and has negligible expression in normal adult liver [6]. Currently, many studies have found that GPC3 expression is increased in HCC tissues [7–9], even though its expression is absent in the hepatocytes of healthy individuals and hepatitis patients. Furthermore, serum GPC3 level is higher in HCC patients than that in healthy individuals and hepatitis patients. Thus, it has been suggested that serum GPC3 is a specific biomarker for HCC [10–27]. But other studies reported conflicting results [28–31]. In the present study, we performed a meta-analysis on the data from all published studies that have evaluated the diagnostic potential of serum GPC3 for HCC and concluded that serum GPC3 is a clinically relevant HCC biomarker.
2. Methods
2.1. Literature Search Strategy
We performed a comprehensive search on the peer-reviewed scientific literatures that are written in English and were published before May 20, 2014, in NCBI PubMed or EMBASE. The following search terms were used: (1) GPC3: glypican-3 and GPC3; and (2) HCC: HCC, hepatocellular carcinoma, liver cancer, liver cell carcinoma, and hepatic cell carcinoma. No restrictions on study design, year of publication, or publication type were set during initial database search. To avoid exclusion of relevant studies, we did not use keywords or indexing terms for diagnostic test accuracy. After filtering the studies based on the criteria listed in the next paragraph, we manually searched the reference lists of selected articles to identify more relevant publications.
2.2. Criteria for Inclusion and Exclusion of Published Studies
The inclusion criteria for articles were as follows: (1) studies investigated the diagnostic accuracy of serum GPC3 for HCC; (2) studies have reported calculable data on sample sizes of HCC and non-HCC patients, true positive (TP), true negative (TN), false positive (FP), and false negative (FN) values; and (3) article is written in English. The exclusion criteria were as follows: (1) studies conducted on animals; (2) studies that evaluated mRNA expression or DNA polymorphisms of GPC3 and did not provide the sensitivity or specificity of using GPC3 as a HCC marker; (3) letters, editorials, expert opinions, and reviews without original clinical data; (4) case reports and studies lacking control groups; and (5) duplicate reports.
2.3. Identification of Eligible Studies and Data Extraction
Initial screening for potentially eligible studies was carried out by author Sheng-Li Yang based on the titles and abstracts of articles. Two authors Sheng-Li Yang and Xiefan Fang independently reviewed and included eligible studies based on the criteria described above. Disagreements were resolved by discussion or consulting with author Zao-Zao Huang. After all the eligible studies were identified, the following characteristics were retrieved from each study: authors, geographic distribution of patients, study design, number of patients, reference test, methods of measurement, cutoff values, and raw data including TP, FP, TN, and FN results.
2.4. Quality Assessment of Included Studies
Authors Sheng-Li Yang and Xiefan Fang independently assessed qualities of the eligible studies using the recommended checklist of Quality Assessment of studies of Diagnostic Accuracy included in Systematic reviews (QUADAS, Cochrane Collaboration). Each of the eleven items in the QUADAS checklist was scored as “yes,” “no,” or “unclear” [13]. Any disagreements in quality assessments were resolved by discussion or consulting with author Zao-Zao Huang.
2.5. Data Analysis
For each study, we calculated sensitivity, specificity, positive likelihood ratios (LR+), and negative likelihood ratios (LR−), as well as their corresponding 95% confidence intervals (95% CIs). The data were visualized as forest plots and receiver operating characteristic curves (ROC). Heterogeneity of the retrieved data from eligible studies was evaluated by using the Q statistics, with a significance level at P < 0.10. The I-square value, a quantitative measurement of inconsistency across different studies [32], was also calculated. I-square value typically ranges from 0 (no observed heterogeneity) to 100% (maximal heterogeneity), and an I-square value ≥ 50% is considered to represent substantial heterogeneity. If heterogeneity across studies was not identified, the fixed-effects model was used for meta-analysis. Otherwise, the random-effects model (DerSimonian-Laird method) was used [33]. Publication bias was measured by Deeks' funnel plot asymmetry test [34]. The meta-analysis was performed by the statistical software STATA version 12.0 (StataCorp LP, College Station, TX, USA), and statistical significance was defined as P value less than 0.05.
2.6. Subgroup Analysis
Because substantial heterogeneity existed in the included studies, we performed subgroup meta-analysis by dividing studies into one group that found that GPC3 has diagnostic value in HCC detection and another group that claimed that GPC3 has no diagnostic value for HCC. The Spearman approach was applied to test if the heterogeneity can be explained by a threshold effect.
3. Results
3.1. Study Selection
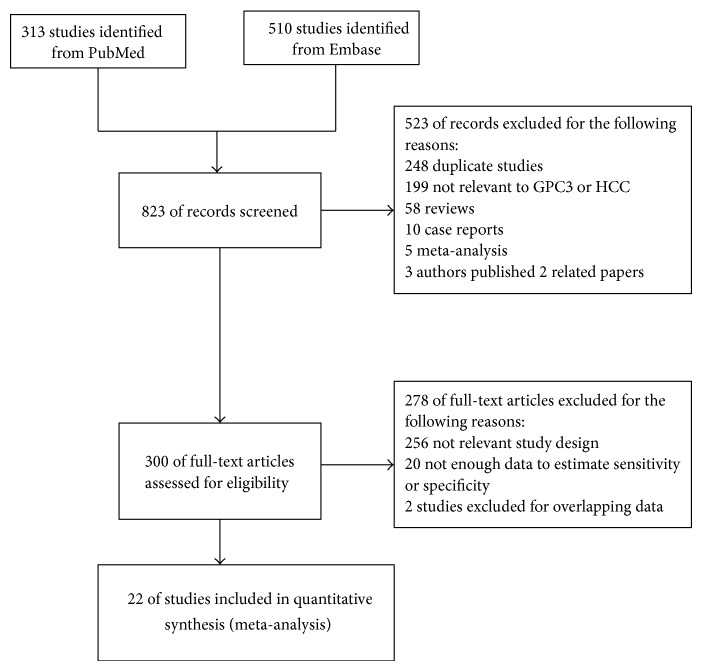
A total of 823 potentially relevant articles were identified by searches in NCBI PubMed and Embase. After reviewing their titles and abstracts, 523 articles, including duplicate studies, case reports, reviews, and comments, were excluded. After reviewing the full texts, 256, 20, and 2 studies were excluded due to irrelevant study design, insufficient data to estimate sensitivity or specificity, and publishing overlapping data, respectively. In the references of the retrieved studies, no additional articles met our inclusion criteria. Finally, twenty-two studies were included for meta-analysis [10–31] (Figure 1). The characteristics of the included studies are shown in Table 1. The eight studies from China mostly enrolled hepatitis B virus- (HBV-) associated HCC patients [10, 13, 14, 17, 18, 20, 22, 28]. The five studies from Egypt mainly had hepatitis C virus- (HCV-) associated HCC patients [12, 15, 16, 19, 21]. The study from the United Kingdom had HCC patients without virus infection [24]. There were differences in what human subjects were used as experimental controls among the included studies. Some studies enrolled healthy individuals as controls [13], some enrolled patients with hepatitis or liver cirrhosis [11, 12, 16, 17, 22–27, 29–31], and others combined both healthy people and patients with hepatitis or liver cirrhosis as one control group [10, 14, 15, 18–21, 28].
Figure 1.
Study selection process.
Table 1.
Characteristic and methodology assessment of the included studies.
| First author, year, country | Characteristics of HCC | Characteristics of controls | GPC3 | ||||
|---|---|---|---|---|---|---|---|
| Assay type | Cut-off value | Antibody for detection | HCC value (ng/mL) | Controls value (ng/mL) | |||
| Yu [10], 2014, China | NA | LC or hepatitis patients; healthy individuals | Chemiluminescent immunoassay | 30 ng/mL | GPC3 8G6 mcAb and 7D11 mcAb (Millipore Corporation) | 108.67 ± 230.04 | 3.99 ± 7.68 |
|
| |||||||
| Lee [11], 2014, Korea | 62.5% were HBV associated | CLD (HCV) patients; 50% had liver cirrhosis | ELISA | 73 ng/mL | ELISA kit (Wuhan Cusabio Biotech) | 75.8 ± 117.5 | 66.4 ± 33.2 |
|
| |||||||
| Badr [12], 2014, Egypt | NA | LC (HCV) patients | ELISA | 240 ng/mL | ELISA kit (Wuhan Uscn) | 551.47 ± 185.25 | 98.23 ± 73.54 |
|
| |||||||
| Li [13], 2013, China | HBV associated HCC | Healthy individuals | ELISA | NA | ELISA kit (usabio Biotech) | 12.63 ± 2.93 for patients with AFP <400 µg/L; 20.20 ± 5.41 for patients with AFP ≥ 400 µg/L |
1.92 ± 0.95 |
|
| |||||||
| Chen [14], 2013, China | NA | LC or hepatitis patients; healthy individuals | ELISA | 25.25 ng/mL | 7C8 and GP9 mcAb (self-made) | 99.94 ± 267.2 | 19.44 ± 50.88 for LC patients; 10.45 ± 46.02 for chronic hepatitis; 4.14 ± 31.65 for healthy controls |
|
| |||||||
| Abdelgawad [15], 2013, Egypt | 67.5% were HCV associated; 17.5% were HBV associated | LC patients; healthy individuals | ELISA | 4.9 ng/mL | ELISA kit (Wuhan Uscn) | 7.7 | 3.24 |
|
| |||||||
| Gomaa [16], 2012, Egypt | 12.9% were HBV associated; 87.1% were HCV associated | LC patients with HCV/HBV | ELISA | 5.41 ng/mL | ELISA kit (BioMosaics) |
8.13 ± 3.25 | 3.14 ± 1.16 |
|
| |||||||
| Wang [17], 2012, China | HBV associated | LC patients with HBV | ELISA | NA | ELISA kit (Wuhan Huamei) |
NA | NA |
|
| |||||||
| Qiao [18], 2011, China | 76.2% were HBV associated; 7.9% were HCV associated | LC (HBV/HCV) or hepatitis patients; healthy individuals | ELISA | 26.8 ng/mL | ELISA kit (USCN life science) |
29.29 ± 17.34 | 12.09 ± 9.69 for LC patients; 9.98 ± 9.60 for chronic hepatitis patients; 5.93 ± 5.46 for healthy controls |
|
| |||||||
| Abd El Moety [19], 2011, Egypt | HCV associated | LC or hepatitis patients; healthy individuals | ELISA | 2.0 ng/mL | ELISA kit (BioMosaics) |
34.63 ± 23.8 | NA |
|
| |||||||
| Zhang [20], 2010, China | NA | LC or hepatitis patients with HCV/HBV; healthy individuals | Immunoassay | 3.10 ng/mL | Self-made | 116.8 ± 98.6 | 24.60 ± 24.01 for LC patients; 6.73 ± 12.2 for hepatitis B patients; 13.67 ± 15.68 for hepatitis C patients; 0.86 ± 1.12 for healthy controls |
|
| |||||||
| Youssef [21], 2010, Egypt | HCV and HBV associated | LC patients with HBV/HCV; healthy individuals |
ELISA | 4.6 ng/mL | ELISA kit (BioMosaics) |
NA | NA |
|
| |||||||
| Liu [22], 2010, China | 84% were HBV associated; 16% were HCV associated | LC patients with HBV/HCV | ELISA | 300 ng/L | ELISA kit (BioMosaics) |
NA | NA |
|
| |||||||
| Tangkijvanich [23], 2010, Thailand | 59% were HBV associated; 11% were HCV associated | LC or hepatitis patients with HBV/HCV | ELISA | NA | Self-made | 46.3 (0–7826.6)■ | 0 (0–43.6)■ |
|
| |||||||
| Beale [24], 2008, UK | 60% had ALD and 40% patients had NAFLD | LC patients with ALD/NAFLD | ELISA | NA | ELISA kit (BioMosaics) |
161.41 ± 422.33 | 125.41 ± 281.05 |
|
| |||||||
| Yoshitaka Hippo [25], 2004, Japan | NA | LC patients | ELISA | 2.0 ng/mL | Self-made | 4.84 ± 8.91 | 1.09 ± 0.74 |
|
| |||||||
| Nakatsura [26], 2003, Japan | 12.1% were HBV associated; 40.9% were HCV associated | LC patients with HBV/HCV/PBC/AIH | ELISA | 10 U/mL | Self-made | NA | NA |
|
| |||||||
| Capurro [27], 2003, Canada | 44.1% were HBV associated; 27.2% were HCV associated; 14.7% had ALD | LC or hepatitis patients with HBV/HCV | ELISA | 117 ng/mL | Self-made 1G12 antibody |
NA | NA |
|
| |||||||
| Wang [28], 2014, China | 84.5% were HBV associated; 9.5% were HBV associated; 3.5% were alcoholic cirrhosis associated | LC or hepatitis (HBV) patients; healthy individuals | ELISA | NA∗ | ELISA kit (usabio Biotech) | 4.55 ± 3.5 | 9.03 ± 4.1 for LC patients; 4.56 ± 3.2 for HBV chronic hepatitis patients; 6.71 ± 1.8 for healthy controls |
|
| |||||||
| Nault [29], 2013, France | With alcoholic cirrhotic, 46.4% were early stage | Alcoholic cirrhotic patients | ELISA | NA∗ | ELISA kit (Wuhan Cusabio Biotech) | 1.4 ± 0.5 (early stage) 4.2 ± 1.8 (advanced stage) |
2.5 ± 1.4 |
|
| |||||||
| Özkan [30], 2011, Turkey | 52% were HBV associated; 23% were HCV associated | LC patients with HBV/HCV/HDV/AIH/Wilson disease/Cryptogenic liver disease; healthy individuals | ELISA | NA∗ | ELISA kit (Wuhan EIAab) |
5.13 ± 22.7 | 5.51 ± 59.2 |
|
| |||||||
| Yasuda [31], 2010, Japan | 16% were HBV associated; 77.5% were HCV associated | CLD patients with HBV/HCV | ELISA | NA∗ | ELISA kit (BioMosaics) |
924.8 (495.2, 1335.6)▲ | 1161.6 (762.0, 1784.0)▲ |
NA: data are not available; HBV: hepatitis B virus; HCV: hepatitis C virus; LC: liver cirrhosis; CLD: chronic liver disease; ALD: alcoholic liver disease; ALC: alcoholic liver cirrhosis; NAFLD: nonalcoholic fatty liver diseases; PBC: primary biliary cirrhosis; AIH: autoimmune hepatitis; ▲mean median (25% and 75% quartiles); ■mean median (ranges); ∗cutoff values were not provided because these studies found serum GPC3 has no correlation with HCC.
3.2. Quality of the Studies
The results of QUADAS quality assessment of the included studies are shown in Figure 2. The quality was not satisfactory. The major biases of the included studies fell in the domains of “representative spectrum,” “index test results blinded,” and “relevant clinical information.” In the domain of “representative spectrum,” only eight studies enrolled both healthy people and patients with hepatitis or liver cirrhosis as controls [10, 14, 15, 18–21, 28]. In the domain of “index test results blinded,” all studies reported the diagnostic standard for HCC; however, no studies have stated whether the index results were blindly interpreted or not. In the domain of “relevant clinical information,” seven studies did not provide enough relevant clinical information of the HCC patients [10, 12, 14, 19, 20, 24, 25].
Figure 2.

Summary of methodological quality of included studies on the basis of review authors' judgments on the 11 items of QUADAS checklist for each study.
3.3. Diagnostic Accuracy of GPC3 for HCC
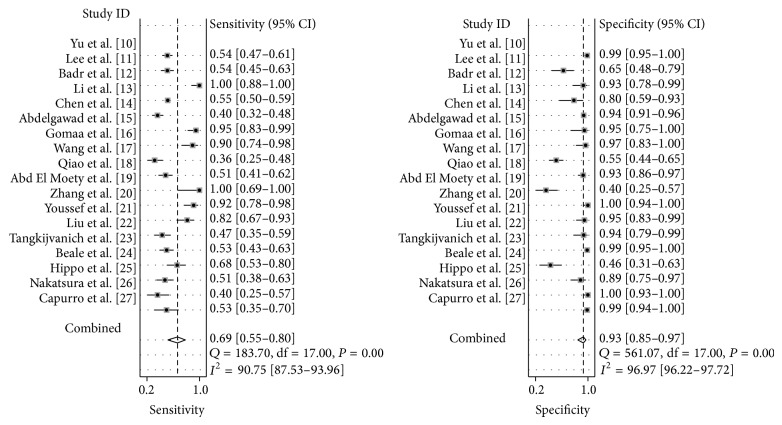
Among the twenty-two studies, eighteen of them have demonstrated that serum GPC3 level is higher in HCC patients than that in control subjects, which include healthy individuals and patients with hepatitis or liver cirrhosis [10–27] (Table 1), whereas four studies have claimed that GPC3 is not a diagnostic marker for HCC because serum GPC3 level is lower in HCC patients than that in patients with liver cirrhosis [28–31]. These four studies did not provide TP, TN, FP, or FN values [28–31] (Table 2). Thus, we divided the studies into two subgroups, and only the subgroup that provided the values of TP, TN, FP, and FN was used for meta-analysis [10–27]. The forest plots of sensitivity (TP rate) and specificity (FP rate) for the 18 studies are shown in Figure 3. The sensitivity and specificity of the studies were plotted in a hierarchical summary receiver operating characteristic graph (SROC, Figure 4). The sensitivity of using GPC3 for HCC diagnosis ranged from 36 to 100%, and the specificity ranged from 42 to 100%. The average sensitivity and specificity were 69% (95% CI was 55–80%) and 93% (95% CI was 85–97%), respectively. The overall diagnostic odds ratio (DOR) and the area under SROC were 31 (95% CI was 11–92) and 0.89 (95% CI was 0.86–0.91), respectively. I-square values of sensitivity and specificity were 90.71 and 97.30, respectively, indicating that substantial heterogeneity existed among the eligible studies. The Spearman correlation coefficient between the logic of sensitivity and the logic of 1-specificity was 0.043 (P = 0.86), indicating that the heterogeneity among eligible studies was not caused by the threshold effect.
Table 2.
Patients enrolled in the selected studies used for meta-analysis.
| Author (ref.) | Case | Control | TP | FP | FN | TN |
|---|---|---|---|---|---|---|
| Yu et al. [10] | 192 | 101 | 104 | 1 | 88 | 100 |
| Lee et al. [11] | 120 | 40 | 65 | 14 | 55 | 26 |
| Badr et al. [12] | 30 | 30 | 30 | 2 | 0 | 28 |
| Li et al. [13] | 605 | 25 | 330 | 5 | 275 | 20 |
| Chen et al. [14] | 155 | 440 | 62 | 27 | 93 | 413 |
| Abdelgawad et al. [15] | 40 | 20 | 38 | 1 | 2 | 19 |
| Gomaa et al. [16] | 31 | 30 | 28 | 1 | 3 | 29 |
| Wang et al. [17] | 78 | 97 | 28 | 44 | 50 | 53 |
| Qiao et al. [18] | 101 | 88 | 52 | 6 | 49 | 82 |
| Abd El Moety et al. [19] | 10 | 40 | 10 | 24 | 0 | 16 |
| Zhang et al. [20] | 36 | 93 | 33 | 0 | 3 | 56 |
| Youssef et al. [21] | 40 | 40 | 33 | 2 | 7 | 38 |
| Liu et al. [22] | 75 | 32 | 35 | 2 | 40 | 30 |
| Tangkijvanich et al. [23] | 100 | 100 | 53 | 1 | 47 | 99 |
| Beale et al. [24] | 50 | 41 | 34 | 22 | 16 | 19 |
| Hippo et al. [25] | 69 | 38 | 35 | 4 | 34 | 34 |
| Nakatsura et al. [26] | 40 | 50 | 16 | 0 | 24 | 50 |
| Capurro et al. [27] | 34 | 91 | 18 | 1 | 16 | 90 |
| Wang et al. [28] | 84 | 173 | NA | NA | NA | NA |
| Nault et al. [29] | 125 | 170 | NA | NA | NA | NA |
| Özkan et al. [30] | 75 | 55 | NA | NA | NA | NA |
| Yasuda et al. [31] | 200 | 200 | NA | NA | NA | NA |
TP: true positive; FP: false positive; TN: true negative; FN: false negative; NA: data are not available.
Figure 3.
Forest plots of sensitivity and specificity of using GPC3 as a diagnostic marker for hepatocellular cancer (HCC) in the eighteen studies included for meta-analysis [10–27].
Figure 4.
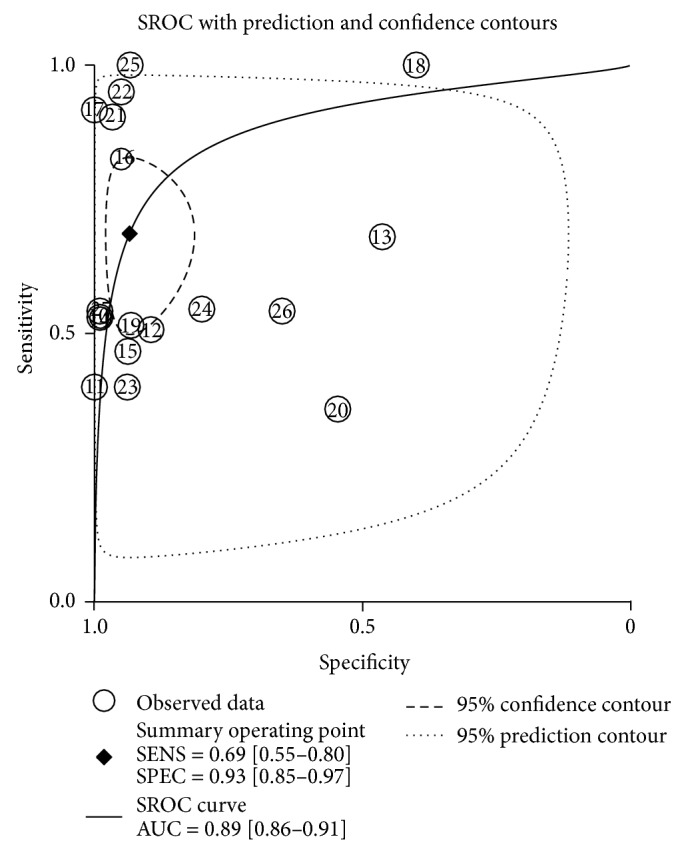
Summary receiver operating characteristic curves (SROC) from the hierarchical summary receiver operating characteristic model generated from the eighteen studies that found that GPC3 is a diagnostic marker for HCC [10–27].
3.4. Publication Bias
We used Deeks' funnel plot asymmetry test to evaluate publication bias among the included studies. The slope coefficient of the regression line had a P value of 0.33 indicating that the data were symmetric and did not have a likelihood of publication bias (Figure 5).
Figure 5.
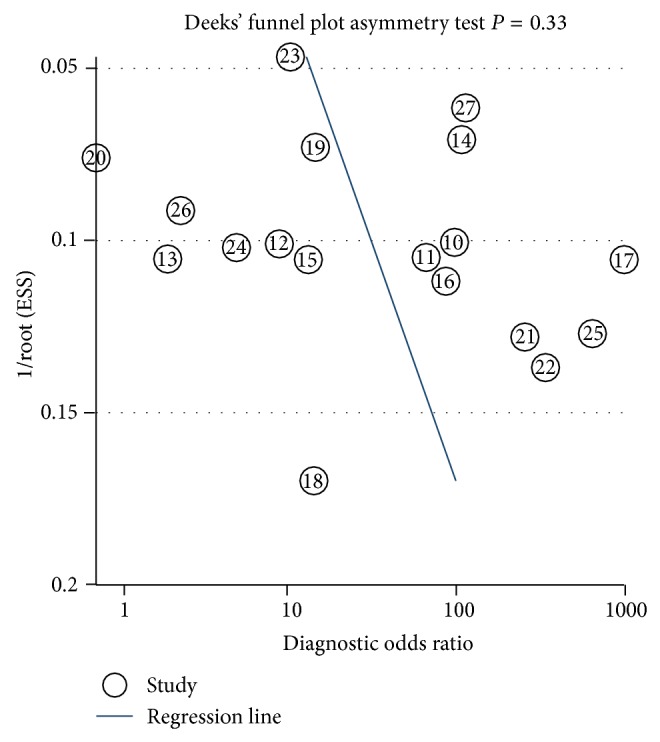
Linear regression test of funnel plot asymmetry. The statistically nonsignificant P value of the slop coefficient indicates symmetry of the data and a low likelihood of publication bias.
4. Discussion
HCC is one of the most lethal malignancies with a survival rate less than 10% and its incidence is increasing worldwide [35]. Specific and sensitive methods are urgently needed to accurately diagnose HCC at an early stage. Detection of serum biomarkers is a simple, rapid, and noninvasive method to diagnose HCC. Currently, several serum biomarkers for HCC, including AFP, AFP-L3, Golgi protein 73 (GP73), and GPC3 have been identified [36]. GPC3 is a 60 kDa cell-surface protein that belongs to the heparan-sulfate proteoglycan family, which contains GPC1, 2, 3, 4, 5, and 6. GPC3 is cleaved by furin between Arg358 and Ser359 to release a 40 kDa protein with the amino (N) terminal and a 30 kDa protein with the membrane-bound carboxyl (C) terminal [37]. The protein derived from the N-terminal of GPC3, also known as soluble GPC3 (sGPC3), is detected in the sera of HCC patients and is a potential serum marker for HCC diagnosis.
In this meta-analysis, we identified twenty-two studies that have investigated the diagnostic accuracy of serum GPC3 for HCC. Eighteen of them demonstrated that GPC3 is an ideal HCC diagnostic marker with pooled sensitivity, specificity, LR+, and LR− of 69%, 94%, 10.50, and 0.34, respectively [10–27]. They have found that serum GPC3 is elevated in HCC patients compared with healthy individuals and patients with hepatitis or liver cirrhosis [10–27]. But four studies have found lowered or equivalent serum levels of GPC3 in HCC patients compared with liver cirrhosis patients [28–31]. With careful comparison, we found that HBV or HCV infection and source of GPC3 antibodies were not the culprits for this discrepancy. Instead, the possible reason is that GPC3 is not able to differentially diagnose HCC and liver cirrhosis, and thus the studies that used liver cirrhosis patients as controls have found conflicting results [28–31].
We found that the infection status of HBV or HCV in HCC and control subjects were different in the included studies. In the studies from Egypt and Japan, more than 60% of the enrolled patients with HCC were infected with HCV, and approximately 15–20% of patients were infected with HBV [12, 15, 16, 31]. In contrast, approximately 85 and 10% of Chinese HCC cases were associated with HBV and HCV infection, respectively [13, 17, 22, 28, 38]. The four studies which held the controversial views were not conducted in the same region, but in France [29], China [28], Japan [31], and Turkey [30]. Therefore, the virus associated with HCC has no apparent influences on the diagnostic accuracy of GPC3.
The assay reagents used to measure serum GPC3 levels differed among the included studies. Some studies used self-made antibodies. For example, Hippo et al. used an antibody that binds to the N-terminal portion of GPC3 cleaved at Arg358 (amino acids 25–358) [25]. Capurro et al. used the anti-GPC3 monoclonal antibody “clone 1G12” that recognizes the last 70 amino acids of the C-terminal of the core protein (amino acids 491–560) [27]. The other studies used commercially available kits from different manufacturers, and their antibodies may target different regions of GPC3 [11, 12, 16–18]. The four conflicting studies used kits from Wuhan Cusabio Biotech, Usabio Biotech, Wuhan EIAab, and BioMosaics [28–31], which were also used in the other studies that found that GPC3 is a useful HCC biomarker [11, 13, 16, 19, 22, 24]. Therefore, the source of antibodies is not the reason that led to inconsistent results among the included studies.
We noticed that one common experimental design of the four conflicting studies is that they enrolled liver cirrhosis patients as control subjects, even though they had various complications, including alcoholic cirrhosis and HBV- or HCV-associated cirrhosis [28–31]. In fact, all four studies observed that serum GPC3 level is higher in liver cirrhosis patients than that in HCC patients [28–31]. Therefore, elevated serum GPC3 level is not a unique feature in HCC and is also seen in liver cirrhosis patients. But because measuring serum GPC3 level is a quick assay, it still could be the first step to screen for HCC. If elevated GPC3 level is detected, further testing should be done to confirm HCC diagnosis.
Considering that the included studies have substantial heterogeneity and part of them held the opposite views, a well-designed prospective study with larger cohorts should be performed to rigorously evaluate the diagnostic accuracy of GPC3 and determine if it has a better diagnostic value compared with other known HCC serum biomarkers. In addition, experimental design should be improved in the following areas: (1) double-blind studies should be designed to avoid bias; (2) the cohorts of healthy individuals, hepatitis patients, and liver cirrhosis patients should be compared as separate groups; (3) the study could use two or more different GPC3 antibodies to measure GPC3 level; (4) it is important to examine the stability of GPC3 during long-term storage. Half of the included studies measured GPC3 level in frozen serum, but it is not sure if GPC3 would be degraded after long-term storage. If the stability of serum GPC3 in long-term storage is not good, the diagnostic performance of serum GPC3 may be greatly affected.
To our knowledge, several studies have performed comprehensive reviews on using serum GPC3 as a diagnostic indicator for HCC [39–41]. However, Huang et al. only provided a protocol for meta-analysis and did not make a conclusion in his report [39]. Xu et al. performed a meta-analysis on merely ten studies and agreed upon that GPC3 is a good diagnostic marker for HCC, and they did not include the conflicting studies that we have found [41]. Liu et al. included twelve studies for their analysis and found inconsistent results among literatures as we did [40]. They suggested that GPC3 has moderate diagnostic accuracy for HCC and additional studies with larger sample size should be done to make a conclusion [40]. Compared with the previous reviews, our meta-analysis included more patients (5931) and studies (22), including those published after 2013. More importantly, we identified that having liver cirrhosis patients as control subjects is the potential cause for the inconsistent results in literatures, whereas race, country, infection with HBV or HCV, or assay reagents are not responsible for the discrepancies. We found that elevated GPC3 level is not unique for HCC but is also found in liver cirrhosis patients. These findings are valuable for future experimental designs to further explore the link between GPC3 and HCC.
5. Conclusion
In summary, our meta-analysis indicates that serum GPC3 level is elevated in HCC patients compared with healthy individuals. But whether GPC3 is an index to differentially diagnose HCC and liver cirrhosis is still uncertain.
Acknowledgments
The authors thank Professor Yi Sun (School of Public Health, Tongji Medical College, Huazhong University of Science and Technology) for her valuable suggestions. This study was supported by the National Natural Science Foundation of China (no. 81272421).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors’ Contribution
Sheng-Li Yang and Xiefan Fang contributed equally to this work.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2012. CA Cancer Journal for Clinicians. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Shin H.-R., Bray F., Forman D., Mathers C., Parkin D. M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International Journal of Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Blechacz B., Mishra L. Hepatocellular carcinoma biology. Recent Results in Cancer Research. 2013;190:1–20. doi: 10.1007/978-3-642-16037-0_1. [DOI] [PubMed] [Google Scholar]
- 4.European Association for the Study of the Liver EASL clinical practice guidelines: management of cholestatic liver diseases. Journal of Hepatology. 2009;51(2):237–267. doi: 10.1016/j.jhep.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Abu El Makarem M. An overview of biomarkers for the diagnosis of hepatocellular carcinoma. Hepatitis Monthly. 2012;12(10, article e6122) doi: 10.5812/hepatmon.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia H.-L., Ye Q.-H., Qin L.-X., Budhu A., Forgues M., Chen Y., Liu Y.-K., Sun H.-C., Wang L., Lu H.-Z., Shen F., Tang Z.-Y., Xin W. W. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clinical Cancer Research. 2007;13(4):1133–1139. doi: 10.1158/1078-0432.CCR-06-1025. [DOI] [PubMed] [Google Scholar]
- 7.Grozdanov P. N., Yovchev M. I., Dabeva M. D. The oncofetal protein glypican-3 is a novel marker of hepatic progenitor/oval cells. Laboratory Investigation. 2006;86(12):1272–1284. doi: 10.1038/labinvest.3700479. [DOI] [PubMed] [Google Scholar]
- 8.Honsová E., Lodererová A., Fraňková S., Oliverius M., Trunečka P. Glypican-3 immunostaining significantly improves histoloqical diagnosis of hepatocellular carcinoma. Casopis Lekaru Ceskych. 2011;150(1):37–40. [PubMed] [Google Scholar]
- 9.Kandil D., Leiman G., Allegretta M., Trotman W., Pantanowitz L., Goulart R., Evans M. Glypican-3 immunocytochemistry in liver fine-needle aspirates : a novel stain to assist in the differentiation of benign and malignant liver lesions. Cancer. 2007;111(5):316–322. doi: 10.1002/cncr.22954. [DOI] [PubMed] [Google Scholar]
- 10.Yu J.-P., Xu X.-G., Ma R.-J., Qin S.-N., Wang C.-R., Wang X.-B., Li M., Li M.-S., Ma Q., Xu W.-W. Development of a clinical chemiluminescent immunoassay for serum GPC3 and simultaneous measurements alone with AFP and CK19 in diagnosis of hepatocellular carcinoma. Journal of Clinical Laboratory Analysis. 2014 doi: 10.1002/jcla.21733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H. J., Yeon J. E., Suh S. J., Lee S. J., Yoon E. L., Kang K., Yoo Y. J., Kim J. H., Seo Y. S., Yim H. J., Byun K. S. Clinical utility of plasma glypican-3 and osteopontin as biomarkers of hepatocellular carcinoma. Gut and Liver. 2014;8(2):177–185. doi: 10.5009/gnl.2014.8.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badr E. A. E., Korah T. E., Ghani A. A., El-Sayed S., Badr S. Role of serum glypican-3 in the diagnosis and differentiation of small hepatocellular carcinoma from hepatitis-C virus cirrhosis. Alexandria Journal of Medicine. 2014;50(3):221–226. doi: 10.1016/j.ajme.2014.01.002. [DOI] [Google Scholar]
- 13.Li B., Liu H., Shang H. W., Li P., Li N., Ding H. G. Diagnostic value of glypican-3 in alpha fetoprotein negative hepatocellular carcinoma patients. African Health Sciences. 2013;13(3):703–709. doi: 10.4314/ahs.v13i3.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M., Li G., Yan J., Lu X., Cui J., Ni Z., Cheng W., Qian G., Zhang J., Tu H. Reevaluation of glypican-3 as a serological marker for hepatocellular carcinoma. Clinica Chimica Acta. 2013;423:105–111. doi: 10.1016/j.cca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Abdelgawad I. A., Mossallam G. I., Radwan N. H., Elzawahry H. M., Elhifnawy N. M. Can glypican3 be diagnostic for early hepatocellular carcinoma among Egyptian patients? Asian Pacific Journal of Cancer Prevention. 2013;14(12):7345–7349. doi: 10.7314/APJCP.2013.14.12.7345. [DOI] [PubMed] [Google Scholar]
- 16.Gomaa A. H. O., Aboraia G., Attia H., Elezawy H., Nafie E., ElBaz S. The diagnostic value of peripheral blood glypican-3 in patients with hepatocellular carcinoma. World Journal of Medical Sciences. 2012;7(2):105–112. [Google Scholar]
- 17.Wang W., Zhao L.-J., Wang Y., et al. Application of HBx-induced anti-URGs as early warning biomarker of cirrhosis and HCC. Cancer Biomarkers. 2012;11(1):29–39. doi: 10.3233/CBM-2012-0261. [DOI] [PubMed] [Google Scholar]
- 18.Qiao S.-S., Cui Z.-Q., Gong L., Han H., Chen P.-G., Guo L.-M., Yu X., Wei Y.-H., Ha S.-A., Kim J. W., Jin Z.-T., Li S., Peng J.-R., Leng X.-S. Simultaneous measurements of serum AFP, GPC-3 and HCCR for diagnosing hepatocellular carcinoma. Hepato-Gastroenterology. 2011;58(110-111):1718–1724. doi: 10.5754/hge11124. [DOI] [PubMed] [Google Scholar]
- 19.Abd El Moety H. A., Abd El Moety A. A., Rostom Y., El Sawy M. Glypican-3 amino terminal marker for early detection of HCC. Hepatology International. 2011;5(1):435. [Google Scholar]
- 20.Zhang Q., Xiao Q., Lin Z., Ying X., Li Z., Lin J.-M. Development of a competitive radioimmunoassay for glypican-3 and the clinical application in diagnosis of hepatocellular carcinoma. Clinical Biochemistry. 2010;43(12):1003–1008. doi: 10.1016/j.clinbiochem.2010.04.074. [DOI] [PubMed] [Google Scholar]
- 21.Youssef M., El-Sharkawy S., Abbas N., et al. Clinical utility of Glypican-3 in hepatocellular carcinoma. International Journal of Integrative Biology. 2010;10(1):41–47. [Google Scholar]
- 22.Liu H., Li P., Zhai Y., Qu C.-F., Zhang L.-J., Tan Y.-F., Li N., Ding H.-G. Diagnostic value of glypican-3 in serum and liver for primary hepatocellular carcinoma. World Journal of Gastroenterology. 2010;16(35):4410–4415. doi: 10.3748/wjg.v16.i35.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tangkijvanich P., Chanmee T., Komtong S., Mahachai V., Wisedopas N., Pothacharoen P., Kongtawelert P. Diagnostic role of serum glypican-3 in differentiating hepatocellular carcinoma from non-malignant chronic liver disease and other liver cancers. Journal of Gastroenterology and Hepatology. 2010;25(1):129–137. doi: 10.1111/j.1440-1746.2009.05988.x. [DOI] [PubMed] [Google Scholar]
- 24.Beale G., Chattopadhyay D., Gray J., Stewart S., Hudson M., Day C., Trerotoli P., Giannelli G., Manas D., Reeves H. AFP, PIVKAII, GP3, SCCA-1 and follisatin as surveillance biomarkers for hepatocellular cancer in non-alcoholic and alcoholic fatty liver disease. BMC Cancer. 2008;8, article 200 doi: 10.1186/1471-2407-8-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hippo Y., Watanabe K., Watanabe A., Midorikawa Y., Yamamoto S., Ihara S., Tokita S., Iwanari H., Ito Y., Nakano K., Nezu J.-I., Tsunoda H., Yoshino T., Ohizumi I., Tsuchiya M., Ohnishi S., Makuuchi M., Hamakubo T., Kodama T., Aburatani H. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Research. 2004;64(7):2418–2423. doi: 10.1158/0008-5472.CAN-03-2191. [DOI] [PubMed] [Google Scholar]
- 26.Nakatsura T., Yoshitake Y., Senju S., Monji M., Komori H., Motomura Y., Hosaka S., Beppu T., Ishiko T., Kamohara H., Ashihara H., Katagiri T., Furukawa Y., Fujiyama S., Ogawa M., Nakamura Y., Nishimura Y. Glypican-3, overexpressed specifically in human hepatocellular carcinoma, is a novel tumor marker. Biochemical and Biophysical Research Communications. 2003;306(1):16–25. doi: 10.1016/S0006-291X(03)00908-2. [DOI] [PubMed] [Google Scholar]
- 27.Capurro M., Wanless I. R., Sherman M., Deboer G., Shi W., Miyoshi E., Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125(1):89–97. doi: 10.1016/S0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Yang H., Xu H., Lu X., Sang X., Zhong S., Huang J., Mao Y. Golgi protein 73, not Glypican-3, may be a tumor marker complementary to α-Fetoprotein for hepatocellular carcinoma diagnosis. Journal of Gastroenterology and Hepatology. 2014;29(3):597–602. doi: 10.1111/jgh.12461. [DOI] [PubMed] [Google Scholar]
- 29.Nault J.-C., Guyot E., Laguillier C., Chevret S., Ganne-Carrie N., N'Kontchou G., Beaugrand M., Seror O., Trinchet J.-C., Coelho J., Lasalle P., Charnaux N., Delehedde M., Sutton A., Nahon P. Serum proteoglycans as prognostic biomarkers of hepatocellular carcinoma in patients with alcoholic cirrhosis. Cancer Epidemiology, Biomarkers & Prevention. 2013;22(8):1343–1352. doi: 10.1158/1055-9965.EPI-13-0179. [DOI] [PubMed] [Google Scholar]
- 30.Özkan H., Erdal H., Koçak E., Tutkak H., Karaeren Z., Yakut M., Köklü S. Diagnostic and prognostic role of serum glypican 3 in patients with hepatocellular carcinoma. Journal of Clinical Laboratory Analysis. 2011;25(5):350–353. doi: 10.1002/jcla.20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuda E., Kumada T., Toyoda H., Kaneoka Y., Maeda A., Okuda S., Yoshimi N., Kozawa O. Evaluation for clinical utility of GPC3, measured by a commercially available ELISA kit with Glypican-3 (GPC3) antibody, as a serological and histological marker for hepatocellular carcinoma. Hepatology Research. 2010;40(5):477–485. doi: 10.1111/j.1872-034X.2010.00624.x. [DOI] [PubMed] [Google Scholar]
- 32.Higgins J. P. T., Thompson S. G., Deeks J. J., Altman D. G. Measuring inconsistency in meta-analyses. British Medical Journal. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DerSimonian R., Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.Deeks J. J., Macaskill P., Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of Clinical Epidemiology. 2005;58(9):882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Altekruse S. F., Mcglynn K. A., Dickie L. A., Kleiner D. E. Hepatocellular carcinoma confirmation, treatment, and survival in surveillance, epidemiology, and end results registries, 1992–2008. Hepatology. 2012;55(2):476–482. doi: 10.1002/hep.24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuzaki R., Karp S. J., Omata M. New serum markers of hepatocellular carcinoma. Seminars in Oncology. 2012;39(4):434–439. doi: 10.1053/j.seminoncol.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Ho M., Kim H. Glypican-3: a new target for cancer immunotherapy. European Journal of Cancer. 2011;47(3):333–338. doi: 10.1016/j.ejca.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka M., Katayama F., Kato H., Tanaka H., Wang J., Qiao Y. L., Inoue M. Hepatitis B and C virus infection and hepatocellular carcinoma in China: a review of epidemiology and control measures. Journal of Epidemiology. 2011;21(6):401–416. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang T.-S., Shyu Y.-C., Turner R., Chen H.-Y., Chen P.-J. Diagnostic performance of alpha-fetoprotein, lens culinaris agglutinin-reactive alpha-fetoprotein, des-gamma carboxyprothrombin, and glypican-3 for the detection of hepatocellular carcinoma: a systematic review and meta-analysis protocol. Systematic Reviews. 2013;2(article 37) doi: 10.1186/2046-4053-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X.-F., Hu Z.-D., Liu X.-C., Cao Y., Ding C.-M., Hu C.-J. Diagnostic accuracy of serum glypican-3 for hepatocellular carcinoma: a systematic review and meta-analysis. Clinical Biochemistry. 2014;47(3):196–200. doi: 10.1016/j.clinbiochem.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Xu C., Yan Z., Zhou L., Wang Y. A comparison of glypican-3 with alpha-fetoprotein as a serum marker for hepatocellular carcinoma: a meta-analysis. Journal of Cancer Research and Clinical Oncology. 2013;139(8):1417–1424. doi: 10.1007/s00432-013-1458-5. [DOI] [PubMed] [Google Scholar]