Abstract
Rationale: Death from infection is a highly heritable trait, yet there are few genetic variants with known mechanism influencing survival during septic shock.
Objectives: We hypothesized that a synonymous coding variant in the IL-1 receptor antagonist gene (IL1RN), rs315952, previously associated with reduced risk for acute respiratory distress syndrome, would be functional and associate with improved survival in septic shock.
Methods: We used a human endotoxin (LPS) model of evoked inflammatory stress to measure plasma IL-1 receptor antagonist (IL1RA) following low-dose Food and Drug Administration–grade LPS injection (1 ng/kg) in 294 human volunteers. RNA sequencing of adipose tissue pre- and post-LPS was used to test for allelic imbalance at rs315952. In the Vasopressin and Septic Shock Trial cohort, we performed a genetic association study for survival, mortality, and organ failure–free days.
Measurements and Main Results: Adipose tissue displayed significant allelic imbalance favoring the rs315952C allele in subjects of European ancestry. Consistent with this, carriers of rs315952C had slightly higher plasma IL1RA at baseline (0.039) and higher evoked IL1RA post-LPS (0.011). In the Vasopressin and Septic Shock Trial cohort, rs315952C associated with improved survival (P = 0.028), decreased adjusted 90-day mortality (P = 0.044), and faster resolution of shock (P = 0.029).
Conclusions: In European ancestry subjects, the IL1RN variant rs315952C is preferentially transcribed and associated with increased evoked plasma IL1RA and with improved survival from septic shock. It may be that genetically determined IL1RA levels influence survival from septic shock.
Keywords: septic shock, polymorphism, functional genetic variant, RNA-seq
At a Glance Commentary
Scientific Knowledge on the Subject
Death from infection is a highly heritable trait, yet there are few replicated genetic variants associated with death from septic shock, and fewer still with known molecular function.
What This Study Adds to the Field
The present study demonstrates that a synonymous IL1RN single nucleotide variant is a site for preferential transcription of one allele in adipose tissue and is associated with higher evoked plasma IL-1 receptor antagonist in response to endotoxin and with increased survival in septic shock.
Septic shock remains a common cause of death in the intensive care unit, with mortality rates as high as 35% (1, 2). Although improved recognition of the syndrome and careful attention to early antibiotic therapy (3) and hemodynamic targets (4) have decreased mortality over time (1), there remains no specific pharmacotherapy for septic shock. Furthermore, among patients meeting criteria for septic shock, there may be unappreciated heterogeneity in molecular pathophysiology (5–7) or genetic predisposition (8, 9) that influences response to treatment or outcome. Several studies have suggested that either high initial or persistent proinflammatory cytokine levels, including IL-1β, in the plasma may correlate with organ dysfunction and death (7, 10, 11).
We previously identified a synonymous coding variant in the IL-1 receptor antagonist gene (IL1RN) associated with lower risk of developing the acute respiratory distress syndrome (ARDS) in three critically ill populations (combined odds ratio, 0.81; P = 4.2 × 10−5) (12). The largest population, with more than 2,000 subjects, had sepsis as the primary risk factor for ARDS (12). The IL1RN gene encodes for IL-1 receptor antagonist protein (IL1RA), the naturally occurring antagonist for IL-1α and IL-1β. IL1RA competes with IL-1α and IL-1β to bind the IL-1 receptor 1 (IL1R1), yet IL1RA does not trigger IL1R1 signaling, and instead acts as a brake on inflammasome activation (13–15).
Among critically ill subjects, we demonstrated that the ARDS low-risk IL1RN single nucleotide polymorphism (SNP) rs315952C associated with higher plasma levels of IL1RA (12), consistent with a hypothesis that genetically determined higher endogenous plasma IL1RA levels might mitigate ARDS risk. Other groups have examined the association between sepsis outcomes and IL1RN variation, and found a variable number of tandem repeat polymorphism known as allele 2 (IL1RN*2) to associate with increased susceptibility to sepsis (16, 17) or increased risk of death from sepsis (11, 18), although the effects of IL1RN*2 on secreted IL1RA protein have been inconsistent (11, 19). In addition, a well-described IL1RN promoter SNP rs4251961C has been consistently associated with decreased IL1RA levels in response to infection or pathogen-associated molecular patterns (20–23). These three IL1RN variants (rs4251961, IL1RN*2 tagged by rs419598, and rs315952) exhibit little to no linkage disequilibrium between one another in either European or African ancestry (EA, AA) populations (24).
Given these findings, we sought to identify the genetic mechanism by which rs315952C might associate with higher evoked plasma IL1RA and to test prior IL1RN candidate SNPs for association with evoked IL1RA. We used intravenous low-dose endotoxin (LPS) as a human experimental model of inflammatory stress to understand the dynamics of IL1RA in response to a standardized insult. In addition, we sought to test whether rs315952C demonstrated any protective associations during septic shock, including improved survival, faster resolution of shock, or reduced time on the ventilator. These complimentary approaches allowed us to investigate the mechanistic effects of rs315952 both in response to a uniform inflammatory stimulus and during clinical septic shock. Our primary hypothesis was that rs315952 would be more strongly associated with innate immunity-evoked IL1RA, and that this would translate into improved survival during septic shock. Some of the results of this study have been previously reported in the form of abstracts (25, 26).
Methods
Study Populations
GENE study.
The Genetics of Evoked response to Niacin and Endotoxemia (GENE) study recruited 294 healthy nonobese subjects (27). The protocol was approved by the University of Pennsylvania Institutional Review Board, had regulatory oversight by the US Food and Drug Administration (LPS: IND 5,984), and was monitored by a National Institute of Health–appointed data safety and monitoring board. Subjects in GENE were admitted to the clinical translational research center inpatient unit for administration of intravenous endotoxin (LPS; 1 ng/kg) and were monitored closely as described (27). Serial blood draws were collected immediately before and 1, 2, 4, 6, 12, and 24 hours post-LPS for plasma. Gluteal adipose biopsy using a liposuction catheter under local anesthesia was performed at baseline and 4 hours post-LPS as described for RNA extraction (28, 29).
VASST cohort.
The Vasopressin and Septic Shock Trial (VASST) was a multicenter, double-blind, randomized controlled trial evaluating vasopressin versus norepinephrine for septic shock (30). The study enrolled 778 patients with septic shock and requiring at least 5 μg/min norepinephrine infusion; details have been published (30, 31). The research ethics boards of all participating institutions approved the trial, and written informed consent was obtained from all patients or their authorized representatives, including permission to perform downstream mechanistic testing. Of 778 patients in the VASST trial, 632 had available DNA and were included in this study (8, 31, 32). A subgroup of subjects, determined by study personnel availability, also had plasma collected at study enrollment (n = 399). Clinical outcomes included mortality at 28 and 90 days; site of infection; Acute Physiology and Chronic Health Evaluation II (APACHE II) score; duration of vasoactive drug infusion; and days free of moderate, severe, or extreme organ failure as defined by the Brussels criteria (33).
Genotyping and Protein Analysis
Genomic DNA was extracted from whole blood using a QIAmp kit (Qiagen, Missaugua, ON, Canada). The GENE study was genotyped with the Illumina (San Diego, CA) Infinium Exome chip, filtered for rs315952, rs4251961, and rs419598 and SNPs within 1 kb of IL1RN. Plasma IL1RA was measured by ELISA (R&D Systems, Minneapolis, MN) in duplicate. The VASST cohort was genotyped using the Human 1M Duo platform (Illumina) and filtered for rs315952 and SNPs within 1 kb of IL1RN. Plasma IL1RA and IL1β levels were measured in VASST in a subgroup of patients by human multiplex kits (EMD Millipore, Billerica, MA) using an antibody-linked magnetic bead assay according to the manufacturer’s recommendations. For RNA sequencing (RNA-seq), RNA was extracted from adipose using RNeasy Lipid Tissue total RNA mini kit (Qiagen, Valencia, CA), and prepared and sequenced as previously described (29, 34). Results were filtered for RNA-seq reads flanking rs315952 (chr2:113890304), and the number of reads carrying each allele (C or T) was counted.
Statistical Analysis
Plasma levels in the GENE population were analyzed by nonparametric methods between genotypes at specific time points and for the area under the IL1RA curve, determined using the trapezoidal rule. Additive (nonparametric trend) and dominant (Wilcoxon rank sum) genetic models were assessed. We adopted a dominant model because there were only 14 homozygous rs315952CC subjects, limiting power in this stratum. To analyze the data across all time points, accounting for large variation in concentration by endotoxin stimulus and for repeated measures within each individual, we quantile-transformed data at each time point and used a linear mixed effects model with an individual-specific random effect to control for the correlations among repeated measures within individuals (35). Analyses were separate by genetic ancestry given significant differences in IL1RN gene structure (36). To test whether our results were influenced by genetic ancestral substructure, we included the first three principal components of genetic ancestry determination along with covariates sex and body mass index in a quantile regression model for peak IL1RA response. We also regressed transformed plasma IL1RA levels on all polymorphic typed SNPs (n = 18) within 1 kb of the IL1RN gene to assess whether the determinants of baseline and evoked IL1RA were distinct.
We used RNA-seq to analyze for allelic imbalance (AI) by quantifying transcription from both paternal and maternal haplotypes using individuals that are heterozygous at the test SNP (37). To test for AI, we compared the number of RNA-seq reads with C versus T allele by a one degree of freedom chi-square goodness of fit test, with the null hypothesis being that 50% of the reads would contain each allele at this locus.
In VASST, survival analysis was performed by Cox proportional hazards methods. Association between genotype and mortality was tested by chi-square and logistic regression, and between genotype and continuous outcomes by linear regression, assuming an additive genetic model and adjusting for APACHE II score and the first three principal components of genetic ancestry determination. Analyses were restricted to EA given IL1RN gene structure and the low numbers of non-EA subjects. Plasma levels were inspected by genotype and analyzed by additive model using analysis of variance with Bonferroni adjustment, or by recessive model using Student t test, on log-transformed values. A recessive model was evaluated given the observed data distribution. For all analyses, a two-sided P value less than 0.05 was considered significant. Additional details including genotyping quality assurances, determination of genetic ancestry, ELISA and multiplex assay characteristics, and power considerations are provided in the online supplement.
Results
IL1RN Variation and the Response to Intravenous Endotoxin
The GENE population is described in Table 1. Because our prior associations with rs315952 were present only for EA subjects (12), we initially focused on this population. The SNP displayed Hardy-Weinberg equilibrium (P = 0.69) and its observed minor allele frequency was 0.27, comparable with HapMap EA populations (36). At baseline, EA carriers of rs315952C had slightly increased plasma IL1RA, and this effect was magnified post-LPS (Table 2), with a peak additive effect at 4 hours (Figure 1). Peak IL1RA response remained associated with rs315952 when adjusting for the first three principal components of genetic ancestry (P = 0.008). At 24 hours post-LPS, IL1RA levels remained significantly associated with rs315952C. In a repeated measures mixed effects model conditioned on the individual (Figure 2), rs315952C was associated with increased IL1RA levels whether analyzed by additive (P = 0.049) or dominant (P = 0.023) models of genetic risk. In a cis quantitative trait locus analysis considering 18 genotyped loci falling in the IL1RN region, rs315952 ranked highest for peak response, third for area under the curve, and 18th for baseline IL1RA (see Table E1 in the online supplement).
Table 1.
GENE Population Clinical Characteristics Stratified by Genotype
| TT (n = 122) | CT (n = 115) | CC (n = 35) | P Value | |
|---|---|---|---|---|
| Age | 25.4 ± 6.7 | 26.1 ± 6.7 | 26.5 ± 7.2 | 0.33 |
| Female sex | 55 (47.8%) | 16 (45.7%) | 19 (76.0%) | 0.33 |
| Body mass index | 23.7 ± 2.8 | 24.1 ± 3.0 | 23.4 ± 2.8 | 0.87 |
| African ancestry | 28 (23.0%) | 47 (40.9%) | 21 (60.0%) | <0.001 |
| European ancestry | 94 (77.1%) | 68 (59.1%) | 14 (40.0%) |
Definition of abbreviation: GENE = Genetics of Evoked response to Niacin and Endotoxemia.
Values shown are mean ± standard deviation or number (proportion). Groups were compared in an additive fashion by linear regression for continuous variables and logistic regression for categorical ones.
Table 2.
rs315952C Is Associated with Higher Baseline and Evoked IL1RA Post-LPS in European Ancestry Subjects
| TT (n = 93) | CT (n = 68) | CC (n = 14) | P Value | Adjusted P Value* | |
|---|---|---|---|---|---|
| Additive model | |||||
| Baseline IL1RA, pg/ml | 101.8 (84.5–146.6) | 120.1 (98.0–157.8) | 108.07 (87.2–141.9) | 0.12 | 0.073 |
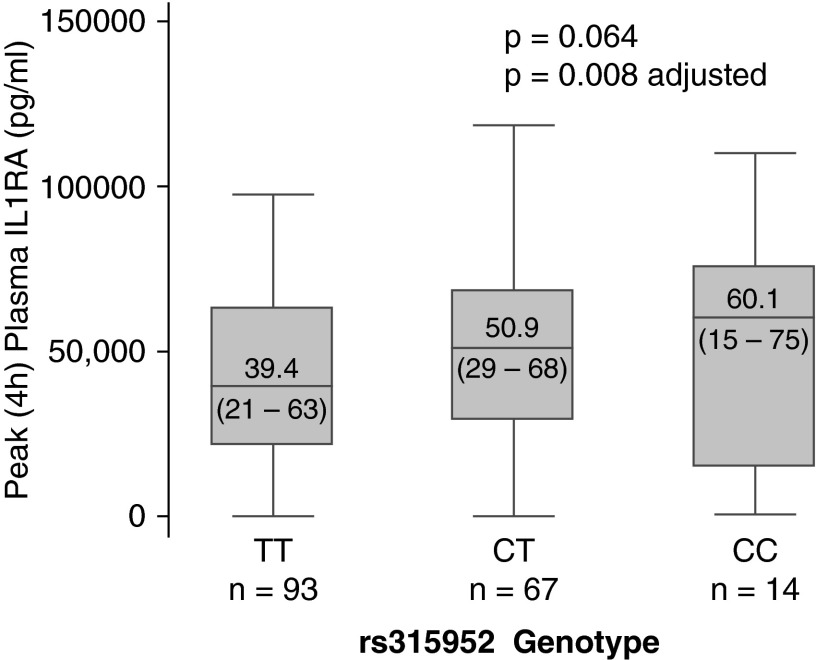
| IL1RA at 4 h, ng/ml† | 39.4 (21.5–63.1) | 50.9 (28.8–68.3) | 60.1 (14.8–75.5) | 0.064 | 0.008 |
| IL1RA at 24 h, pg/ml | 232.8 (183.0–286.1) | 272.2 (221.7–338.1) | 232.2 (184.6–347.9) | 0.064 | 0.016 |
| AUC IL1RA, ng/ml† | 51.4 (26.9–86.1) | 64.3 (38.2–90.8) | 74.5 (21.5–99.3) | 0.11 | 0.013 |
| Dominant model | |
||||
| Baseline IL1RA, pg/ml | 101.8 (84.5–146.6) | 117.3 (94.5–157.4) |
0.032 | 0.039 | |
| IL1RA at 4 h, ng/ml† | 39.4 (21.5–63.1) | 52.2 (28.8–68.8) |
0.044 | 0.011 | |
| IL1RA at 24 h, pg/ml | 232.8 (183.0–286.1) | 268.6 (216.8–338.8) |
0.009 | 0.015 | |
| AUC IL1RA, ng/ml† | 51.4 (26.9–86.1) | 66.0 (37.9–92.4) | 0.075 | 0.026 | |
Definition of abbreviation: AUC = area under the IL1RA curve.
Median (interquartile range) values are shown. Peak IL1RA response was at 4 hours post-LPS, and no shift in peak response was observed by genotype.
Given the low number of homozygous CC individuals, we collapsed CT and CC for a dominant model and analyzed by rank sum test or quantile regression adjusting for sex and body mass index.
Additive genetic models were tested by nonparametric trend and adjusted for sex, body mass index, and the first three components of genetic ancestry using quantile regression.
Change in scale to ng/ml for peak response and area under the IL1RA curve.
Figure 1.
Peak LPS-evoked plasma IL1RA increases with increasing copies of rs315952C. Adjusted for sex and body mass index.
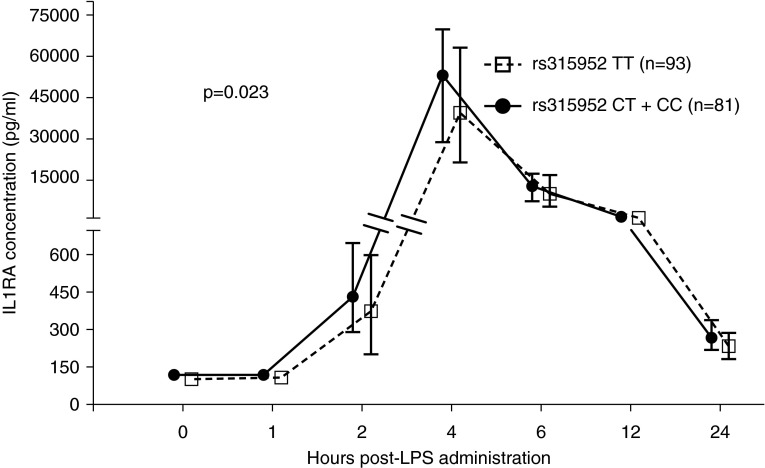
Figure 2.
Carriers of rs315952C have increased LPS-evoked plasma IL1RA levels. At each time point, the median (dot or square) and interquartile range (whiskers) are shown, stratified by rs315952C allele carriage. The y axis is interrupted to allow discrimination at the earlier time points, when the concentration of plasma IL1RA was 100-fold lower. Repeated measures analysis was performed by linear mixed effects model with an individual-specific random effect and with quantile transformation of the data at each time point.
We tested previously reported IL1RN variants for association with baseline and evoked IL1RA. The IL1RN promoter SNP rs4251961, previously associated with lower IL1RA levels (21, 23), associated with lower baseline IL1RA levels but not with altered peak IL1RA response or area under the IL1RA curve (see Table E2). In contrast, rs419598, a tag for IL1RN*2 with perfect linkage disequilibrium in European populations (r2 = 1.0) (12, 38), showed no association with baseline or evoked IL1RA (see Table E3). As shown in Table E1, the local determinants of LPS-evoked plasma IL1RA were highly distinct from those determining baseline plasma IL1RA. Results for AA GENE subjects (n = 94) are presented in the online supplement (see Table E4). No association between rs315952C and plasma IL1RA levels was observed in AA subjects.
Because rs315952C is a synonymous coding SNP in the terminal exon of IL1RN and not previously shown to fall within a transcription factor binding site (39, 40), we hypothesized that AI might be the genetic mechanism responsible for higher plasma IL1RA levels (37). Using RNA obtained from adipose biopsy at baseline and 4 hours post-LPS in nine subjects heterozygous for rs315952C/T (chr2:113890304), we performed RNA sequencing (RNA-seq) at a median read depth of 414 million (range, 298–492 million) reads per sample. Results for EA subjects are shown in Table 3. Of nine heterozygous subjects, five had fewer than 20 reads at this locus and were excluded, leaving two EA subjects and two AA subjects (see Table E5) for the AI analysis. As anticipated, IL1RN was up-regulated in adipose post-LPS, with log2(fold change) = 1.71, P = 5.0 × 10−5. Furthermore, in both EA subjects, rs315952 demonstrated strong AI favoring the C allele at baseline and this increased post-LPS, with the most dramatic instance being a 80–20% imbalance post-LPS (P = 2.1 × 10−36).
Table 3.
RNA-Seq Analysis at rs315952 (chr2:113890304) in Adipose Tissue for Heterozygous European Ancestry Subjects Indicates Strong Allelic Imbalance Favoring the C Allele
| Pre-LPS |
Post-LPS |
|||||||
|---|---|---|---|---|---|---|---|---|
| C Allele Counts | T Allele Counts | Proportion C Reads | P Value | C Allele Counts | T Allele Counts | Proportion C Reads | P Value | |
| A | 112 | 36 | 0.7568 | 4.18 × 10−10 | 358 | 91 | 0.7973 | 2.10 × 10−36 |
| B | 163 | 98 | 0.6245 | 5.74 × 10−5 | 289 | 198 | 0.5934 | 3.73 × 10−5 |
Of four European Ancestry subjects with adipose RNA and heterozygous (C/T) at rs315952, only subjects “A” and “B” met our filtering criteria of greater than or equal to 20 reads at this locus. The expected proportion of reads containing the C allele was 0.50, or 50%. Both subjects demonstrated strong allelic imbalance favoring the C allele (bold) both at rest and post-LPS.
rs315952 in the VASST Septic Shock Cohort
Characteristics of the VASST cohort subjects with available genotyping are shown in Table 4; rs315952 displayed Hardy-Weinberg equilibrium (P = 0.86). Nonsurvivors of septic shock were older and had higher APACHE II scores in addition to other organ failures. Baseline clinical characteristics were largely similar across genotype groups with CC homozygotes being slightly younger and displaying lower APACHE II scores, as shown in Table E6.
Table 4.
Characteristics of the VASST Population with Available DNA for Genotyping
| 90-d Nonsurvivors (n = 287) | 90-d Survivors (n = 345) | P Value | |
|---|---|---|---|
| Age | 63.5 ± 16 | 57.7 ± 17 | <0.001 |
| Female | 118 (41.1%) | 144 (41.7%) | 0.72 |
| Ancestry | |||
| European | 232 (80.8%) | 298 (86.4%) | 0.065 |
| Asian | 24 (8.4%) | 17 (4.9%) | 0.10 |
| African | 9 (3.1%) | 5 (1.5%) | 0.18 |
| Site of infection | |||
| Lung | 129 (45.0%) | 147 (42.6%) | 0.56 |
| Abdomen | 73 (25.4%) | 97 (28.1%) | 0.39 |
| Other | 85 (29.6%) | 101 (29.3%) | |
| Infectious pathogen | |||
| Gram-positive bacteria | 79 (27.5%) | 110 (31.9%) | 0.32 |
| Gram-negative bacteria | 55 (19.2%) | 83 (24.1%) | 0.19 |
| Fungal or viral | 38 (13.2%) | 41 (11.9%) | 0.40 |
| Not identified | 131 (46.8%) | 143 (41.5%) | 0.18 |
| APACHE II | 28.0 ± 9 | 25.3 ± 7 | <0.001 |
| Randomized to vasopressin | 141 (49.1%) | 185 (53.6%) | 0.26 |
| Plasma available | 164 (57%) | 235 (68%) | 0.004 |
| Acute organ failure | |||
| Lung | 286 (99.7%) | 334 (96.8%) | 0.032 |
| Kidney | 246 (85.7%) | 161 (46.7%) | <0.001 |
| Liver | 236 (82.2%) | 121 (35.1%) | <0.001 |
| Coagulation | 269 (96.4%) | 289 (87.3%) | <0.001 |
| Central nervous system | 269 (93.7%) | 289 (83.8%) | <0.001 |
| Days alive and free of vasopressors, 28 d | 0 (0–9) | 23 (19–25) | <0.001 |
| Days alive and free of ventilator, 28 d | 0 (0–4) | 18 (10–23) | <0.001 |
Definition of abbreviations: APACHE = Acute Physiology and Chronic Health Evaluation; VASST = Vasopressin and Septic Shock Trial.
Variables are displayed as mean ± standard deviation, median (25–75 percentile range), or as number (percentage). Site of infection and infectious pathogen were not exclusive and thus the total may exceed 100%. Acute organ failures and organ failure–free days were defined by Brussels criteria within the first 28 days. Comparisons were made by t test, Wilcoxon rank sum test, chi-square, or Fisher exact test as appropriate. PF ratio = ratio of PaO2 to FiO2.
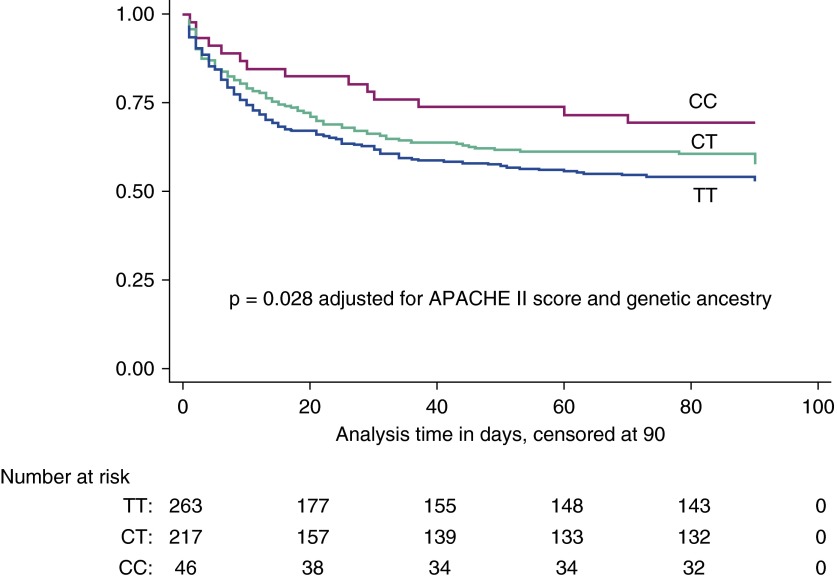
Survival curves for EA subjects stratified by rs315952 genotype are shown in Figure 3. The rs315952 genotype satisfied the proportional-hazards assumption (Schoenfeld residual test, P = 0.53) and demonstrated a reduced hazard of death with increasing copies of the C allele (hazard ratio, 0.80; 95% confidence interval, 0.65–0.99; P = 0.038). This result was unchanged by adjustment for APACHE II score and for the first three components of the genetic ancestry multidimensional scaling analysis (hazard ratio, 0.79; 95% confidence interval, 0.64–0.98; P = 0.028). In addition, rs315952C was associated with decreased 90-day adjusted mortality (hazard ratio, 0.75; 95% confidence interval, 0.51–0.99; P = 0.044) and increased days alive and free of cardiovascular system failure (P = 0.041) (see Table E7). Ventilator-free days were higher (P = 0.061) with increasing copies of the rs315952C allele, although this result was not statistically significant (see Table E7). Eighty SNPs within 1 kb of IL1RN were genotyped on the Illumina 1M platform (24). We performed logistic regression of 90-day mortality with all 80 IL1RN SNPs assuming an additive model of genetic risk, and rs315952C was the fifth most significantly associated P value, at P = 0.057, as displayed in Figure E1.
Figure 3.
By Cox proportional hazards regression, rs315952C is associated with improved 90-day survival among European ancestry subjects of the Vasopressin and Septic Shock Trial cohort in an additive fashion (P = 0.038 unadjusted, P = 0.028 adjusted for Acute Physiology and Chronic Health Evaluation [APACHE] II score and genetic ancestry). The y axis indicates the proportion alive. Magenta line = CC homozygotes; green line = CT heterozygotes; blue line = TT homozygotes.
Initial plasma levels of IL-1β and IL1RA were available for 399 subjects, 51% of the overall cohort (see Table E8). Plasma IL-1β (P = 0.038) was lower for homozygous carriers of rs315952CC, whereas we were unable to demonstrate significant difference in plasma IL1RA (P = 0.19). The pair-wise correlation between plasma IL1RA and IL-1β levels was very strong, with r2 = 0.90.
Discussion
Variation of genetic structure across human ancestral populations has been shown to bear strong marks of selection, where evolutionary pressures have maintained the presence of variant alleles at increased frequency (41–45). Cytokine genes have notably diverse genetic structures across global populations (46), which may be attributable to evolutionary forces including injury or infection shaping human genetic diversity. It may be that by understanding genetic risk factors for death during sepsis, we will uncover sepsis endotypes that may have unique pathophysiology or may respond differentially to specific therapy. We investigated whether rs315952C may identify a genetically determined endotype of septic shock with hypothesized more efficient transcription of IL1RN, higher plasma IL1RA, and improved survival.
To study the genetic contribution to evoked plasma IL1RA more precisely, we turned to an experimental model of inflammatory stress with intravenous LPS, ensuring that each subject received an identical stimulus. Low-dose LPS reliably induced a 100-fold increase in evoked plasma IL1RA. In this system, we replicated the association between rs315952C and increased plasma IL1RA levels (47), and this effect was most pronounced at the peak inflammatory response compared with baseline. Interestingly, the previously studied genetic variants affecting IL1RA response, rs4251961 and the VNTR tagged by rs419598, did not influence peak evoked IL1RA, and only rs4251961 associated with resting plasma IL1RA levels. We interpret these results as implicating rs315952 as an important locus for regulating evoked inflammation, such as might occur during septic shock.
Our RNA sequencing analyses implicate adipose tissue AI favoring transcription of the C allele in EA individuals and suggest that rs315952 is a functional SNP. Adipose tissue strongly expresses IL1RN and is a significant source of plasma IL1RA (48). Body mass index explains a significant proportion of the observed variance in baseline plasma IL1RA (27, 47). In addition, adipose explants secrete IL1RA in response to LPS (48), making adipose a relevant tissue to investigate. However, our group has previously shown dramatic tissue specificity to basal and LPS-evoked gene expression (34), whereas others have shown tissue specificity to AI responses within the same individual (49, 50). The AI that we detected in two EA individuals was statistically impressive, yet the sample size is small. In the future, it will be important to test for similar imbalance in transcription in monocytes and neutrophils, cell types we believe to substantially contribute to plasma IL1RA during sepsis, and to confirm these findings in a larger population. Indeed, optimally such work would test for AI in leukocytes harvested directly from patients with septic shock, and would confirm more efficient IL1RN transcription and higher plasma IL1RA levels in comparably timed blood samples.
To confirm the significance of rs315952C as a functional variant, we tested the SNP’s association with outcomes following septic shock. In the VASST cohort, we demonstrate an association between carriage of the C allele and improved survival, and faster resolution of shock, a direction of effect consistent with our prior results (12). We also report for the first time that homozygous carriers of rs315952C (CC) demonstrate lower plasma IL-1β level, consistent with the hypothesis that reduced IL-1β–driven signaling may improve outcomes during sepsis.
We did not identify a consistent relationship between rs315952C and plasma IL1RA in VASST, in contrast to the findings in GENE and in our and other’s prior publications (12, 47). Although this was surprising, it may be that the timing of plasma draw in VASST, with a median of 12-hour duration of septic shock (30), a late manifestation of a severe systemic inflammatory response, influenced results. Our previous report used plasma primarily drawn in the emergency department with either severe sepsis or severe trauma (12). If the C allele of rs315952 is more efficiently transcribed, as our RNA-seq data suggest, it may be that earlier up-regulation of IL1RA acts as a brake on subsequent IL-1β–driven self-induction (51), and might explain a lower IL-1β and IL1RA level observed at a later period because IL-1β also regulates IL1RA (14, 52). Previous human and primate studies report that exogenous IL1RA therapy lowers plasma IL-1β and even IL-6 levels, demonstrating the potential for IL1RA to have a broad-based modulation of the cytokine inflammatory response (53, 54). Although this is an attractive theory, it does not explain the observed differences between homozygous and heterozygous carriers of the C allele, because one would predict that each C allele would lower IL-1β in an additive fashion. Several other differences in the populations studied may account for the observed results, including differences between a clinical trial population and observational cohorts, differences between trauma- and sepsis-induced inflammatory responses, or between sepsis and septic shock. Ideally, we would use a prospective cohort of patients with sepsis with uniform repeat blood and leukocyte mRNA sampling to replicate the associations observed in GENE.
Our results in both the GENE and VASST populations implicate rs315952 as a variant modifying IL1RA response and septic outcomes in EA populations, with attenuated or no effect in non-EA subgroups. In GENE, AA subjects demonstrated no association between rs315952C and plasma IL1RA levels, and AI at the SNP is either absent or slightly favors the T allele. It may be that we were underpowered to detect a difference in AA populations, but it is also striking how the AI analysis yielded very discordant results for the two ancestries. Although our data suggest this locus is a cis regulatory variant or enhancer in EA subjects (49), the region is not an area of known enhancer function by the VISTA Enhancer database (55), nor is it predicted to be a transcription factor binding site by the ChIP-seq experiments performed by the Encyclopedia of DNA Elements project (39). A potential explanation for how ancestral IL1RN gene structure might result in different allele-specific results at our locus would be that rs315952 alters binding of the CCCTC-binding factor, a regulator of chromatin and transcription factor binding. Prior work has established that CCCTC-binding factor binding can vary in an allele-specific manner (56–58), and the divergent linkage disequilibrium across chromosome 2q13 in ancestral populations might influence overall conformation in a population-specific manner. Additional studies to understand chromosome conformation at this locus will be important to pursue.
Our investigations had some limitations. We performed multiple tests in the GENE population centered on the hypothesis that rs315952 would associate with increased evoked IL1RA, and our multiple testing may have inflated the overall type I error. The associations we observed between rs315952 and plasma IL1RA levels would not withstand a stringent Bonferroni adjustment for multiple testing; however, they are not truly independent tests. Because the analyses were conducted in a previously completed observational study and clinical trial, our sample size was fixed and may only provide limited power to detect modest genetic effects on complex traits. In GENE, we were powered to detect a difference in half of one standard deviation in means for plasma IL1RA, whereas in VASST, our minimal detectable relative risk for genotype on mortality was 1.49 (59). For both populations, these are moderate to large effect sizes. Our RNA-seq data, while compelling, involved a small number of individuals. Our power limitation was more potent in the AA population in the GENE study, and the VASST cohort lacked sufficient non-EA subjects to make inferences in African or Asian populations. In addition, we acknowledge the inability of a low-dose LPS injection to precisely model the complexity of septic shock, yet we believe the controlled nature of the LPS challenge is ideal for studying evoked response to a uniform stimulus. Furthermore, there are ample data that the inflammatory response to severe infections and to LPS share many features (60–63). We attempted to measure IL-1β in the plasma of all GENE subjects but for many subjects, the level of plasma IL-1β seemed to be at the limits of detection at peak response (4 h), and undetectable at other times. Thus, we were unable to test whether increased peak IL1RA response with rs315952C results in lower IL-1β post-LPS.
Previous clinical trials of recombinant IL1RA, anakinra, for severe sepsis failed to achieve a large reduction in mortality (54, 64, 65). Given the associations of rs315952C with improved survival in the VASST cohort, with potential preferential transcription, and with increased evoked plasma IL1RA, attenuating IL-1β remains an attractive potential treatment paradigm, particularly if it were possible to predict which patients might be more likely to respond to such an intervention. Anakinra may display pharmacogenomic variation in response to treatment for rheumatoid arthritis (66); it is unknown whether response to this drug when used to decrease mortality in sepsis may also have varied by genotype. Beyond genetic variation, it may be that clinical factors result in sepsis endotypes that differ in their degree of inflammasome activation (6). Alternatively, a more successful intervention may need to combine anti–IL-1β therapy with strategies to block IL-18 (67, 68), or to mitigate vascular permeability (69). Despite recent disappointments in the ability of pharmacologic interventions to improve sepsis mortality (70), we remain optimistic that with improved understanding of sepsis endotypes, effective therapies may yet emerge.
Footnotes
The GENE project was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant UL1TR000003, NIH-NHLBI SCCOR Project grant (P50-HL-083799), and R01-HL-113147 (M.P.R. and M.L.) and by the Penn Genome Frontiers Institute under a grant with the Pennsylvania Department of Health (which disclaims responsibility for any analyses, interpretations, or conclusions) (M.P.R.). The VASST study was supported by the Canadian Institute of Health MCT-44152 (J.A.R.). In addition, N.J.M. is supported by National Institutes of Health HL102254; J.F.F. is supported by a postdoctoral fellowship grant from the American Heart Association (12POST11840017). M.P.R. is also supported by HL111694, DK090505, HL108636, and HL107643. J.D.C. is supported by HL115354, HL087115, HL096845, HL113252, and HL114626.
Author Contributions: N.J.M., J.F.F., J.D.C., K.R.W., and M.P.R. contributed to conception and design of this study. J.F.F., P.N.P., C.X., J.H.B., J.A.R., K.R.W., and M.P.R. contributed to the acquisition of data. All authors contributed to the analysis and interpretation of data. N.J.M. drafted the manuscript, and all authors critically revised it for intellectual content and approved the final version.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201403-0586OC on August 4, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Kumar G, Kumar N, Taneja A, Kaleekal T, Tarima S, McGinley E, Jimenez E, Mohan A, Khan RA, Whittle J, et al. Milwaukee Initiative in Critical Care Outcomes Research Group of Investigators. Nationwide trends of severe sepsis in the 21st century (2000-2007) Chest. 2011;140:1223–1231. doi: 10.1378/chest.11-0352. [DOI] [PubMed] [Google Scholar]
- 2.Annane D, Aegerter P, Jars-Guincestre MC, Guidet B CUB-Réa Network. Current epidemiology of septic shock: the CUB-Réa Network. Am J Respir Crit Care Med. 2003;168:165–172. doi: 10.1164/rccm.2201087. [DOI] [PubMed] [Google Scholar]
- 3.Kumar A, Ellis P, Arabi Y, Roberts D, Light B, Parrillo JE, Dodek P, Wood G, Kumar A, Simon D, et al. Cooperative Antimicrobial Therapy of Septic Shock Database Research Group. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest. 2009;136:1237–1248. doi: 10.1378/chest.09-0087. [DOI] [PubMed] [Google Scholar]
- 4.Rivers E, Nguyen B, Havstad S, Ressler J, Muzzin A, Knoblich B, Peterson E, Tomlanovich M Early Goal-Directed Therapy Collaborative Group. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med. 2001;345:1368–1377. doi: 10.1056/NEJMoa010307. [DOI] [PubMed] [Google Scholar]
- 5.Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, Meyer K, Checchia PA, Lin R, Shanley TP, et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med. 2011;39:2511–2517. doi: 10.1097/CCM.0b013e3182257675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maslove DM, Tang BM, McLean AS. Identification of sepsis subtypes in critically ill adults using gene expression profiling. Crit Care. 2012;16:R183. doi: 10.1186/cc11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinsky MR, Vincent JL, Deviere J, Alegre M, Kahn RJ, Dupont E. Serum cytokine levels in human septic shock. Relation to multiple-system organ failure and mortality. Chest. 1993;103:565–575. doi: 10.1378/chest.103.2.565. [DOI] [PubMed] [Google Scholar]
- 8.Nakada TA, Russell JA, Wellman H, Boyd JH, Nakada E, Thain KR, Thair SA, Hirasawa H, Oda S, Walley KR. Leucyl/cystinyl aminopeptidase gene variants in septic shock. Chest. 2011;139:1042–1049. doi: 10.1378/chest.10-2517. [DOI] [PubMed] [Google Scholar]
- 9.Man M, Close SL, Shaw AD, Bernard GR, Douglas IS, Kaner RJ, Payen D, Vincent JL, Fossceco S, Janes JM, et al. Beyond single-marker analyses: mining whole genome scans for insights into treatment responses in severe sepsis. Pharmacogenomics J. 2013;13:218–226. doi: 10.1038/tpj.2012.1. [DOI] [PubMed] [Google Scholar]
- 10.Casey LC, Balk RA, Bone RC. Plasma cytokine and endotoxin levels correlate with survival in patients with the sepsis syndrome. Ann Intern Med. 1993;119:771–778. doi: 10.7326/0003-4819-119-8-199310150-00001. [DOI] [PubMed] [Google Scholar]
- 11.Arnalich F, López-Maderuelo D, Codoceo R, Lopez J, Solis-Garrido LM, Capiscol C, Fernandez-Capitán C, Madero R, Montiel C. Interleukin-1 receptor antagonist gene polymorphism and mortality in patients with severe sepsis. Clin Exp Immunol. 2002;127:331–336. doi: 10.1046/j.1365-2249.2002.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer NJ, Feng R, Li M, Zhao Y, Sheu CC, Tejera P, Gallop R, Bellamy S, Rushefski M, Lanken PN, et al. IL1RN coding variant is associated with lower risk of acute respiratory distress syndrome and increased plasma IL-1 receptor antagonist. Am J Respir Crit Care Med. 2013;187:950–959. doi: 10.1164/rccm.201208-1501OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 14.Granowitz EV, Vannier E, Poutsiaka DD, Dinarello CA. Effect of interleukin-1 (IL-1) blockade on cytokine synthesis. II. IL-1 receptor antagonist inhibits lipopolysaccharide-induced cytokine synthesis by human monocytes. Blood. 1992;79:2364–2369. [PubMed] [Google Scholar]
- 15.Granowitz EV, Clark BD, Vannier E, Callahan MV, Dinarello CA. Effect of interleukin-1 (IL-1) blockade on cytokine synthesis. I. IL-1 receptor antagonist inhibits IL-1-induced cytokine synthesis and blocks the binding of IL-1 to its type II receptor on human monocytes. Blood. 1992;79:2356–2363. [PubMed] [Google Scholar]
- 16.Zapata-Tarrés M, Arredondo-García JL, Rivera-Luna R, Klünder-Klünder M, Mancilla-Ramírez J, Sánchez-Urbina R, Vázquez-Cruz MY, Juárez-Villegas LE, Palomo-Colli MA. Interleukin-1 receptor antagonist gene polymorphism increases susceptibility to septic shock in children with acute lymphoblastic leukemia. Pediatr Infect Dis J. 2013;32:136–139. doi: 10.1097/INF.0b013e31827566dd. [DOI] [PubMed] [Google Scholar]
- 17.Fang XM, Schröder S, Hoeft A, Stüber F. Comparison of two polymorphisms of the interleukin-1 gene family: interleukin-1 receptor antagonist polymorphism contributes to susceptibility to severe sepsis. Crit Care Med. 1999;27:1330–1334. doi: 10.1097/00003246-199907000-00024. [DOI] [PubMed] [Google Scholar]
- 18.Ma P, Chen D, Pan J, Du B. Genomic polymorphism within interleukin-1 family cytokines influences the outcome of septic patients. Crit Care Med. 2002;30:1046–1050. doi: 10.1097/00003246-200205000-00015. [DOI] [PubMed] [Google Scholar]
- 19.Danis VA, Millington M, Hyland VJ, Grennan D. Cytokine production by normal human monocytes: inter-subject variation and relationship to an IL-1 receptor antagonist (IL-1Ra) gene polymorphism. Clin Exp Immunol. 1995;99:303–310. doi: 10.1111/j.1365-2249.1995.tb05549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiner AP, Wurfel MM, Lange LA, Carlson CS, Nord AS, Carty CL, Rieder MJ, Desmarais C, Jenny NS, Iribarren C, et al. Polymorphisms of the IL1-receptor antagonist gene (IL1RN) are associated with multiple markers of systemic inflammation. Arterioscler Thromb Vasc Biol. 2008;28:1407–1412. doi: 10.1161/ATVBAHA.108.167437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wurfel MM, Gordon AC, Holden TD, Radella F, Strout J, Kajikawa O, Ruzinski JT, Rona G, Black RA, Stratton S, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med. 2008;178:710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrol ED, Payton A, Payne D, Miyajima F, Chaponda M, Mankhambo LA, Banda DL, Molyneux EM, Cox H, Jacobson G, et al. The IL1RN promoter rs4251961 correlates with IL-1 receptor antagonist concentrations in human infection and is differentially regulated by GATA-1. J Immunol. 2011;186:2329–2335. doi: 10.4049/jimmunol.1002402. [DOI] [PubMed] [Google Scholar]
- 23.Tekola Ayele F, Doumatey A, Huang H, Zhou J, Charles B, Erdos M, Adeleye J, Balogun W, Fasanmade O, Johnson T, et al. Genome-wide associated loci influencing interleukin (IL)-10, IL-1Ra, and IL-6 levels in African Americans. Immunogenetics. 2012;64:351–359. doi: 10.1007/s00251-011-0596-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer NJ, Patel PN, Rushefski M, Shashaty MG, Feng R, Christie JD, Reilly MP, Ferguson JF.IL1rn coding variant associates with decreased ARDS risk and increased plasma ILl1ra. Presented at the Pittsburgh International Lung Conference. October 5, 2012, Pittsburgh, PA
- 26.Meyer NJ, Russell JA, Christie JD, Walley KR. IL1rn coding SNP is associated with decreased interleukin-1 beta and decreased mortality in the VASST septic shock cohort [abstract] Am J Respir Crit Care Med. 2013;187:A1010. [Google Scholar]
- 27.Ferguson JF, Patel PN, Shah RY, Mulvey CK, Gadi R, Nijjar PS, Usman HM, Mehta NN, Shah R, Master SR, et al. Race and gender variation in response to evoked inflammation. J Transl Med. 2013;11:63. doi: 10.1186/1479-5876-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah R, Hinkle CC, Haris L, Shah R, Mehta NN, Putt ME, Reilly MP. Adipose genes down-regulated during experimental endotoxemia are also suppressed in obesity. J Clin Endocrinol Metab. 2012;97:E2152–E2159. doi: 10.1210/jc.2012-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Ferguson JF, Xue C, Silverman IM, Gregory B, Reilly MP, Li M. Evaluating the impact of sequencing depth on transcriptome profiling in human adipose. PLoS ONE. 2013;8:e66883. doi: 10.1371/journal.pone.0066883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell JA, Walley KR, Singer J, Gordon AC, Hébert PC, Cooper DJ, Holmes CL, Mehta S, Granton JT, Storms MM, et al. VASST Investigators. Vasopressin versus norepinephrine infusion in patients with septic shock. N Engl J Med. 2008;358:877–887. doi: 10.1056/NEJMoa067373. [DOI] [PubMed] [Google Scholar]
- 31.Nakada TA, Russell JA, Boyd JH, McLaughlin L, Nakada E, Thair SA, Hirasawa H, Oda S, Walley KR. Association of angiotensin II type 1 receptor-associated protein gene polymorphism with increased mortality in septic shock. Crit Care Med. 2011;39:1641–1648. doi: 10.1097/CCM.0b013e318218665a. [DOI] [PubMed] [Google Scholar]
- 32.Nakada TA, Russell JA, Boyd JH, Aguirre-Hernandez R, Thain KR, Thair SA, Nakada E, McConechy M, Walley KR. beta2-Adrenergic receptor gene polymorphism is associated with mortality in septic shock. Am J Respir Crit Care Med. 2010;181:143–149. doi: 10.1164/rccm.200903-0332OC. [DOI] [PubMed] [Google Scholar]
- 33.Bernard G. The Brussels score. Sepsis. 1997;1:43–44. [Google Scholar]
- 34.Liu Y, Ferguson JF, Xue C, Ballantyne RL, Silverman IM, Gosai SJ, Serfecz J, Morley MP, Gregory BD, Li M, et al. Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler Thromb Vasc Biol. 2014;34:902–912. doi: 10.1161/ATVBAHA.113.303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 36.Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, et al. International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heap GA, Yang JHM, Downes K, Healy BC, Hunt KA, Bockett N, Franke L, Dubois PC, Mein CA, Dobson RJ, et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum Mol Genet. 2010;19:122–134. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Minkelen R, de Visser MCH, Houwing-Duistermaat JJ, Vos HL, Bertina RM, Rosendaal FR. Haplotypes of IL1B, IL1RN, IL1R1, and IL1R2 and the risk of venous thrombosis. Arterioscler Thromb Vasc Biol. 2007;27:1486–1491. doi: 10.1161/ATVBAHA.107.140384. [DOI] [PubMed] [Google Scholar]
- 39.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grossman SR, Shlyakhter I, Karlsson EK, Byrne EH, Morales S, Frieden G, Hostetter E, Angelino E, Garber M, Zuk O, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 42.Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 2009;19:711–722. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris EE, Meyer D. The molecular signature of selection underlying human adaptations. Am J Phys Anthropol. 2006;131:89–130. doi: 10.1002/ajpa.20518. [DOI] [PubMed] [Google Scholar]
- 44.Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Dyke AL, Cote ML, Wenzlaff AS, Land S, Schwartz AG. Cytokine SNPs: Comparison of allele frequencies by race and implications for future studies. Cytokine. 2009;46:236–244. doi: 10.1016/j.cyto.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luotola K, Pietilä A, Alanne M, Lanki T, Loo B-M, Jula A, Perola M, Peters A, Zeller T, Blankenberg S, et al. Health 2000, FINRISK97, and AIRGENE Study Groups. Genetic variation of the interleukin-1 family and nongenetic factors determining the interleukin-1 receptor antagonist phenotypes. Metabolism. 2010;59:1520–1527. doi: 10.1016/j.metabol.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Juge-Aubry CE, Somm E, Giusti V, Pernin A, Chicheportiche R, Verdumo C, Rohner-Jeanrenaud F, Burger D, Dayer JM, Meier CA. Adipose tissue is a major source of interleukin-1 receptor antagonist: upregulation in obesity and inflammation. Diabetes. 2003;52:1104–1110. doi: 10.2337/diabetes.52.5.1104. [DOI] [PubMed] [Google Scholar]
- 49.Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet. 2010;11:533–538. doi: 10.1038/nrg2815. [DOI] [PubMed] [Google Scholar]
- 50.Morcos L, Ge B, Koka V, Lam KC, Pokholok DK, Gunderson KL, Montpetit A, Verlaan DJ, Pastinen T. Genome-wide assessment of imprinted expression in human cells. Genome Biol. 2011;12:R25. doi: 10.1186/gb-2011-12-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinarello CA, Ikejima T, Warner SJ, Orencole SF, Lonnemann G, Cannon JG, Libby P. Interleukin 1 induces interleukin 1. I. Induction of circulating interleukin 1 in rabbits in vivo and in human mononuclear cells in vitro. J Immunol. 1987;139:1902–1910. [PubMed] [Google Scholar]
- 52.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 53.Fischer E, Marano MA, Van Zee KJ, Rock CS, Hawes AS, Thompson WA, DeForge L, Kenney JS, Remick DG, Bloedow DC, et al. Interleukin-1 receptor blockade improves survival and hemodynamic performance in Escherichia coli septic shock, but fails to alter host responses to sublethal endotoxemia. J Clin Invest. 1992;89:1551–1557. doi: 10.1172/JCI115748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fisher CJ, Jr, Slotman GJ, Opal SM, Pribble JP, Bone RC, Emmanuel G, Ng D, Bloedow DC, Catalano MA IL-1RA Sepsis Syndrome Study Group. Initial evaluation of human recombinant interleukin-1 receptor antagonist in the treatment of sepsis syndrome: a randomized, open-label, placebo-controlled multicenter trial. Crit Care Med. 1994;22:12–21. doi: 10.1097/00003246-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 55.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McDaniell R, Lee B-K, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ong C-T, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verlaan DJ, Berlivet S, Hunninghake GM, Madore A-M, Larivière M, Moussette S, Grundberg E, Kwan T, Ouimet M, Ge B, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gauderman WJ, Morrison J.Quanto v1.2.4 [updated 2009 May; accessed 2014 Aug 22]. Available from: http://biostats.usc.edu/Quanto.html [Google Scholar]
- 60.Andreasen AS, Krabbe KS, Krogh-Madsen R, Taudorf S, Pedersen BK, Møller K. Human endotoxemia as a model of systemic inflammation. Curr Med Chem. 2008;15:1697–1705. doi: 10.2174/092986708784872393. [DOI] [PubMed] [Google Scholar]
- 61.Visser T, Pillay J, Pickkers P, Leenen LP, Koenderman L. Homology in systemic neutrophil response induced by human experimental endotoxemia and by trauma. Shock. 2012;37:145–151. doi: 10.1097/SHK.0b013e31823f14a4. [DOI] [PubMed] [Google Scholar]
- 62.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. Inflamm and Host Response to Injury Large Scale Collab. Res. Program. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 63.Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Hayden DL, Hennessy L, Moore EE, Minei JP, et al. Inflammation and Host Response to Injury Large-Scale Collaborative Research Program. A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fisher CJ, Jr, Dhainaut JF, Opal SM, Pribble JP, Balk RA, Slotman GJ, Iberti TJ, Rackow EC, Shapiro MJ, Greenman RL, et al. Recombinant human interleukin 1 receptor antagonist in the treatment of patients with sepsis syndrome: results from a randomized, double-blind, placebo-controlled trial. Phase III rhIL-1ra Sepsis Syndrome Study Group. JAMA. 1994;271:1836–1843. [PubMed] [Google Scholar]
- 65.Opal SM, Fisher CJ, Dhainaut JF, Vincent JL, Brase R, Lowry SF, Sadoff JC, Slotman GJ, Levy H, Balk RA, et al. Confirmatory interleukin-1 receptor antagonist trial in severe sepsis: a phase III, randomized, double-blind, placebo-controlled, multicenter trial. The Interleukin-1 Receptor Antagonist Sepsis Investigator Group. Crit Care Med. 1997;25:1115–1124. doi: 10.1097/00003246-199707000-00010. [DOI] [PubMed] [Google Scholar]
- 66.Camp NJ, Cox A, di Giovine FS, McCabe D, Rich W, Duff GW. Evidence of a pharmacogenomic response to interleukin-l receptor antagonist in rheumatoid arthritis. Genes Immun. 2005;6:467–471. doi: 10.1038/sj.gene.6364228. [DOI] [PubMed] [Google Scholar]
- 67.Vanden Berghe T, Demon D, Bogaert P, Vandendriessche B, Goethals A, Depuydt B, Vuylsteke M, Roelandt R, Van Wonterghem E, Vandenbroecke J, et al. Simultaneous targeting of IL-1 and IL-18 is required for protection against inflammatory and septic shock. Am J Respir Crit Care Med. 2014;189:282–291. doi: 10.1164/rccm.201308-1535OC. [DOI] [PubMed] [Google Scholar]
- 68.Opal SM. Dual inhibition of interleukin-1β and interleukin-18: a new treatment option for sepsis? Am J Respir Crit Care Med. 2014;189:242–244. doi: 10.1164/rccm.201312-2292ED. [DOI] [PubMed] [Google Scholar]
- 69.London NR, Zhu W, Bozza FA, Smith MCP, Greif DM, Sorensen LK, Chen L, Kaminoh Y, Chan AC, Passi SF, et al. Targeting Robo4-dependent Slit signaling to survive the cytokine storm in sepsis and influenza. Sci Transl Med. 2010;2:23ra19. doi: 10.1126/scitranslmed.3000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Opal SM, Laterre PF, Francois B, LaRosa SP, Angus DC, Mira JP, Wittebole X, Dugernier T, Perrotin D, Tidswell M, et al. ACCESS Study Group. Effect of eritoran, an antagonist of MD2-TLR4, on mortality in patients with severe sepsis: the ACCESS randomized trial. JAMA. 2013;309:1154–1162. doi: 10.1001/jama.2013.2194. [DOI] [PubMed] [Google Scholar]