Abstract
The Big Data movement in computer science has brought dramatic changes in what counts as data, how those data are analyzed, and what can be done with those data. Although increasingly pervasive in the business world, it has only recently begun to influence clinical research and practice. As Big Data draws from different intellectual traditions than clinical epidemiology, the ideas may be less familiar to practicing clinicians. There is an increasing role of Big Data in health care, and it has tremendous potential. This Demystifying Data Seminar identifies four main strands in Big Data relevant to health care. The first is the inclusion of many new kinds of data elements into clinical research and operations, in a volume not previously routinely used. Second, Big Data asks different kinds of questions of data and emphasizes the usefulness of analyses that are explicitly associational but not causal. Third, Big Data brings new analytic approaches to bear on these questions. And fourth, Big Data embodies a new set of aspirations for a breaking down of distinctions between research data and operational data and their merging into a continuously learning health system.
Keywords: automatic data processing, data mining, data collection, information systems, multilevel analyses
There is an emergent movement in computer science, the business world, and clinical research under the banner of “Big Data.” Its maximalist proponents argue that it is a fundamentally new approach that will transform and accelerate health care, correct decades of erroneous research, and reshape both clinical science and the business of caring for patients. Its critics often argue that it is newfangled dressing being put on the same old mistakes, yet again confusing “bigness” with “truth.”
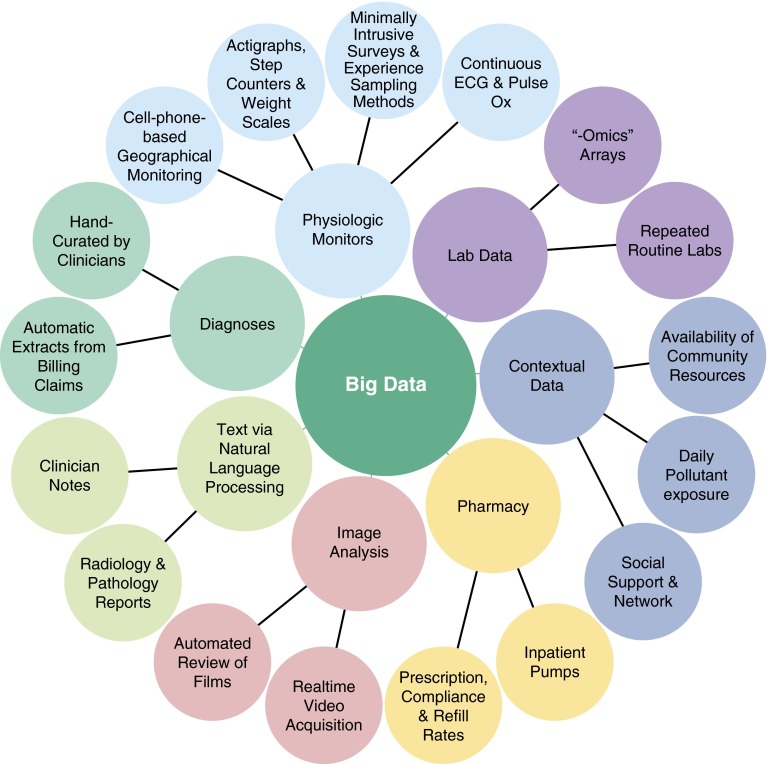
Figure 1.
Some sources of Big Data.
It is our contention that there is much rhetoric obscuring fundamentally important changes in the face of clinical research and, increasingly, clinical operations. Yet, those changes build on centuries of rigorous intellectual discipline. This Demystifying Data Seminar attempts to limit such rhetoric and identify four main strands within the Big Data movement (Table 1). We explicitly seek to explain Big Data in terms of those epidemiologic approaches more familiar to most clinicians. To make our biases clear: we believe these new Big Data approaches are a core part of our future armamentarium, and it is essential to integrate Big Data into our search for knowledge and improved clinical practice.
Table 1.
Four different ways in which a project may be “Big Data”
| Material: inclusion in analyses of data from new sources |
| Question: focus on the usefulness of locally stable associations and correlations even in the absence of causal evidence |
| Analytic method: new, often nonlinear tools for pattern recognition from computer science and other fields |
| Aspirations: close integration of routine clinical information systems to allow bidirectional flow between research and operations |
Big Data’s Success Outside of Medicine
Big Data has already made substantial changes in the world outside the hospital. Spurred by rapid advances in technology and the ubiquity of digital devices, our daily routines have been radically altered. Staying abreast of expansive networks of family, friends, colleagues, and our favorite personalities requires only a brief tour of digital walls, timelines, and feeds; real-time Big Data applications filter so that the individualized relevant content rises to the surface. Finding the fastest route to a new location takes a few taps on GPS-enabled mapping devices; along the way, crowd-sourced Big Data warns of traffic hotspots. Picking a movie no longer requires calling your friends for their advice or reading diverse critics; streaming media sites deliver personalized recommendations directly to your nearest screen. Along with that content comes a barrage of real-time, tailored advertisements based on our increasingly clear digital profiles. And, Google Now promises to provide us “the right information at just the right time.” Eric Topol suggests that the digital revolution outside the hospital is now priming the health-care industry for the “creative destruction of medicine.”
Big Data as New Material
At the heart of Big Data is the recognition of the newly ubiquitous base of interoperable digital computing underlying nearly all work being done in the modern health-care setting. Simply put, it is really cheap to put computers in everything, and so they are now there, from ventilators and health records to pumps and watches. This means that all sorts of things that were once expensive to record now trivially create electronic records of their own actions. These records can, at feasible cost, be aggregated into datasets. In the terms of Big Data proponents, a huge number of our actions have been “data-fied”—they now leave electronic traces. The technology to integrate these diverse data streams into unified datasets is increasingly mature and straightforward.
This changes the perspective on what the raw material for clinical studies should be. In the past, collecting data meant having a highly trained professional carefully identify and select exactly each measurement of a variable. Furthermore, it meant careful specification of the data to be collected to allow clear informed consent. Now it is possible to rapidly have a computer program simply accumulate those measurements from laboratory values, physiologic monitors, pharmacy disbursals, ventilators—and soon gene chips and other “-omics”-based arrays. As corporations move to further “data-fy” information, such as cell phone location information, the possibilities for data become increasingly richer. (But at the same time, we must note that the privacy and appropriate ethical controls, particularly when such data-fication is done for profit, are by no means fully clear or uniformly instituted.)
Even more exciting, there is the prospect of moving from structured data (think: data where the stuff already is in a coherent column in a spreadsheet) to so-called unstructured data (think: a big stack of progress notes written by clinicians). A key promise of Big Data is that the use of unstructured data that could include free-text clinical documentation (notes, reports, e-mails, social media) and even media files (pathology and radiology images, cardiac catheterization films, Google Glass images, videos). For the first time, the computational analytics and infrastructure are becoming available to tap into the vast amounts of information locked within unstructured data. Although such tools have existed previously in one-off forms, they have not been accessible in a scaleable technologic solution. As a result, Big Data now offers the hope of being able not only to collate and comb through millions of clinical notes but also to make sense of them automatically using techniques of natural language processing.
This leads to a number of changes in the material used as data, each with trade-offs. The benefit of Big Data is getting greatly more observations on many more variables for the same amount of time and money spent acquiring them. Furthermore, almost all the work is setting up the equipment to collect the first observation—the dataset can continue growing for very little additional work. The loss is that such measurements are done without the direct supervision of a human for each and every value, with the consequent loss of potential error correction. (Not, of course, that human-based measurement recording was ever perfect!) Big Data is a move from data where each point is carefully collected to an approach where data streams are curated. Big Data is also a move from data that were sought intentionally to an approach that capitalizes on the ubiquitous data being generated in the course of routine clinical care or by other processes. As a result, the data that are produced are rarely clean, in the sense of a finalized epidemiological dataset, but rather can be made clean enough.
Big Data maximalists argue that this is a qualitative disjuncture in the amount, granularity, and approach to data collection compared with what has gone before. Others might prefer to emphasize the continuity. Health services researchers have been examining Medicare claims for decades to derive fundamental insights into both epidemiology and health-system function. Such Medicare data are not created for research but instead are the electronic residue of the massive financial transactions that power the American health-care market. In that sense, compared with hand-collected clinical study data, Big Data has been around for years. Nonetheless, there are undeniably more and more data being stored and linked than ever before, and many people are actively engaged in this process, both within and without the label of Big Data.
Big Data as New Questions
Traditionally, clinical epidemiology and most of social science have given first priority to the task of identifying causal mechanisms. Observational data, at their best, were sought to reveal the same sort of strong mechanistic truths that experimental scientists have sought by other means.
In contrast, much of the Big Data movement is unapologetic that their interest is in locally stable associations and correlations. By this we mean that Big Data is particularly suited to finding patterns of association that have been true for “a while” and seem likely to be true for “a while longer.” Exactly how long “a while” is, they point out, depends on what one wants to do with that correlation. Because Big Data analytics are often designed to be continually updated, such instability is not problematic—if the association starts to change, it is argued, ongoing analytics will detect that change and report it. Big Data, in this sense, emphasizes providing current and radically up-to-date information over only enduring but potentially dated information.
Big Data therefore strongly challenges traditional clinical epidemiology about when a causal model is truly necessary. And in this way, the causation-obsessed stereotype of clinical epidemiology is revealed as a facade, because of course there has always been a strong interest in prognostic models within clinical epidemiology. Instead, the role of Big Data is to really foreground when “mere association” is not something to apologize for but rather precisely what we need. Big Data foregrounds that there are at least four different sorts of scientific questions that can be asked, and gives renewed prestige to the first three:
-
1.
Prognostic questions: Widely recognized in both clinical epidemiology and Big Data, these are questions of the form: “When a patient presents with characteristics X, Y, and Z, what is their probability of having outcome W”? Clinical epidemiology has long known that prognostic questions are best approached as associational questions, albeit with a debate as to the extent to which a causal foundation increases either clinician acceptance or long-term durability of the model. Big Data builds on the complexity of such questions but also emphasizes that similar questions can be asked on potentially much smaller scales, such as: “When I turn down the Pressure Support by 2 on a patient with an acute exacerbation of chronic obstructive pulmonary disease, how rapidly does the Pco2 equilibrate?” In general, prognostic questions require temporally stable associations, not underlying causal models.
-
2.
Predictive questions: Whereas prognostic questions emphasize what is going to happen, predictive questions ask what will happen if something different is done. Most commonly, predictive questions are studies that seek to ask which sorts of patients are most likely to have a favorable response (or an unwanted side effect) to a particular therapy. Wonderful examples of such predictive studies have been done in oncology, where predictive models now routinely guide decisions on chemotherapy. They play an important role in detecting heterogeneity of treatment effect—when a given therapy works differently in some subpopulations than in others. In general, predictive questions require temporally stable associations, not underlying causal models. Informal versions of such predictive models are ubiquitous in clinical medicine, often in the form of pearls passed from generation to generation of house-officer (e.g., “age + blood urea nitrogen = Lasix dose”). It is worth noting that the distinction between prognostic and predictive questions hinges on content knowledge and is not always recognized by Big Data scientists.
-
3.
Patterning questions: “How often do patients with acute myocardial infarction have classic substernal chest pain?” “Is there a constellation of symptoms that reliably diagnose ARDS?” Such questions have been at the heart of descriptive epidemiology, but they are increasingly Big Data’s forte. Big Data excels at pattern recognition based on a diverse set of measures—more formally, finding patterns in high-dimensional problem spaces. As discussed in the next section, Big Data has brought powerful new tools to clinical research to search for such patterns of association. In our opinion, the relative crudeness of such tools in clinical research in the past has led us to systematically undervalue such patterning questions. In general, patterning questions require temporally stable associations, not underlying causal models.
-
4.
Prescriptive questions: In contrast to the three previous sorts of questions, some questions are asked precisely because a bedside clinician wants to know how to change his/her behavior. That is, I want to know: “Should I start giving patients like X drug Y?” For these sorts of questions, conventional observational data absent a causal model are inadequate—they do not distinguish between (1) patients who get drug Y because they were doing better already, (2) patients who get better because of drug Y, and (3) patients for whom drug Y does nothing but who were getting otherwise better care and just happened to also get drug Y. These problems of selection and confounding by indication are not problems of dataset size but of the fundamental and often unobservable structure of decision making. In almost all cases, clinicians must demand causal evidence, because of the grave risk—repeatedly documented in critical care’s hasty jumps to adopt new therapies—of real harm if correlation is confused with causation for prescriptive questions.
The Big Data movement has brought important new tools to bear on all of these questions. Importantly, Big Data has encouraged a clarity of conversation that let us move beyond facile gestures to the “hierarchy of evidence” to appropriately match the application to data needs.
Big Data as New Analytic Methods
The basic tools of biostatistics as taught in medical school are the 2 × 2 table, simple distributional statistics (Chi-square and t tests), and multivariable regression. Such tools evolved in a regime where data were expensive and computation was hard, often being done by pen-and-paper by principal investigators themselves. In such a world, efficiency was prized, and fixed distributions were valued as useful approximations that saved work. Big Data analytic methods come from a tradition where data are cheap and computation is readily available. It is important to recognize that there is no single Big Data analytic method. Rather, we are witnessing an infusion of tools from computer science and engineering of various degrees of maturity and appropriateness, often lumped under the terms “machine learning,” “data mining,” or “data science.”
Unifying these Big Data techniques is that, in general, they are designed to let patterns emerge from the data. This contrasts with hypothesis testing in traditional clinical epidemiology; strict constructionists sometimes interpret hypothesis testing to mean that only a single prespecified hypothesis can ever be validly tested in any given look at the data.
In approaching Big Data analytic methods, Gilbert and colleagues (2010) propose that a useful distinction exists between approaches that privilege a single response variable as the target to be explained as opposed to those approaches that seek patterns among an array of variables, with none a priori more important than others (Table 2). Wang and Krishnan (2014) also provide an extensively referenced review of specific Big Data techniques used in clinical medicine. Interested readers are referred to these manuscripts for detailed citations; here our goal is to provide an overview.
Table 2.
Alternative data mining analytic methods
| Privilege a Single Variable | No Focal Variable | Changes over Time | |
|---|---|---|---|
| Example questions | “What are the predictors of death in patients with ARDS?” | “Are there important subtypes of severe sepsis?” | “What are the different ways in which patients develop ARDS and multiorgan dysfunction during their illness?” |
| Standard clinical epidemiologic tools | Regression discriminant analysis | Principal components analysis | None |
| Big Data tools | Neural networks | Cluster analysis (many methods including k-means clustering, nearest neighbor, spectral clustering) | Trajectory analysis |
| Genetic algorithms | |||
| Support vector machines | |||
| Tree-based methods (including classification and regression trees) |
Tools that privilege a single response variable are familiar in traditional statistics such as regression (where the response variable is the “outcome” variable or “y” variable). There are several Big Data approaches here, and there is active debate in the literature as to how to determine which approach is best suited to which problem. Many Big Data tools work on the basis of evolutionary learning; the most familiar may be artificial neural networks. In such a model, the analyst connects a series of inputs (the variables to be considered) to a set of interconnected “black boxes.” This software is then exposed to a learning regime, where each set of inputs is paired with a known outcome. (This would be called a derivation dataset in traditional epidemiology.) Connections within the black boxes that are associated with the correct outcome are strengthened; those that are associated with an incorrect outcome are weakened. This is done iteratively in a process that draws metaphorically on early synaptic pruning and strengthening until a stable network evolves. This network can then be switched from a “learning” phase to a “doing phase,” whereby it is given inputs and offers a prediction about the outcome.
There are, of course, a host of details about how exactly one structures the “black boxes” and strengthens or weakens connections in any given model. Conceptually similar models also draw on evolutionary metaphors, whereby a series of random “genetic algorithms” compete against each other and mutate to develop an approach to integrating inputs that most consistently predicts the outcomes during the learning stage. Other methods in this domain include “support vector machines” and “decision trees.” In each case, they are typically looking for nonlinear interactions between variables to allow consistent prediction of an outcome variable.
Other Big Data approaches do not privilege any particular variables, but instead seek “clusters” within an array of variables. Such cluster analysis, including principal component analyses, have long traditions in biostatistics and statistical genetics. Big Data has brought a renewed attention to such models, particularly with their extraordinary usefulness in large-scale genetic studies. Conceptually, clustering models try to identify consistent clumps in the variables, within which individuals are quite similar across an array of different variables. Clumps are then clumped together based on their similarity, again and again. The data are often presented in dendrograms (familiar to any student of evolution), so that the analyst can use some criteria (sometimes judgment, sometimes formalized as so-called “information criteria”) to decide what the useful level of aggregation is for a given problem.
A hybrid set of techniques are known as trajectory models. Such models privilege changes over time and seek to explore the dynamics of change in one or many variables together over time. In some trajectory models, a group of a priori trajectories are specified beforehand, and the analysis tries to see how many individuals clump nearest to each of these set options. In other cases, the trajectory models can allow a freer exploration for the best set of shapes for the trajectories over time—and then having found them, clump the individuals into groups best fitting each archetypal trajectory.
Because of the vast number of other fields in which Big Data–like projects are occurring—essentially any field that wishes to use computers to improve or replace the judgment of humans—these tools are constantly growing. There have, however, been numerous controversies about particular results, such as the failure to replicate Google Flu’s early successes. The development of new statistical methods is an area of ongoing and exciting research.
A difference has evolved between Big Data and traditional epidemiology in aesthetic approach to selecting analytic methods. In epidemiology, it is traditional to prespecify one’s sole primary analytic method, to avoid having one’s hypothesis testing “contaminated” by post hoc knowledge of the data. In contrast, in many Big Data applications, analysis proceeds akin to a methodological “bake-off.” Output from multiple models and approaches are compared against one another with a post hoc decision made to tailor the best method to the specific problem at hand. Although a single model may be ultimately chosen for implementation, the comparative performance of different model inputs and outputs can inform the “winner’s” final design. In fact, the final model selected for implementation may often be a more parsimonious or transparent model, even though its performance may be somewhat worse, depending on the constraints of the application and end user.
Across any of these analytic methods, traditional clinical epidemiological concerns about overfitting and external generalizability apply, and prudent clinicians may demand empiric verification before use in the same way that they would of any tool.
Big Data as New Aspirations
Finally—and perhaps most centrally to its appeal—Big Data has been increasingly viewed as the effector arm to the goal of a continuously learning health system. This agenda has been articulated by the Institute of Medicine across 15 volumes published by the National Academies Press under the aegis of their “Roundtable on Value and Science-Driven Health Care.” The goal of a learning health system, in brief, is to create a bidirectional rapid flow between the research arm and the operations arm of health-care systems. In such a world, every interaction with every patient becomes a data point that can be studied to discover new physiologic truths, evaluate the safety of current practice, and inform future clinical decisions and systems design. In this capacity, Big Data offers new opportunities to enhance this virtuous cycle by accelerating the pace of research and discovery while also improving the quality and depth of operational decision making. At the same time, such intensive monitoring also means that there are extensive computerized decision supports and protocols in place and that new research findings can be rapidly translated into improved protocols and better bedside decisions. In such a system, the current problems of persistent quality gaps, medical errors, and long lag times of diffusion are shortened or abolished. Similarly, every patient is learned from, so that no unnecessary suffering occurs before incrementally better ways to do things are put in place.
In such a world, Big Data fosters a seamless transition from Big Answers (one-time analyses to answer specific scientific questions and report findings) to Big Opportunities (ongoing analyses that impact patient outcomes or reduce costs at the point of care). One-shot Big Data will discover new answers and associations and will get papers published. But real-time Big Data, it is hoped, will be what propels our ability to intervene at the bedside or to handle surges and shifts in patient acuity without demanding heroic efforts from individual clinicians. As such, the “Bigness” depends on how quickly a decision maker needs the data. Analyzing 1 billion rows of data in 1 month is simply not that hard anymore. Analyzing a mere 1 million rows of new data, but doing so every 10 seconds to get data to intensive care unit clinicians to support decision making—that is where fundamentally new computational approaches are evolving. And, in this context, novel technologies that allow for rapid and dynamic visualization of large streams of data, it is hoped, can vastly improve high-quality decision making.
In its generalities, the advantages of such a Big Data–infused system are inarguable. Suffice it so say, however, that there are substantial challenges in its implementation. These challenges are assuredly not merely technical problems of linking all the data together or even of building continuous data-mining systems to examine all that data. Instead, there are fundamental challenges here, as in any sociotechnical system, of developing work routines that effectively integrate these new technologies. Examples of failure to integrate potentially revolutionary technology are legion. Within health care, there are certainly cautionary tales from even the comparatively mundane challenges of implementing computerized order entry and clinician alerts from physiologic monitors and smart pumps.
Conclusions
In sum, then, Big Data is an important but diverse intellectual movement seeking to bring new technologies of data acquisition, data integration, and data analysis into clinical research, hospital operations, and clinical practice. These trends will only accelerate for the foreseeable future, as they build on decades of others doing exactly those same things. Big Data will not solve fundamental challenges of either logical inference or of human behavior. But Big Data will continue to provide new knowledge and decision-making support for an array of real and pressing clinical problems.
Acknowledgments
Acknowledgment
The authors thank Jeremy Kahn for his thoughts on organizing the “Analytics” section and participants at the 34th Annual International Symposium on Intensive Care and Emergency Medicine Roundtable on Evidence-Based Care: New Directions for stimulating conversations on earlier versions of these ideas.
Footnotes
Supported by National Institutes of Health grant R21AG044752 and Veterans Affairs Health Services Research and Development grant IIR 11-109.
This work does not necessarily represent the views of the U.S. Government or the Department of Veterans Affairs.
Author Contributions: T.J.I. conceived and designed the review and drafted the manuscript. V.L. provided critical revision of the manuscript for important intellectual content, including significant rewriting of sections.
Author disclosures are available with the text of this article at www.atsjournals.org.
Recommended Reading
- Celi LA, Mark RG, Stone DJ, Montgomery RA. “Big data” in the intensive care unit: closing the data loop. Am J Respir Crit Care Med. 2013;87:1157–1160. doi: 10.1164/rccm.201212-2311ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert K, Sànchez-Marrè M, Codina VC. Choosing the right data mining technique: classification of methods and intelligent recommendation. In:Swayne DA, Yang W, Voinov AA, Rizzoli A, Filatova T.Proceedings of the iEMSs Fourth Biennial Meeting: International Congress on Environmental Modelling and Software (iEMSs 2010). International Environmental Modelling and Software Society. July 2010, Ottawa, Canada [Google Scholar]
- Hidalgo C.Saving Big Data from big mouths. SA Forum. Scientific American. 2014. [accessed 2014. Apr 29]Available from: http://www.scientificamerican.com/article/saving-big-data-from-big-mouths/
- Lazer D, Kennedy R, King G, Vespignani A. Big data. The parable of Google Flu: traps in big data analysis. Science. 2014;343:1203–1205. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- Mayer-Schönberger V, Cukier K.Big data. New York: Houghton Mifflin Harcourt; 2012 [Google Scholar]
- Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman LW, Moody G, Heldt T, Kyaw TH, Moody B, Mark RG. Multiparameter Intelligent Monitoring in Intensive Care II: a public-access intensive care unit database. Crit Care Med. 2011;39:952–960. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol E.The creative destruction of medicine: how the digital revolution will create better health care. New York: Basic Books; 2012 [Google Scholar]
- Wang W, Krishnan E. Big data and clinicians: a review on the state of the science. JMIR Medical Informatics. 2014;2:1–11. doi: 10.2196/medinform.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]