Abstract
The interaction between the receptor FLT3 (FMS-like tyrosine kinase-3) and its ligand FL leads to crucial signalling during the early stages of the commitment of haematopoietic stem cells. Mutation or over-expression of the FLT3 gene, leading to constitutive signalling, enhances the survival and expansion of a variety of leukaemias and is associated with an unfavourable clinical outcome for acute myeloid leukaemia (AML) patients. In this study, we used a murine cellular model for AML and primary leukaemic cells from AML patients to investigate the molecular mechanisms underlying the regulation of FLT3 gene expression and identify its key cis- and trans-regulators. By assessing DNA accessibility and epigenetic markings, we defined regulatory domains in the FLT3 promoter and first intron. These elements permit in vivo binding of several AML-related transcription factors, including the proto-oncogene MYB and the CCAAT/enhancer binding protein C/EBPα, which are recruited to the FLT3 promoter and intronic module, respectively. Substantiating their relevance to the human disease, our analysis of gene expression profiling arrays from AML patients uncovered significant correlations between FLT3 expression level and that of MYB and CEBPA. The latter relationship permits discrimination between patients with CEBPA mono- and bi-allelic mutations, and thus connects two major prognostic factors for AML.
Keywords: AML, C/EBP alpha, FLT3, MYB, regulation
INTRODUCTION
Acute myeloid leukaemia (AML) represents a biologically and clinically heterogeneous disorder characterised by a clonal expansion of haematopoietic progenitor cells blocked in their maturation. About 50% of AMLs present cytogenetic abnormalities of various types that can be used to categorise the patients into different risk groups. The remaining cytogenetically normal AML (CN-AML) can be subdivided based on molecular markers, such as mutations in the genes encoding the FLT3 receptor, C/EBPα, NPM1, IDH1 and IDH2, C-KIT, RAS or WT1.1
Constituting some of the most frequent mutations found in CN-AML, mutations in the FLT3 gene generally associate with an inferior outcome. The FMS-like tyrosine kinase-3 (FLT3), a member of the Type III RTK subfamily, is a membrane-bound tyrosine kinase receptor, which together with its ligand FL, has an important role in the normal development of stem cells and the immune system.2,3 The FLT3 protein is commonly mutated in AML, with 20–30% of cases carrying an internal tandem duplication within the juxtamembrane domain (FLT3-ITD) and 8–12% a single point mutation at position 835 in the kinase domain (FLT3-D835), leading to constitutive signalling and enhanced cell survival and proliferation. While the presence of the FLT3-ITD mutation significantly correlates with adverse clinical outcome, the prognostic significance of FLT3 single point mutations remains unclear (see4 for review). By themselves, FLT3-ITD mutant proteins are sufficient to induce a myeloproliferative disorder, but the development of FLT3-ITD AML may require additional mutations that impair haematopoietic differentiation.5
In this respect, the functional cooperation between FLT3-ITD and mutation of the CCATT/enhancer binding protein alpha (C/EBPα) may be of particular significance in CN-AML.6,7 C/EBPα is a leucine-zipper transcription factor that has a pivotal role in granulocyte and neutrophil development8 but also act as a tumour suppressor in the haematopoietic system.9 As the result of an alternative translation initiation, C/EBPα is produced in two major isoforms, p42 (42kDa) and p30 (30kDa). Mutations of the CEBPA gene are reported in 5–14% of patients with AML and can be largely categorised into two types: the N-terminal stop mutations that result in the expression of the p30 isoform from the mutated allele and the C-terminal inframe insertions/deletions within the basic region leucine zipper DNA binding domain.10,11 In most cases, CEBPA mutations are bi-allelic with one allele harbouring the N-terminal mutation and the other the C-terminal, while the homozygous N- or C-terminal mutations are relatively rare.12 Several studies have demonstrated that bi-allelic mutations of the CEBPA gene give a favourable prognosis for AML patients,13 although this benefit appears to be nullified in the presence of FLT3 activating mutations.14 When coexisting, FLT3-ITD mutations may cooperate in leukaemogenesis by over-riding the differentiation impairment brought about by C/EBPα bi-allelic mutations and promote the formation of committed myeloid progenitors, the templates for leukaemia-initiating cells.6
In the absence of FLT3 mutation, the auto-activation of the receptor was also reported in patients with high expression level of FLT3, making this feature another poor prognosis marker.15 As might be expected, the combination of both FLT3-ITD mutation and high-level expression associates with a dismal outcome for patients. Paralleling FLT3 activating mutations, constitutive signalling through FLT3 overexpression may cooperate with other mutations in leukaemogenesis. For instance, such collaboration was suggested with RUNX1 mutation in AML with chromosome 13 trisomy.16 In light of its clinical relevance, it is surprising that, aside from recent evidence indicating a role for HOXA9 and MEIS1,17,18 the mechanisms regulating FLT3 expression in AML remains largely undefined.
In order to investigate the regulators of FLT3 transcription in AML, we made use of a murine-cultured model for CN-AML and bone marrow samples from CN-AML patients. We identified two enhancers regulating FLT3 expression by allowing recruitment of several AML-related transcription factors. Among these, we identified the transcription factor MYB, which is increasingly implicated as a key component of leukaemia maintenance and oncogene addiction, and C/EBPα as keys regulators of FLT3 expression in AML. Furthermore, we showed a direct relationship between CEBPA mutation status and FLT3 expression level and highlighted an important molecular connection between these factors in CN-AML cells.
MATERIALS AND METHODS
Cell line
FMH9 cells were cultured in RPMI medium supplemented with 10% foetal bovine serum, 50 ng/ml stem cell factor, 5 ng/ml granulocyte-macrophage colony-stimulating factor, 5 ng/ml interleukin-3 and 5 ng/ml interleukin-6. All the cytokines were purchased from Peprotech EC (London, UK).
AML patient samples
Collected at diagnosis, bone marrow samples of de novo AML patients were obtained through the Central England Haemato-oncology Research Biobank and used for Ficoll-Plaque separation of white mononucleated cells.
X-ChIP
X-ChIP assays were performed as previously described,19 using antibodies from Santa Cruz Biotechnology (Dallas, TX, USA) Upstate Ltd (Merck Millipore, Billerica, MA, USA) (MYB) or locally produced antibodies (H3K9ac, H4K8ac).
Nuclease hypersensitive site mapping
Cell nuclei, prepared in 1 ml digestion buffer (15 mm Tris-HCl pH7.5, 15 mm NaCl, 60 mm KCl, 5 mm MgCl2, 300 mm glucose, 0.5 mm EGTA, 0.1%NP40), were digested 10 min at 37 °C with DNase I (0–60units). The reaction was terminated by adding 330 μl of 100 mm EDTA/4% SDS. RNA and proteins were sequentially digested at 37 °C with 100 μg RNaseA (1 h) and 100 μg proteinase K (overnight). Extracted DNA was used as template for quantitative-PCR reactions (primers listed in Supplementary Table S1).
Patient profiling arrays
AML patient CEL files were analysed using MAS (background), INVARIANT SET (normalisation), MAS (pm correct) and LIWONG (summary) methods as recommend for gene coexpression analysis.20 Intensities were mean centred to account for undesirable variations (Supplementary Figure S2). The respective scaling factors are listed in Supplementary Table S2. For transcripts represented by multiple probes on the array, the average value of the different probes was determined. The log10 values of the gene expression from all 46 CN-AML CEBPA wild-type profiling arrays in the data set GSE15210 were used for the calculation of the Pearson correlation coefficient. Within the larger GEO data set GSE14468 containing a mix of CN and abnormal AML cases, 58 profiling arrays from CN-AML of M0 to M3 subtypes (according the French-American-British classification system) with no CEBPA mutations were identified and used for this study as well as seven CN-AML with a CEBPA monoallelic N-terminal stop mutation and 25 CN-AML arrays of patients with CEBPA bi-allelic mutations.
Plasmids and transfection method
In total, 5 × 106 FMH9 cells were electroporated using the Amaxa transfection kit (Biosystems, Warrington, UK) according to the manufacturer’s instructions. The expression vectors for the wild-type form of C/EBPα and MYB as well as the correspondent small hairpin RNA (shRNA) were purchased from Origene (Cambridge, UK). The C/EBPα mutant constructs (Supplementary Figure S3), kindly provided by Toshio Kitamura, were as described.7
RESULTS
The FMH9 cell line recapitulates AML cell features
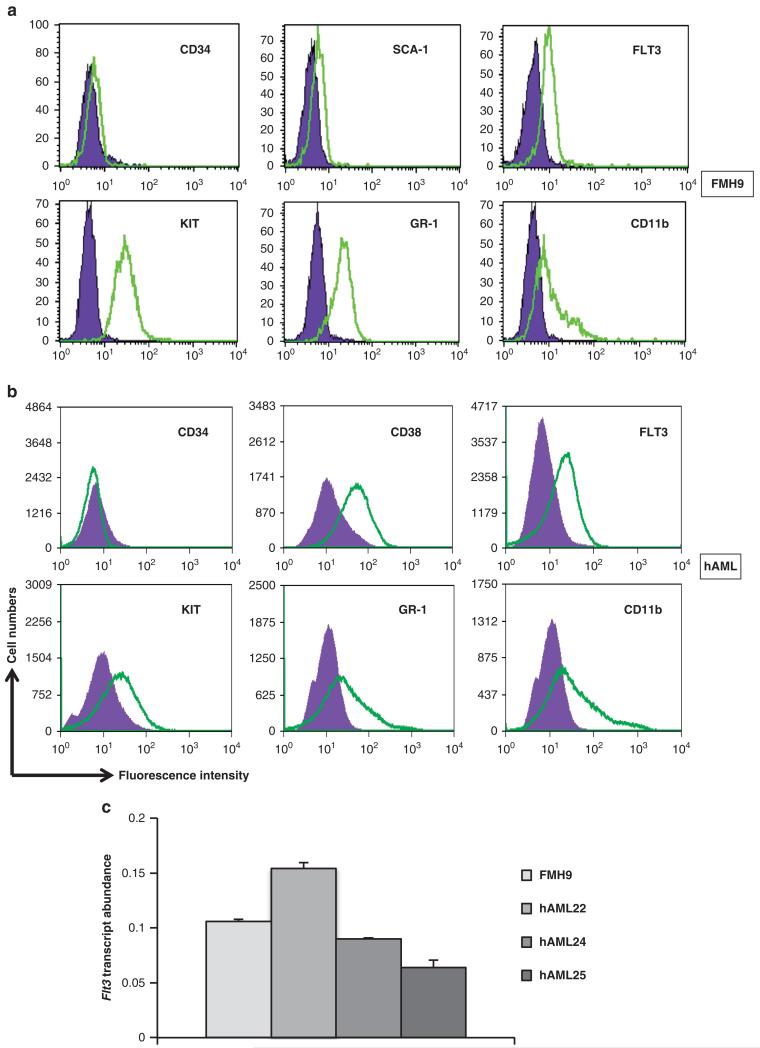
To define the mechanism underlying FLT3 gene regulation in leukaemia, we used a murine cell line modelling CN-AML. The FMH9 line was obtained from primary bone marrow progenitors cotransduced with HOXA9 and MEIS1, a combination causal for AML.18,21,22 Parallel morphological and flow cytometric analyses with patient cells confirmed the FMH9 AML-like phenotype (Figure 1a). Expressing the surface markers GR-1, CD11b, KIT and FLT3, the transformed murine myeloblastic FMH9 cells mimic a CD14 + /CD11b + /CD38 + /c-Kit + /Flt3 + leukaemic fraction of CN-AML patient bone marrow (Figure 1b). Crucially, mRNA quantification indicated similar levels of FLT3 expression in FMH9 and AML patient cells (Figure 1c).
Figure 1. FMH9 cells and AML cells flow cytometric characterisation.
(a) (b) Staining with labelled antibodies against stem cell-related antigens (KIT, SCA-1, CD34, FLT3) and lineage specific markers (GR-1, CD11b) are overlaid against matched isotype controls. (c) FLT3 mRNA quantification by quantitative-PCR, normalised against the 18S house keeping gene results.
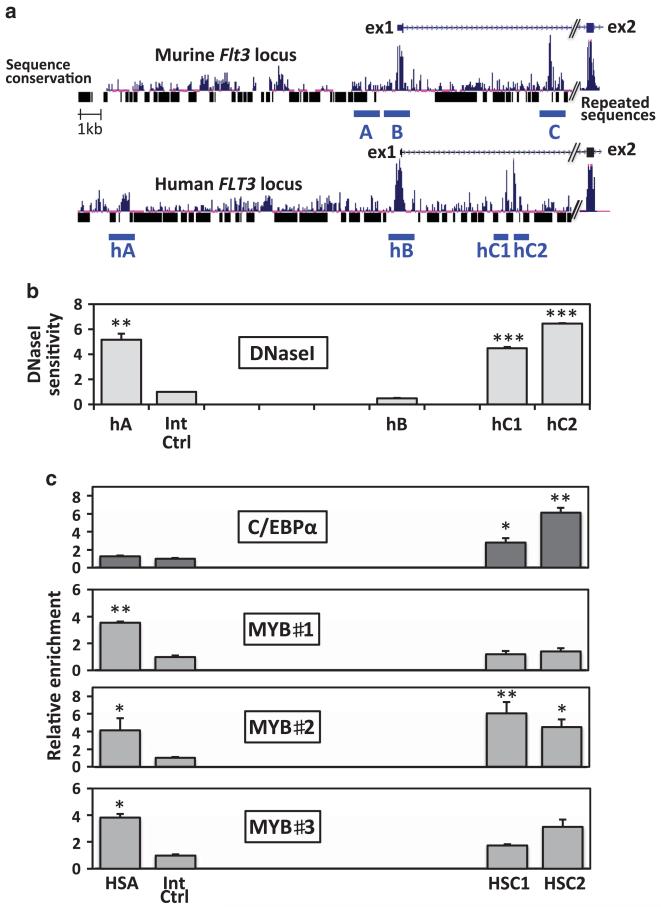
Flt3 cis-regulatory modules lie in the promoter and first intron
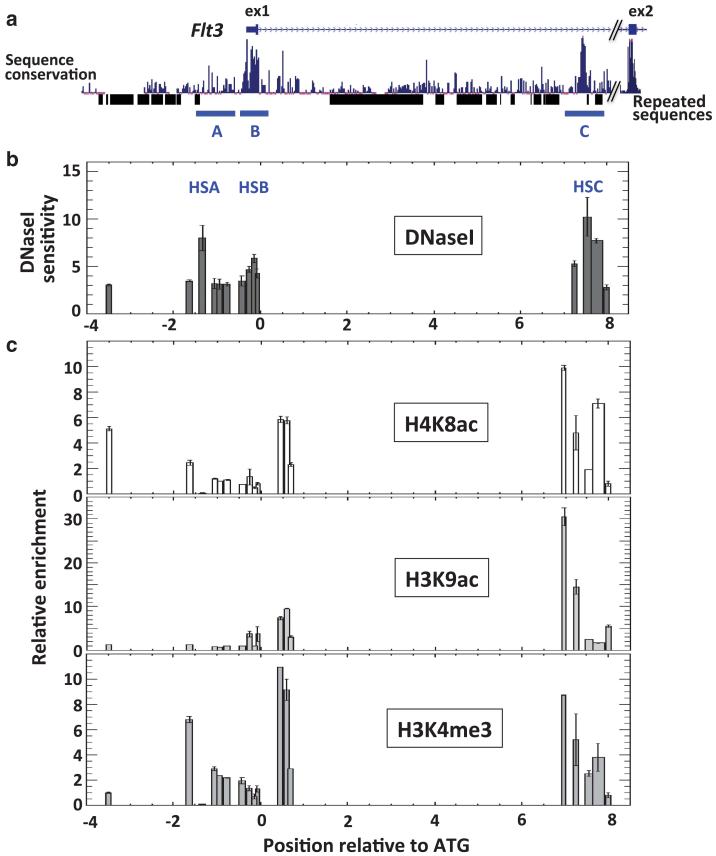
The Flt3 gene is characterised by an abundance of repeated sequences and an overall low degree of cross-species sequence conservation. On the basis that crucial cis-regulatory domains are preserved across species, we focussed on scrutinising the regions of sequence conservation. Three of these regions (A, B and C) locate to the regions surrounding the promoter, the Flt3 transcription start site and the first intron (Figure 2). Using our murine model for CN-AML, we assayed DNaseI sensitivity across these domains to locate sites of nuclease hypersensitivity, symptomatic of protein-DNA interaction. This analysis highlighted three hypersensitive sites at – 1.4 kb (HSA), –0.15 kb (HSB) and + 7.5 kb (HSC) relative to the Flt3 initiating codon (Figure 2b). To further assess their potential relevance with regards to Flt3 transcriptional regulation, we tested for the presence of histone marks that associate with regulatory domains, and found H4K8ac, H3K9ac and H3K4me3 in the vicinity of each domain (Figure 2c).
Figure 2. Flt3 regulatory modules.
(a) Schematic representation of the Flt3 promoter and first intron (filled boxes indicate exon1 and 2). The level of sequence conservation between seven mammalian species and the position of repeated sequences are indicated below. The global illustration is aligned to the lower (b) and bottom (c) plots. (b) DNaseI HSS mapping at the Flt3 locus. Ratios between quantitative-PCR results from partially digested and untreated samples (2–ΔCt) reflect the extent of nuclease sensitivity across regions covered by PCR amplicons. (c) Histone mark distribution at the Flt3 locus in the murine FMH9 AML model. Relative enrichments are determined against the control immunoglobulin G X-ChIP material.
HOXA9, MEIS1, MYB and C/EBPα bind to the Flt3 regulatory modules in the FMH9 AML-line
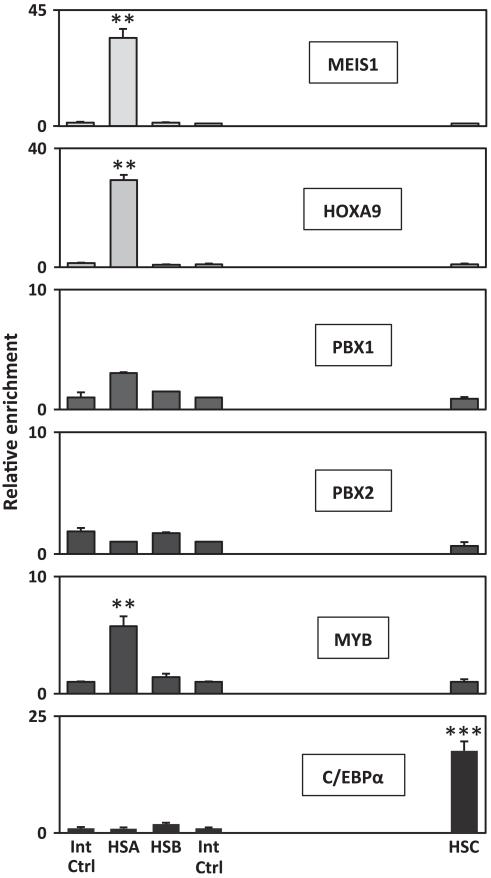
As a first step towards the identification of potential trans-regulators, the sequences encompassing the different hypersensitive sites were screened for the presence of transcription factor binding sites that are conserved across species. This analysis identified several consensus sites for the binding of leukaemia-associated factors such as MYB, C/EBPα and HOX-TALE partner proteins (Supplementary Figure S1). To assess the possible recruitment of factors to the Flt3 locus, we performed X-ChIP assays, using antibodies directed against the different candidate trans-regulators and analysed the immunoprecipitated material by quantitative-PCR at the sites of hypersensitivity and two control regions (Figure 3). In agreement with the previous works,17,18 we identified direct binding of HOXA9 and MEIS1 to the Flt3 promoter in the AML-like FMH9 line, which ectopically expresses both factors. Interestingly, these data also show direct recruitment of MYB and C/EBPα to the Flt3 promoter and enhancer module, respectively.
Figure 3. Detection of transcription factor binding at the sites of nuclease hypersensitivity.
Relative enrichments were determined against the immunoglobulin G control material by quantitative-PCR at the location of the hypersensitive regions (HSA to C at and – 1.45, – 0.15 + 7.5 kb relative to the Flt3 initiating codon) and normalised against two internal control regions (at 3.5 and – 0.27 kb from the ATG). Error bars represent the s.e. of – the mean. All plots are representative of a minimum of three independent experiments used to determine the two-tailed P-value by paired t-test: ***00.001 and **00.01.
FLT3 expression correlates with that of HOXA9, MEIS1, MYB and CEBPA in CN-AML
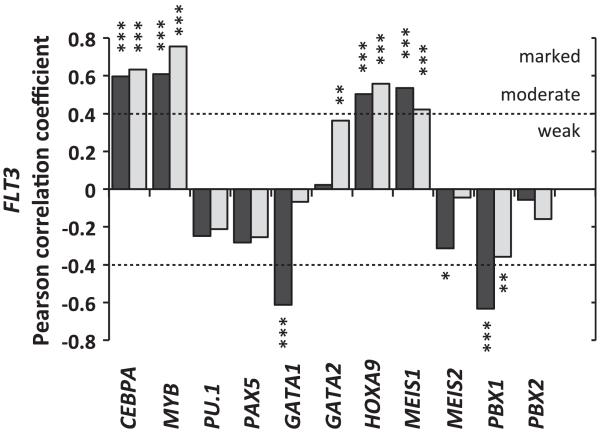
The abundance of the aforementioned factors and their potential influence on FLT3 expression were next evaluated in CN-AML. For this purpose, we made use of publicly available data sets of AML patient profiling arrays and assessed the relationship between the expression of FLT3 and its predicted regulators. Evaluating gene expression codependency, the Pearson correlation coefficients were calculated between the logarithmic expression of FLT3 and the different candidate factors. As C/EBPα is included in this list, the analysis was restricted to patients with no CEBPA mutations, summing 46 and 58 CN-AML arrays within two independent sets (Geo data sets GSE15210 and GSE14468, respectively). Remarkably, despite the biological heterogeneity and variety of genomic mutations included within and between the sets, the analysis showed that FLT3 RNA expression is linked to that of CEBPA, MYB, MEIS1 and HOXA9 in CN-AML (Figure 4). Together with our finding that all four proteins are recruited to the Flt3 promoter and intronic regions in the FMH9 line, these data strongly suggest that MYB and C/EBPα have a determining role alongside HOXA9 and MEIS1 in the control of FLT3 expression in CN-AML.
Figure 4. Expression codependency between FLT3 and its potential transcriptional regulators in CN-AML measured by the Pearson correlation test.
The correlation coefficients were determined using log expression values from two sets of arrays summing 46 and 58 CN-AML patients without CEBPA mutations. Statistical significance ***<0.001 **<0.01 and *<0.05.
MYB and C/EBPα bind to the human FLT3 promoter and intronic elements in CN-AML cells
To confirm that our findings using the FMH9 line were representative of the human disease condition, we reproduced the nuclease assays and X-ChIP experiments using cells from different CN-AML patients. As shown in Figure 5a, the human FLT3 locus has a large number of insertions compared with the murine locus. However, the three identified regions are found conserved at –15.3 kb, –0.37 kb and + 6.2 kb from the human FLT3 initiating codon. The intronic region C was split into two regions (hC1 and hC2) by the presence of a repeated sequence. At these locations, DNaseI hypersensitive sites were only consistently found in regions A and C in CN-AML patient cells (Figure 5b). In agreement with our findings using the murine line, X-ChIP experiments demonstrated the binding of MYB and C/EBPα in human AML to regions A and C on the FLT3 locus, respectively (Figure 5c).
Figure 5. C/EBPα and MYB bind the FLT3 locus in human AML cells.
(a) Schematic comparison of the murine and human FLT3 promoter and first intron (filled boxes represent exons) with indication of sequence conservation and repeated sequence locations. (b) DNaseI hypersensitivity mapping at the human FLT3 locus in CN-AML patients. Plotted for the human sequence orthologs of the murine HSA, HSB, HSC and a control region, ratios between quantitative-PCR results from partially digested and untreated samples are representative of four experiments. (c) Detection of MYB and C/EBPα in vivo binding at the sites of nuclease hypersensitivity, located at – 15.3 kb (hA), –0.37 kb (hB) and + 6.2 kb (hC1/C2) from the human FLT3 initiating codon. Relative enrichments were determined against the immunoglobulin G control material and normalised against the control region (at –6.95 kb from the ATG). Error bars represent the s.e. of the mean. C/EBPα ChIP plots are representative of three independent experiments used to determine the two-tailed P-value by paired t-test. MYB ChIP plots represent patient-specific experiments for which the P-values were calculated using replicate measurements. P-value indications ***<0.001, **<0.01 and *<0.05.
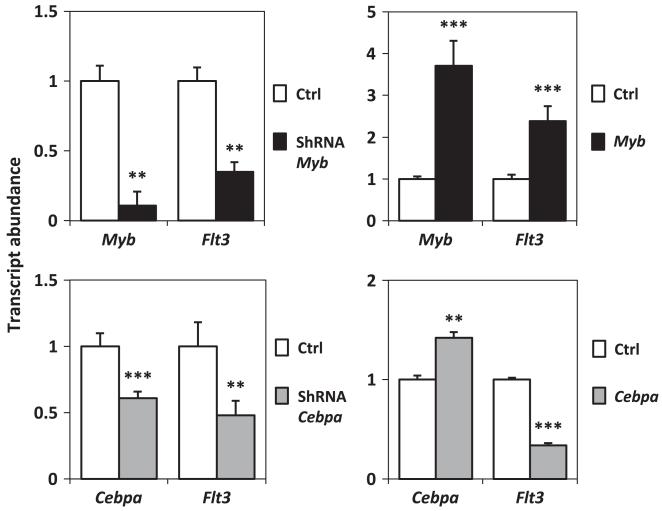
MYB and C/EBPα expression levels influence Flt3 expression
To investigate the extent to which MYB and C/EBPα contribute to the regulation of Flt3 transcription, we manipulated their levels through shRNA knockdown and over-expression in the murine FMH9 AML model. Both shRNA-mediated silencing and over-expression of MYB and C/EBPα were achieved by transient transfection, 24 h after which Flt3 RNA levels were measured by quantitative-PCR. Reduction or over-expression of MYB led to a parallel down- and upregulation of Flt3 expression in the AML-like cells (Figure 6 upper panels). In contrast, while C/EBPα silencing also led in a decrease in Flt3 RNA levels, enforced C/EBPα expression only resulted in a moderate elevation of its transcript accompanied by a marked downregulation of Flt3 expression (Figure 6 lower panel). This latter result suggests that regulation of Flt3 expression could depend on strict C/EBPα activity thresholds in AML.
Figure 6. Silencing and ectopic expression of MYB and C/EBPα in FMH9 cells.
Transcript abundances were determined by quantitative-PCR in cells transfected with shRNA and expression vectors, normalised against the Gapdh house keeping gene PCR result and compared with the correspondent empty vector controls. Results are representative of three or four independent experiments. Statistical significance ***<0.001 and **<0.01.
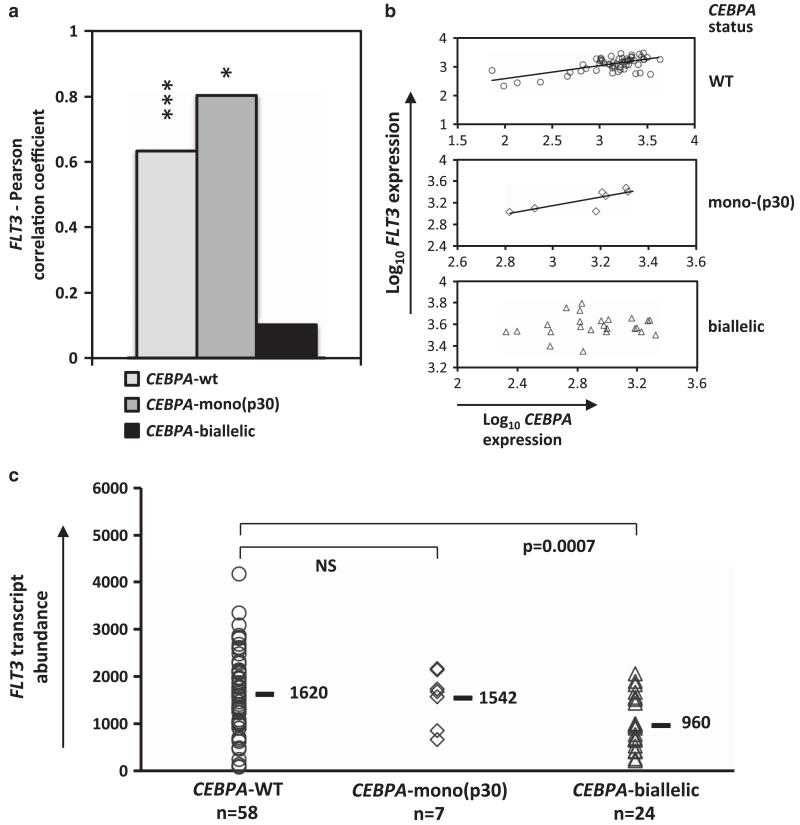
FLT3 expression is affected in CN-AML with CEBPA bi-allelic mutation
The latter finding raised the question of the impact that CEBPA mutations may have on FLT3 expression regulation in CN-AML. To address this question, we assessed and compared the relationship between FLT3 and CEBPA expression in patients with different CEBPA status. Pearson correlations were determined using expression profiling arrays of CN-AML with wild-type CEBPA, CEBPA monoallelic N-terminal stop mutation (expressing the C/EBPα p30 isoform) and CEBPA bi-allelic mutations. The results revealed that the log linear relation between FLT3 and CEBPA transcript expression were conserved in patients with a CEBPA mono-allelic N-terminal stop mutation but lost in the presence of CEBPA bi-allelic mutations (Figures 7a and b). As the loss of this relationship would be consistent with an impairment of C/EBPα-mediated FLT3 transcriptional regulation, we proceeded to evaluate how FLT3 expression may be affected in this subgroup of patients by plotting and comparing FLT3 transcript levels in patients with different CEBPA status (Figure 7c). This analysis showed that CEBPA bi-allelic mutations associate with a decrease in the median expression level of FLT3.
Figure 7.
(a) Pearson correlation test between FLT3 and CEBPA transcript levels applied to CEBPAWT, CEBPASM and CEBPADM patient arrays. Statistical significance: ***<0.001 and *<0.05. (b) Scatter plots and trend line fitting between FLT3 and CEBPA expression in the different subgroups (c) FLT3 mRNA levels in AML patients with wild-type mono- or bi-allelic CEBPA mutations. Median values are indicated. Differential statistical significances are indicated through P-values or non-significance sign NS.
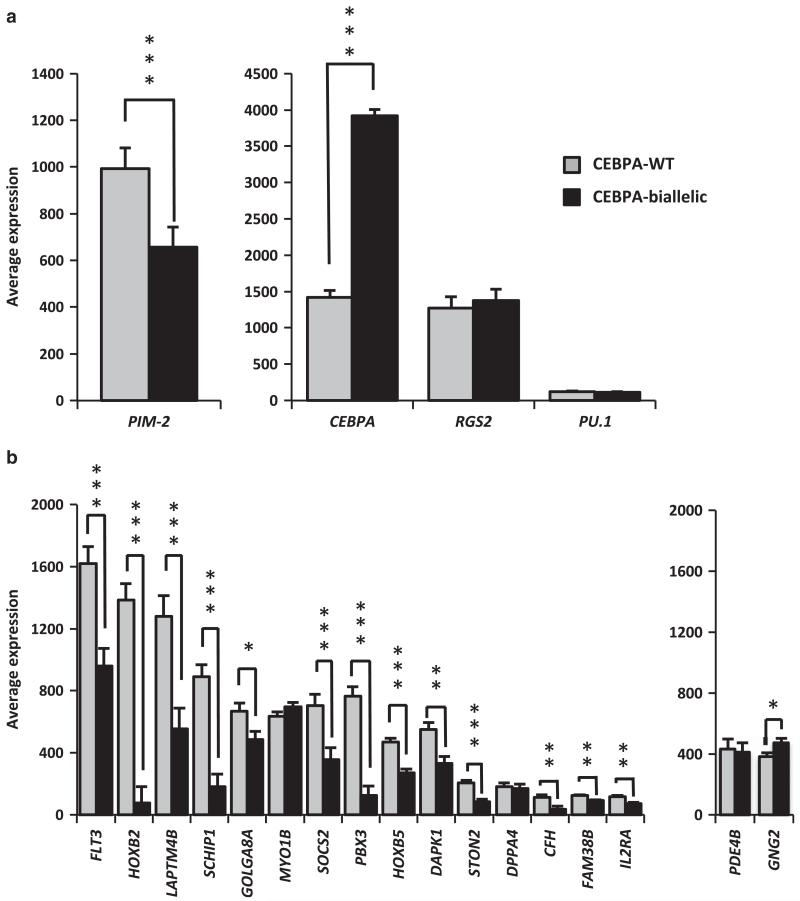
The signature of FLT3 constitutive activation is largely inverted in CN-AML with CEBPA bi-allelic mutation
To test whether this reduced FLT3 expression could affect the receptor function and assess how this may compare with the effects of the FLT3-ITD activating mutation, we analysed the expression of genes that were shown to be downstream targets of FLT3-ITD signalling (Figure 8a) and components of the FLT3-ITD molecular signature in CN-AML (Figure 8b). These encompass (i) genes that were shown to be upregulated (PIM-1) or repressed (CEBPA, RGS2 and PU.1) by the FLT3-ITD signalling pathway23,24 (Figure 8a); and (ii) genes that participate in the signature of the FLT3-ITD mutation, predicting an adverse clinical outcome in patients presenting a combination of elevated (genes listed Figure 8b, left panel), and reduced (PDE4A, GNG2) transcript levels.25 The comparison of the RNA abundance between CN-AML patients with wild type or bi-allelic mutations of CEBPA revealed that most genes were differently expressed in the two groups. Associating with CEBPA mutations, variations in PIM-2 and CEBPA expression were inversed to their response to FLT3-ITD signalling (Figure 8a). Strikingly, the FLT3-ITD molecular signature, as determined against the wild-type receptor cases,25 was largely inverted in CN-AML patients with CEBPA bi-allelic mutations when compared with their wild-type counterparts (Figure 8b).
Figure 8. Patients with wild-type and bi-allelic CEBPA mutations differentially express genes that are part of the signature of the constitutively activated mutant receptor FLT3-ITD.
Bars represent the average expression in AML patients with wild-type and bi-allelic CEBPA mutations and error bars depict the s.e. of the mean. Tested genes were previously shown to be up-(left panel) or downregulated (right panel) in response to FLT3-ITD signalling (a) or are elements of the FLT3-ITD signature in CN-AML (b). Statistical significance: ***<0.001, **<0.01 and *<0.05.
DISCUSSION
Using a murine myeloid leukaemia model and bone marrow cells from AML patients, our study highlights the role of the transcription factors MYB and C/EBPα in the regulation of FLT3 expression in the leukaemia context.
The cooperative activities of MYB and C/EBPα in activating the promoter of myeloid genes have been previously reported.26,27 However, FLT3 constitutes a target of particular significance for both proteins in the context of AML. Having a determinant role in the maintenance and function of the haematopoietic stem cell compartment,28 MYB activity also associates with a broad spectrum of haematological malignancies, including CML, T-ALL and AML.29-31 Notably, MYB was reported to have a critical role for the transforming potential of the AML-inducing fusion protein MLL-ENL.31 However, its function and target genes in the leukaemia context remain poorly understood. Here, by identifying the FLT3 receptor gene as a direct target of MYB in AML cells, our study places MYB activity upstream to that of FLT3 and thus gives a first indication as to how the proto-oncogene MYB may influence the proliferation and survival of AML cells.
Our work suggests that, together with HOXA9 and MEIS1, MYB and C/EBPα are important elements of the combinatorial binding of leukaemia-related transcription factors that regulate FLT3 expression. The role of HOXA9 and MEIS1 in FLT3 regulation, and their recruitment to its promoter, had been previously reported.18 In addition, the recent study of Huang et al.17 considered the existence of enhanceosomes containing these proteins and hinted at the wider relevance of these types of complexes in the regulation of myeloid and leukaemia-related target genes. By analysing the over representation of conserved transcription factor binding sites in the vicinity of HoxA9 and Meis1 binding locations, the authors identified both C/EBPα and MYB as candidate components of these enhanceosomes and suggested a contribution of PU.1. This member of the ETS family of transcription factors was shown to regulate FLT3 expression in dendritic cells32 and to cooperate with C/EBPα and MYB in regulating the transcription of the neutrophil elastase gene.26 Thus, one may expect that a similar collaborative mechanism may underlie the regulation of FLT3 expression. However, we found no evidence of PU.1 binding to the Flt3 locus in the CN-AML murine model or any indication of a relation between PU.1 and FLT3 expression using the human AML micro-array data. Therefore, in contrast with HOXA9, MEIS1, MYB and C/EBPα, PU.1 does not appear to be a crucial regulator of FLT3 expression in the context of CN-AML cells.
The fact that C/EBPα participates in FLT3 regulation is probably the most clinically relevant finding of the present study, as it constitutes a direct link between two major molecular prognostic markers for AML. The individual importance of both FLT3 and CEBPA status for AML prognosis are well documented.33,34 So far, a few hints of a connection between these molecular markers have been given by recent studies showing that the favourable outcome associated with C/EBPα bi-allelic mutations was cancelled out by the presence of FLT3-ITD mutations,6,14 that FLT3-ITD signalling inhibits C/EBPα-differentiating function by promoting its phosphorylation,35 and that C/EBPα with a C-terminal mutation collaborates with FLT3-ITD in inducing AML.7 Such collaborations may rely on the ability of FLT3 signalling to support myeloid commitment of the expanding CEBPA mutated cells.6 Our analysis of profiling arrays from AML patients with differing CEBPA status links FLT3 expression and C/EBPα activity. We find that CEBPA bi-allelic mutations associate with lower levels of FLT3 transcript in AML patients. In a bid to assess the influence of C/EBPα mutated forms on Flt3 expression, we introduced N- and C-terminal mutants, singly or in combination, in the FMH9 cell-line (Supplementary Figure S3). This approach failed to induce statistically significant changes in Flt3 expression, possibly due to the presence of the wild-type form. Similarly, analysis of the profiling arrays did not reveal significant differences in FLT3 expression between patients with-wild type or mono-allelic CEBPA mutations. Conversely, global impairment of C/EBPα activity through shRNA silencing resulted in Flt3 downregulation. Our cell-line based results also suggested that a tight control on C/EBPα expression at the cellular level might prevent any sizeable over-expression. Arguing for the need to maintain such stability, small increases in Cebpa also resulted in reduced Flt3 RNA levels. The explanation for this could lie within a delicate equilibrium between some constituents of the leukaemic transcriptional network. Alternatively, the apparent decrease in Flt3 expression may reflect a change in the cellular context. In fact, small changes in C/EBPα expression may induce granulocytic differentiation of transformed cells, as previously demonstrated in U937 cells.36 In the context of the disease, it is possible that the lower levels of FLT3 in cells from patients with CEBPA bi-allellic mutations may prevent auto-phosphorylation of the FLT3 receptor that occurs when the FLT3 RNA level exceeds 200 000 copies/mg RNA.15 This hypothesis would be consistent with the findings that CEBPA bi-allelic mutations,13,37 or more generally loss of DNA-binding activity,38 associate with a positive prognosis. As this benefit is lost in the presence of FLT3 activating mutations, it would be very interesting to test this theory by assessing whether auto-activation of the FLT3 receptor is less frequent in AML with CEBPA bi-allelic mutations and how this may contribute to the favourable prognosis associated with this status. Here, we analysed the expression level of genes that constitute the molecular signature of CN-AML with FLT3-ITD mutation.25 With reference to CN-AML with wild-type CEBPA, we found that patients with CEBPA bi-allelic mutations present an inverse expression pattern to that of FLT3-ITD patients. Being regulated in an opposite manner by the wild-type and mutated receptors signalling cascades,23,24 CEBPA itself is a notable downstream target. Its elevated expression in patients with bi-allelic mutations further undermines the idea of a FLT3-ITD-related signalling cascade in these patients. Taken together, our results support the hypothesis that the presence of bi-allelic CEBPA mutations may reduce FLT3-mediated leukemogenic signals.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Central England Haemato-oncology Research Biobank for providing the AML patient samples, Robert Slany (Erlangen, Germany) for providing the FMH9 cell line, Toshio Kitamura (Tokyo, Japan) for providing the expression vector for C/EBPα mutants and Roger Bird (Birmingham, UK) for his help with cell sorting. This work was supported by the Leukaemia and Lymphoma Research and the Wellcome Trust.
Footnotes
CONFLICT OF INTEREST: The authors declare no conflict of interest.
REFERENCES
- 1.Gregory TK, Wald D, Chen Y, Vermaat JM, Xiong Y, Tse W. Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J Hematol Oncol. 2009;2:23. doi: 10.1186/1756-8722-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drexler HG. Expression of FLT3 receptor and response to FLT3 ligand by leukemic cells. Leukemia. 1996;10:588–599. [PubMed] [Google Scholar]
- 3.Rosnet O, Buhring HJ, deLapeyriere O, Beslu N, Lavagna C, Marchetto S, et al. Expression and signal transduction of the FLT3 tyrosine kinase receptor. Acta Haematol. 1996;95:218–223. doi: 10.1159/000203881. [DOI] [PubMed] [Google Scholar]
- 4.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: still challenging after all these years. Blood. 2010;116:5089–5102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 5.Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 6.Reckzeh K, Bereshchenko O, Mead A, Rehn M, Kharazi S, Jacobsen SE, et al. Molecular and cellular effects of oncogene cooperation in a genetically accurate AML mouse model. Leukemia. 2012;26:1527–1536. doi: 10.1038/leu.2012.37. [DOI] [PubMed] [Google Scholar]
- 7.Kato N, Kitaura J, Doki N, Komeno Y, Watanabe-Okochi N, Togami K, et al. Two types of C/EBPalpha mutations play distinct but collaborative roles in leukemo-genesis: lessons from clinical data and BMT models. Blood. 2011;117:221–233. doi: 10.1182/blood-2010-02-270181. [DOI] [PubMed] [Google Scholar]
- 8.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasemann MS, Damgaard I, Schuster MB, Theilgaard-Monch K, Sorensen AB, Mrsic A, et al. Mutation of C/EBPalpha predisposes to the development of myeloid leukemia in a retroviral insertional mutagenesis screen. Blood. 2008;111:4309–4321. doi: 10.1182/blood-2007-06-097790. [DOI] [PubMed] [Google Scholar]
- 10.Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19:329–334. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- 11.Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 12.Pabst T, Mueller BU. Transcriptional dysregulation during myeloid transformation in AML. Oncogene. 2007;26:6829–6837. doi: 10.1038/sj.onc.1210765. [DOI] [PubMed] [Google Scholar]
- 13.Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol. 2010;28:570–577. doi: 10.1200/JCO.2008.21.6010. [DOI] [PubMed] [Google Scholar]
- 14.Renneville A, Boissel N, Gachard N, Naguib D, Bastard C, de Botton S, et al. The favorable impact of CEBPA mutations in patients with acute myeloid leukemia is only observed in the absence of associated cytogenetic abnormalities and FLT3 internal duplication. Blood. 2009;113:5090–5093. doi: 10.1182/blood-2008-12-194704. [DOI] [PubMed] [Google Scholar]
- 15.Ozeki K, Kiyoi H, Hirose Y, Iwai M, Ninomiya M, Kodera Y, et al. Biologic and clinical significance of the FLT3 transcript level in acute myeloid leukemia. Blood. 2004;103:1901–1908. doi: 10.1182/blood-2003-06-1845. [DOI] [PubMed] [Google Scholar]
- 16.Dicker F, Haferlach C, Kern W, Haferlach T, Schnittger S. Trisomy 13 is strongly associated with AML1/RUNX1 mutations and increased FLT3 expression in acute myeloid leukemia. Blood. 2007;110:1308–1316. doi: 10.1182/blood-2007-02-072595. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–398. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang GG, Pasillas MP, Kamps MP. Persistent transactivation by meis1 replaces hox function in myeloid leukemogenesis models: evidence for co-occupancy of meis1-pbx and hox-pbx complexes on promoters of leukemia-associated genes. Mol Cell Biol. 2006;26:3902–3916. doi: 10.1128/MCB.26.10.3902-3916.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorvellec M, Dumon S, Maya-Mendoza A, Jackson D, Frampton J, Garcia P. B-Myb is critical for proper DNA duplication during an unperturbed S phase in mouse embryonic stem cells. Stem Cells. 2010;28:1751–1759. doi: 10.1002/stem.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harr B, Schlotterer C. Comparison of algorithms for the analysis of Affymetrix microarray data as evaluated by co-expression of genes in known operons. Nucleic Acids Res. 2006;34:e8. doi: 10.1093/nar/gnj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuki M, Schwable J, Steur C, Choudhary C, Agrawal S, Sargin B, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101:3164–3173. doi: 10.1182/blood-2002-06-1677. [DOI] [PubMed] [Google Scholar]
- 24.Schwable J, Choudhary C, Thiede C, Tickenbrock L, Sargin B, Steur C, et al. RGS2 is an important target gene of Flt3-ITD mutations in AML and functions in myeloid differentiation and leukemic transformation. Blood. 2005;105:2107–2114. doi: 10.1182/blood-2004-03-0940. [DOI] [PubMed] [Google Scholar]
- 25.Bullinger L, Dohner K, Kranz R, Stirner C, Frohling S, Scholl C, et al. An FLT3 gene-expression signature predicts clinical outcome in normal karyotype AML. Blood. 2008;111:4490–4495. doi: 10.1182/blood-2007-09-115055. [DOI] [PubMed] [Google Scholar]
- 26.Oelgeschlager M, Nuchprayoon I, Luscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol. 1996;16:4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burk O, Mink S, Ringwald M, Klempnauer KH. Synergistic activation of the chicken mim-1 gene by v-myb and C/EBP transcription factors. EMBO J. 1993;12:2027–2038. doi: 10.1002/j.1460-2075.1993.tb05852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA. 2009;106:21689–21694. doi: 10.1073/pnas.0907623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lidonnici MR, Corradini F, Waldron T, Bender TP, Calabretta B. Requirement of c-Myb for p210(BCR/ABL)-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood. 2008;111:4771–4779. doi: 10.1182/blood-2007-08-105072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 31.Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carotta S, Dakic A, D’Amico A, Pang SH, Greig KT, Nutt SL, et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Meshinchi S, Appelbaum FR. Structural and functional alterations of FLT3 in acute myeloid leukemia. Clin Cancer Res. 2009;15:4263–4269. doi: 10.1158/1078-0432.CCR-08-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pabst T, Mueller BU. Complexity of CEBPA dysregulation in human acute myeloid leukemia. Clin Cancer Res. 2009;15:5303–5307. doi: 10.1158/1078-0432.CCR-08-2941. [DOI] [PubMed] [Google Scholar]
- 35.Radomska HS, Alberich-Jorda M, Will B, Gonzalez D, Delwel R, Tenen DG. Targeting CDK1 promotes FLT3-activated acute myeloid leukemia differentiation through C/EBPalpha. J Clin Invest. 2012;122:2955–2966. doi: 10.1172/JCI43354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113:3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fos J, Pabst T, Petkovic V, Ratschiller D, Mueller BU. Deficient CEBPA DNA binding function in normal karyotype AML patients is associated with favorable prognosis. Blood. 2011;117:4881–4884. doi: 10.1182/blood-2010-11-320747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.