Abstract
Schizophrenia is a major psychiatric disorder that lacks a unifying neuropathology, while currently available pharmacological treatments provide only limited benefits to many patients. This review will discuss how the field of neuroepigenetics could contribute to advancements of the existing knowledge on the neurobiology and treatment of psychosis. Genome-scale mapping of DMA methylation, histone modifications and variants, and chromosomal loopings for promoter-enhancer interactions and other epigenetic determinants of genome organization and function are likely to provide important clues about mechanisms contributing to dysregulated expression of synaptic and metabolic genes in schizophrenia brain, including the potential links to the underlying genetic risk architecture and environmental exposures. In addition, studies in animal models are providing a rapidly increasing list of chromatin-regulatory mechanisms with significant effects on cognition and complex behaviors, thereby pointing to the therapeutic potential of epigenetic drug targets in the nervous system.
Keywords: epigenome, DNA methylation, epigenetic drug target, histone modification, psychosis, prefrontal cortex
Abstract
La esquizofrenia es un importante trastorno psiquiátrico que carece de una neuropatología única, y que ios tratamientos farmacológicos disponibles en la actualidad solo aportan beneficios limitados para muchos pacientes. Esta revisión discute cómo el campo de la neuroepigenética podría contribuir a los avances del conocimiento existente sobre la neurobiología y el tratamiento de las psicosis. Es probable que el mapeo a gran escala del genoma de la metilación del ADN, las variantes y modificaciones de la histona, y los lazos cromosómicos para las interacciones entre el reforzador y el promotor, y otros determinantes epigenéticos de la organización y función del genoma proporcionen pistas importantes acerca de los mecanismos que contribuyen a la mala regulación de la expresión de los genes sínápticos y metabólicos en el cerebro de pacientes con esquizofrenia, incluyendo los potenciales vínculos con los riesgos genéticos subyacentes a la arquitectura y a las exposiciones ambientales. Además, los estudios en modelos animales están aportando una lista rápidamente creciente de mecanismos reguladores de la cromatina con efectos significativos en la cognición y en conductas complejas, lo que apunta al potencial terapéutico de fármacos epigenéticos para blancos en el sistema nervioso.
Abstract
La schizophrénie, trouble psychiatrique majeur, manque d'une neuropathologie unifiée, les médicaments actuellement disponibles n'offrant que des bénéfices limités à de nombreux patients. Cet article étudie la façon dont la neuroépigénétique pourrait faire progresser les connaissances actuelles en neurobiologie et en thérapeutique de la psychose. La cartographie à l'échelle du génome de la méthylation de l'ADN, des modifications et des variantes de l'histone, des boucles chromosomiques des interactions promoteur-activateur et d'autres déterminants épigénétiques de l'organisation et de la fonction du génome sont probablement des pistes importantes menant aux mécanismes participant à l'expression dérégulée des gènes métaboliques et synaptiques au sein du cerveau schizophrène, y compris les liens éventuels avec l'architecture sous-jacente du risque génétique et les expositions à l'environnement. De plus, des études de modèles animaux fournissent une liste exponentielle de mécanismes de régulation de la chromatine ayant des effets significatifs sur la cognition et les comportements complexes, suggérant donc des cibles médicamenteuses épigénétiques à potentiel thérapeutique dans le système nerveux.
Introduction
Schizophrenia (SCZ) is a psychiatric disorder defined by positive symptoms such as delusions, hallucinations, and disorganized thought, and negative symptoms such as anhedonia (inability to experience pleasure), social withdrawal, and apathy. SCZ reduces the lifespan of an affected individual on average by 15 years, with cardiovascular disease and suicide among the chief causes for increased mortality.1-3 In addition, the mainstay of antipsychotic intervention is medicinal treatment targeting dopaminergic, serotonergic, and monoaminergic receptor systems,4,5 but the majority of patients still experience an incomplete response to treatment.6,7 Currently prescribed antipsychotics exert therapeutic effects on psychosis in up to approximately 75% of patients, but it is the cognitive impairment which is often the more disabling and persistent feature of schizophrenia.8 It has been a challenge to promote rational drug development in SCZ, mainly because of the lack of a unifying neuropathology9,10 and a complex genetic risk architecture,11,12 which so far have defied any narrowly defined signaling pathways or molecular mechanisms representing the majority of affected cases.
This review will outline how neuroepigenetic approaches13—broadly defined as the study of chromatin structure and function in the developing and adult nervous system, including its role for neuronal and behavioral plasticity—could advance our knowledge on SCZ pathophysiology and the underlying genetic risk architecture and pave the way for novel treatment approaches. Epigenetic marks, such as DNA cytosine methylation and histone modifications and variants, could be viewed as a “molecular bridge” by which myriads of external (“environmental”) or internal factors mold and shape the nascent genetic material throughout the entire lifespan of a brain cell.14 While a comprehensive discussion on epi- (Greek for “over,” “above”) genetic regulation in the nervous system would be beyond the scope of this review (the reader is referred to recent handbooks and special journal volumes in this field, see refs 14-17) we have now clearly entered a period with a heightened level of enthusiasm for epigenetic approaches in neurology and psychiatry, and SCZ research is no exception to this trend. This phenomenon is due to a coalescence of multiple factors: First, there is knowledge that many epigenetic markings remain “plastic” throughout all periods of brain development and aging, with ongoing and highly dynamic regulation even in neurons and other differentiated cells. Second, some of the chromatin-modifying drugs—histone deacetylase inhibitors are a well-known example—exert profound effects on brain metabolism and behavior in the animal model.18-21 Third, monogenetic disorders associated with widespread chromatin defects in brain cover a much wider continuum of neurological disease than previously thought, ranging from neurodevelopmental defects of early life to adult onset psychosis and dementia.22 And fourth, there is the emerging concept of transgenerational epigenetic inheritance, including early evidence for a role of environmental conditions and nutrition, as well as the physical and emotional health of a parent, as potential factors modulating the epigenetic state at the site of brain-relevant genes in the offspring.23
Epigenetic regulation in the brain—basic principles
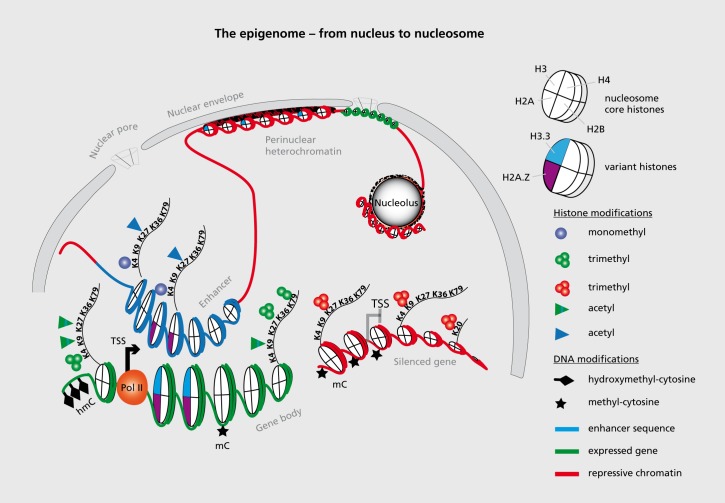
This section is limited to a very brief discussion of epigenetic markings that have been implicated in SCZ (discussed in the next section). The elementary unit of chromatin in the eukaryote cell is the nucleosome, or 146 bp of genomic DNA wrapped around an octamer of core histones, connected by linker DNA and linker histones. The collective set of covalent DNA and histone modifications and variant histones provide the major building blocks for the “epigenome,” or the epigenetic landscapes that define the organization of the genomic material into many tens of thousands of transcriptional units, clusters of condensed chromatin and other features that are differentially regulated in different cell types and developmental stages in a multicellular organ such as brain (Figure 1).24-28
Figure 1. Basic building blocks of the epigenome. The epigenome of a eukaryote (a cell with a well-defined nuclear membrane) is comprised of DNA modifications, including (but not limited to) cytosine methylation and hydroxymethylation, and a large number of site and residue-specific histone modifications and histone variants, only some of which are shown in this figure as representative examples. These molecular building blocks largely define the epigenetic landscapes that organize genomic DNA, often in a locus-specific fashion into active transcriptional units (green) including promoter and enhancer sequences (blue) and condensed chromatin including silenced genes (red). These epigenetic signatures are thought to distinguish between various cell types and developmental stages sharing the same genome.24,25 Many heterochromatic sequences are tethered to the nuclear envelope and pore complex, and also enriched at the periphery of the nucleolus (an intranuclear compartment for ribosomal biogenesis). A representative subset of histone variants and site-specific lysine (K) residues at histone H3 and H4 N-terminal tail that are potentially modified by methylation and/or acetylation, two types of covalent modifications among many others (see text). TSS, transcription start site; Pol, polymerase; hm, hydroxymethylation.
The bulk of DNA modifications exist as cytosine methylation (m) and hydroxymefhylation (hm).29 The mC5 and hmC5 markings show a differential (but not mutually exclusive) pattern of genomic occupancy. The hmC5 mark broadly correlates with local gene expression levels30,31 while methyl-cytosine (mC5) markings, particularly when positioned around the 5' end of genes is thought to function primarily as negative regulator of transcription.32,33 The regulation of chemical histone modifications is even more complex than the DNA methylation discussed above, and it is now thought that there are far more than 100 amino acid residue-specific post-translational modifications (PTMs) in a typical vertebrate cell,34 including mono (me1), di (me2)- and tri (me3) methylation, acetylation, and crotonylation, poly adenosine triphosphate (ADP)-ribosylation and small protein (ubiquitin, small ubiquitin-like modifier—SUMO) modification of specific lysine residues, as well as arginine (R) methylation and “citrullination,” serine (S) phosphorylation, tyrosine (T) hydroxylation, and several others.34-36 It is thought that multiple combinatorial sets of histone PTMs contribute to functional chromatin states that differentially define gene proximal promoters and gene bodies as opposed to enhancer and other regulatory sequences, condensed heterochromatin, and the “insulator” sequences that compartmentalize and provide boundaries for these various domains of chromatin (Figure 1).16 Proteins associated with the regulation of histone PTM are sometimes referred to as “writers”, or “erasers,” or “readers,” essentially differentiating between the process of establishing or removing a mark as opposed to its docking functions for chromatin remodeling complexes that regulate transcription, or induce and maintain chromatin condensation.36-38 In addition to these chemical modifications of the genomic DNA and the nucleosomal histones, other types of epigenetic regulation include histone variants (H3.1, H3.3, H2A.X, H2A.Z, etc) which differ from the canonical histones (H3/H4/H2A/H2B) only at very few amino acid positions, but robustly affect nucleosome stability and compaction.39 In addition there is “supranucleosomar” or higher-order “chromatin” regulation, which, at least in the nervous system, has barely been explored until now. For example, chromosomal loopings provide scaffolds that enable distal regulatory enhancer or silencer elements positioned potentially hundred kilobases apart from a gene, to physically interact directly with that gene's promoter sequences at the transcription start site.40
Epigenetic studies in SCZ postmortem brain and peripheral tissues—past and future
There can be little doubt that despite of the lack of a unifying neuropathology, many cases of SCZ are affected by gene expression alterations in the cerebral cortex and other brain regions, often including transcripts important for oligodendrocyte function and myelination,41-46 or inhibitory and excitatory neurotransmission,47-59 among others. It is almost always unclear whether these transcriptional changes are directly related to the underlying etiology or secondary events in the pathophysiology of disease. Given that transcriptional mechanisms are tightly linked to the chromatin remodeling and histone modification machinery in the nucleus,60,61 it would come as no surprise if some of the genes affected by altered expression in SCZ brain showed concomitant changes in the epigenetic architecture of their promoters, enhancers, repressor elements, and other regulatory sequences. To date, the majority of studies were focused on the quantification of DNA methylation (as a repressive mark) at candidate gene promoters, with some of the early work focused on the REELIN glycoprotein, the catechyl-O-methyltrans ferase COMT, the SOX10 developmental transcription factor.62-64 A few studies have measured changes in promoter-bound nucleosomal histone modifications, including histone acetylation and methylation.65,66 Interestingly, DNA methylation and histone modification changes at some of the promoters with altered epigenetic status in SCZ postmortem brain, including REELIN, Glutamate decarboxylase (GAD) 1 (encoding GAD67 GABA synthesis enzyme) and BDNF (brain-derived neurotrophic factor) were also found in lymphocyte extracts from patients,67-70 which if independently confirmed would warrant further examination as potential epigenetic biomarkers.
At the time of writing, however, very few studies have pursued DNA methylation or histone modification changes in SCZ on a genome-wide scale in brain tissue or peripheral cells,67,71-74 and none of these studies has harnessed the full power of modern (“next-generation”) sequencing technology that provides a near unbiased view of the distribution of an epigenetic mark across the entire genome.75 These modern epigenomic mapping technologies, when applied in conjunction with whole genome sequencing of specific individuals, are expected to inform on epigenetic alterations that could be driven by the underlying genetic risk architecture.76 As an illustrative example for the potential benefits when epigenome mappings are combined with genotyping, consider a recent report on risk-associated genetic variants for the autoimmune disorder multiple sclerosis, which showed a striking enrichment for regulatory sequences subject to distinct epigenetic decorations in immune cells, with disease-associated chromatin signatures that specifically affected promoters and enhancer elements.77 Given that, according to recent genome-scale studies conducted in cerebral and cerebellar cortex of control subjects, many hundreds of DNA methylation sites are significantly affected by single nucleotide polymorphisms (SNPs) and variants, some of which separated from the methylation site by more than one megabase.78,79 From this, there can be little doubt that in SCZ too, a significant portion of the epigenetic risk architecture is likely to be ultimately driven by the underlying genetic risk variants. Importantly, many of the DNA polymorphisms—according to some estimates, several thousand SNPs each could contribute a small but nonetheless significant SCZ risk80,81—do not change protein coding sequence and do not locate to exonic sequence. Therefore, cell-type specific epigenome mappings in normal and diseased human brain will be among the few options currently available to illuminate the functional and biological significance for many of these disease-relevant DNA polymorphisms.81
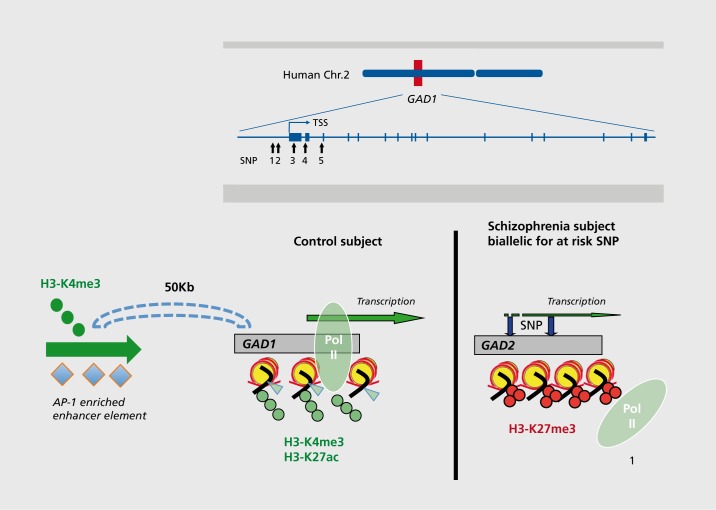
The potential benefits of including genotype information when analyzing epigenetic alterations in brain (or peripheral tissues) of specific cases diagnosed with SCZ also became apparent in some of the aforementioned candidate gene studies. The GAD1 promoter (chr. 2q31), which regulates GAD67 γ-aminobutyric acid (GABA) synthesis enzyme expression, could serve as an illustrative example. The GAD67 transcript is downregulated in cerebral and cerebellar cortex of a significant portion of subjects diagnosed with schizophrenia, depression, or autism, and this type of alteration may contribute to desynchronization of cortical networks and cognitive dysfunction due to defective GABAergic inhibition.50,82-87 Interestingly, a haplotype (a group of neighboring SNPs that are in linkage dysequilibrium with each other) positioned within few Kb from the GAD1 transcription start site confers genetic risk for accelerated loss of frontal lobe gray matter88,89 and, via epistatic interaction with catechol-o-methyl-transferase (COMT) alleles regulating synaptic dopamine, modulates prefrontal GABA levels.90 Notably, subjects with schizophrenia who are biallelic for this GAD1 promoter-associated risk haplotype, in striking contrast to cases with the protective alleles, show a significant deficit in prefrontal GAD67 transcript together with a shift in the epigenetic decoration of the surrounding chromatin, with loss of a facilitative histone methylation marking (histone H3 trimethyl-lysine 4) and excess of a repressive mark, histone 113 trimethyllysine 27 (Figure 2).66 Interestingly, these disease-associated changes in local chromatin templates at specific gene promoters apparently are accompanied by additional alterations in higher order chromatin structures, because decreased GAD1/GAD67 expression in SCZ prefrontal cortex (PFC) is accompanied by a weakening of a long range promoter-enhancer loop that normally interconnects regulatory sequences positioned 50Kb upstream of GAD1 with the gene's transcription start site and proximal promoter.91 Therefore, some of the risk-associated DNA polymorphisms at regulatory noncoding sequences could impact not only the epigenetic status of local chromatin structures but even exert “long-range” effects and impact epigenetic regulation of sequences that are positioned many kilobases further up- or downstream (Figure 2). Hence, one could expect that future epigenetic studies in SCZ postmortem brain will increasingly harness genotype information to explore whether epigenetic changes at the site of regulatory noncoding sequences are affected by underlying genetic variation related to disease risk81 or working memory and other cognitive functions often compromised in psychosis.92
Figure 2. Multiple layers of epigenetic dysregulation for the GAD1 promoter in SCZ prefrontal cortex, (top) A haplotype, comprised of at least five single-nucleotide polymorphisms within a few Kb from glutamate decarboxylase (GAD)1 transcription start site confers genetic risk for childhood-onset schizophrenia and accelerated loss of gray matter88 and is associated with decreased GAD1 gene expression in cerebral cortex of subjects on the psychosis spectrum. The at-risk haplotype, through yet unknown mechanisms, is in diseased individuals associated with decreased GAD1 gene expression, and a shift from open chromatin with high levels of the permissive marks, histone H3trimethyl-lysine 4 (H3K4me3) and H3-acetyl-lysine 27 to a more repressive state with the open marks H3K4me3 and H3K27ac replaced by a restrictive mark, H3K27me3. As a result, there are lower levels of transcription factors and phospho-activated RNA II polymerase (POI) at the proximal portions of the GAD1 gene.65,66 In addition to these changes in the epigenetic architecture of the GAD1 promoter, there are additional alterations in higher order chromatin. These include a chromosomal loop formation that physically connects enhancer sequences 50 kilobases upstream of the GAD1 gene with the GAD1 promoter and transcription start sites. These regulatory sequences are enriched with AP-1 (activating protein 1) transcription factor binding site and likely to promote GAD1 gene expression. In the PFC of some subjects with SCZ, there is a significant decrease in the GAD1 promoter-enhancer interaction frequency.91 .
Epigenetics and “gene x environment” interactions
It is obvious that the very concept of epi- (“over,” “above”) genetics lends itself towards molecular models for “gene x environment” interactions in the field of biological psychiatry and virtually any other field of biomedical research.93 From a heuristic perspective, the idea that myriads of external or internal factors could leave a long-lasting molecular imprint in the genome of our brain cells is extremely appealing to the neurobiological models of SCZ and related disorders, which often are viewed as neurodevelopmental in origin but cannot be fully explained by genetic risk. Some of the well-established risk factors, such as maternal infection during prenatal development with various types of viruses, pathogenic bacteria, or parasites, are estimated to play a significant role (“population-attributable risk”) in more than 30% of SCZ cases.94 Indeed, symptoms of psychosis may not surface until early adulthood, but a multitude of primary disease mechanisms could have operated as early as the prenatal period and in infancy.95 Furthermore, DNA and histone methylation mappings in the developing human cerebral cortex suggest that neuronal epigenomes specifically (and to some degree the non-neuronal constituents of cortex as well) are in the prenatal period and early childhood subject to presumably preprogrammed waves of DNA hydroxymethylation and methylation and histone H3K4 lysine (de) methylation at thousands of loci. In contrast, changes during the subsequent phases of maturation and aging are comparatively minor in relation to these earlier periods.78,96-99 Thus, there is considerable potential for “epigenetic plasticity”, particularly during the critical periods of human cortical development. Whether or not adverse environmental influences operating during these early time windows could indeed result in lasting and maladaptive “imprints” in our brain cells' chromatin is difficult to test. However, evidence from postmortem studies is in support of the hypothesis that early life experience may indeed leave a lasting epigenetic imprint in the human brain. For example, abnormal neuronal expression and DNA methylation of the NR3C1 glucocorticoid receptor distinguishes suicide victims who experienced childhood abuse from those who did not (100) and there is evidence other additional genes and loci are epigenetically altered in adult brain after exposure to early life trauma.101-103 In addition, some of the genes that become frequently dysregulated in the cortex of adult SCZ, including aforementioned GAD1/GAD67 GABA synthesis gene,104 are highly regulated across the extended period of prefrontal development, with expression levels ramping up slowly from the prenatal period at least until early adolescence,66,105 with dynamic changes in promoter-bound DNA methylation and histone methylation and acetylation continuing across the entire lifespan.65,66,96 This would, just as in the case of the aforementioned glucocorticoid receptor gene, indicate heightened epigenetic vulnerability of the GAD1 gene in early life. Indeed, this hypothesis received recent report from animal studies, because in the adult male rat, hippocampal Gad1 expression and open chromatin-associated histone acetylation are positively influenced by the level of maternal care in the postnatal period, while repressive Gadl -promoter DNA methylation was negatively influenced.106 Likewise, prenatal exposure to the alkylating and antimitotic agent methylazoxymethanol (MAM) causes decreased Gadl expression and histone H3K4 methylation in adult rat prefrontal cortex,107 a finding that is of interest given that similar changes were observed in clinical samples.66,108 Other types of prenatal adverse events, such as excessive maternal immune activation and activation of cytokine signaling, could result in prefrontal cortex of adult offspring in widespread changes of the GABAergic transcriptome, including Gad1,109 together with altered expression and epigenetic regulation of other SCZ susceptibility genes, including Disrupted-in-Schizophrenia 1 (DISC1) in the prefrontal cortex of adult offspring.110,111 These types of epigenetic vulnerability may extend to other brain regions and an even wider developmental period. For example, it had recently been reported that stress in adolescent animals could, in the context of a Disc1 mutation, result in altered DNA methylation at the tyrosine hydroxylase gene promoter in dopaminergic projection neurons of the ventral midbrain.112 Furthermore, a number of genes epigenetically dysregulated in SCZ could be modulated, even in a fully matured brain. For example, in the cortex of adult mice, the synthetic nicotinic acetylcholine receptor agonist varenicline, like nicotine in doses comparable to those reported in heavy smokers, reduces DNA methylation load at the Gad1 gene promoter, thereby increasing Gad1 expression.113,114 These findings, taken together, leave little doubt that environmental factors are likely to impact proper epigenetic regulation of SCZ-relevant genes across the entire lifespan.
Chromatin regulators linked to SCZ via evidence from genetics and functional genomics
To date, mutations and structural variants in perhaps up to 50 genes, each encoding a different chromatin regulator, have been linked to a wide range of neurodevelopmental syndromes, including rare monogenic forms of autism.115 Importantly, however, chromatin defects in brain were traditionally considered static lesions of early development that occurred in the context of rare genetic syndromes, but it is now clear that mutations and maladaptations of the epigenetic machinery cover a much wider continuum, including adult-onset neurodegenerative disease.22,116,117 Thus, there is a small but rapidly growing list of cases diagnosed with schizophrenia who harbor mutations in genes encoding chromatin regulators, including methyl-DNA binding proteins, histone modifiying enzymes and transcription factors. Thus, mutations and changes in the amino acid sequence of the neurodevelopmental susceptibility gene Methyl-CpG-binding protein 2 (MECP2, best known as the “Rett Syndrome” gene) are thought play causal roles in some SCZ cases.118,119 Furthermore, gene duplication of the histone methyltransferase KMT1D/EHMT1 or the MYTL1 transcription factor have been linked to some cases with SCZ.120,121
Such types of mono- or oligogenic forms of psychosis due to mutations in gene encoding a chromatin regulator are believed to be very rare and certainly not representative for the large majority of subjects on the SCZ spectrum. However, biological pathway analyses, after combining a diverse group of datasets, including risk loci from genome-wide association (GWAS) and copy number variant (CNV) studies and transcriptomics from diseased brain tissue, point to a broader contribution of the chromatin and nucleosome assembly machinery to the genetic risk architecture and neurobiology of schizophrenia.122,123 This includes the major histocompatibility (MHC) locus, spanning 4 to 7.6 Mb on chromosome 6p21.32-p22.2, and as one of the most intensely explored regions of the human genome, it has been consistently implicated in SCZ genetics as early as 1974.124,125 Interestingly, in a recent study on wholegenome gene expression profiles in lymph oblastoid cell lines (LCLs) from 413 SCZ cases and 446 controls, multiple histone variants encoded within the MHC region, including HIST1H2BD, HIST1H2BC, HIST1H2BH, HIST1H2BG and HIST1H4K, emerged among the top differentially expressed transcripts in the disease cohort126 and are likely to take part in complex chromosomal loopings that define local genome architectures at this locus in brain cells.127
Epigenetic therapies for SCZ?
Antipsychotics are the mainstay of the pharmacological treatment of SCZ, but the majority of patients show an incomplete response with an unfavorable disease course.128 Much of the problem revolves around the negative and cognitive symptoms that are responsible for the debilitating effects of SCZ and often do not respond to pharmacological treatment.129 It remains to be seen whether or not the knowledge gained by the field of neuroepigenetics will contribute towards improved treatment options in the future. Interestingly, both typical antipsychotics acting as dopamine D2 receptor antagonists and atypicals with a more mixed receptor profile affect DNA methylation and histone modification levels in cerebral cortex and striatum, two key nodes in the neural circuits of psychosis.19,130-132 It is currently unclear whether these observations, which are mostly of a correlative nature, indeed would indicate a critical role of chromatin regulatory mechanisms for antipsychotic drug action. In the following, inhibition of histone deacetylase activity will be discussed as one of the potential avenues for new antipsychotic drug research. As further discussed below, there are significant challenges that need to be overcome before such type of epigenetic therapy would be given serious considerations.
Histone acetylation is associated with a more flexible and “open” chromatin state, thereby facilitating gene expression; one example of this is that it enables enhancer and other regulatory sequences separated from a gene target site by thousands of kilo- or even megabases, to engage with distant promoters in chromosomal loop formations.133 Histone acetylation is regulated by the opposing effects of histone acetyltansferases (HATs) and deacetylases (HDACs). There are at least 18 different HDACs encoded in the human genome, which are commonly divided into four classes, based on their equivalents in yeast.134 Class I includes HDAC1, 2, 3, and 8, class II/IIa include HDAC 4,5,6,7,9, and 10, and HDAC11 is the sole representative of class IV (134); all these HDACs are defined by a zinc ion site in the catalytic binding pocket which also explains why many classical HDAC inhibitor (HDACi) drugs, including short-chain fatty acids (eg, butyrates), related compounds (eg, sodium valproate), and trichostatin A,134 have a broad profile and act on multiple HDACs (note that the clinically effective doses of valproate, as a mood stabilizer and anticonvulsant, are below those required to induce histone hyperacetylation in brain).135 Class III HDACs, which are also known as sirtuins, are defined by a different catalytic site, without the zinc ion but with nicotineamide dinucleotide (NAD+) as an essential cofactor.134 Most, or perhaps all of the HDACs are thought to target various nuclear and cytoplasmic non-histone proteins for deacetylation.134
Interestingly, expression of the class I histone deacetylase, HDAC1 was increased (on average 30% to 50%) in the prefrontal cortex and hippocampus of multiple SCZ postmortem brain cohorts.84,136-138 Therefore, abnormal HDAC1 expression in corticolimbic circuitry is a type of molecular pathology representative for a significant portion of subjects with SCZ. Furthermore, overexpression of Hdac1 in young adult mouse prefrontal cortex resulted in robust impairments in working memory, increased repetitive behaviors and abnormal locomotor response profiles in novel environments, in conjunction with dysregulated expression of more than 300 transcripts, including several that are located in the MHC risk locus on chromosome 6p21.3-22.1.138 Interestingly, Hdac1 expression becomes successively downregulated during the course or postnatal development, which could point to a neurodevelopmental etiology for the observed excessive HDAC1 expression in adult SCZ.138 Interestingly, Hdac2 which like Hdac1 is a class I HDAC (see above) has also recently been implicated in SCZ. Specifically, overexpression of Hdac2 in a mouse prefrontal cortex resulted in SCZ-like phenotypes, including diminished prepulse inhibition.19 On the other hand, conditional deletion of Hdac2 in postnatal forebrain neurons resulted in improved attentional set-shifting in the adult,139 which would suggest that alterations in expression or activity of HDAC2 result in very complex brain phenotypes, dependent on cell type and developmental stage.
These findings would suggest that drug-induced inhibition of neuronal and/or glial HDACs could result in a therapeutic effect for SCZ. However, there are significant challenges to explore this hypothesis in a clinical context. As discussed in in a recent review,135 there are newly developed potent HDAC inhibitor drugs (HDACi) either approved or in clinical trials, such as the benzamide-based MS-275 (tradename Entinostat), which crosses the blood-brain barrier and when orally administered indeed exerts a therapeutic effect in preclinical models of traumatic brain injury and neurodegeneration.140,141 However, these drugs, which are mostly used as anticancer agents, broadly inhibit multiple HDAC isoforms, lack CNS specificity, and, while not directly cytotoxic, nonetheless exhibit a safety profile that would mandate additional investigations prior to any experimental use in psychiatric patients.135 In addition, animal studies suggest that HDACi potentially augment therapeutic effects of atypical antipsychotic drugs19,142 and antidepressants.143-146 However, such types of combination treatments and polypharmacy would require even stricter safety criteria as compared with single drug regimens. We have argued that it may be premature to initiate trials with HDACi or other epigenetic drug targets in SCZ, but given that this is a rapidly evolving field, pending the availability of HDACi with favorable safety profiles, such trials should then given serious consideration.135 Interestingly, the human genome also encodes 50 proteins containing “bromodomains,” which essentially recognize and bind acetylated histone lysine residues147 and are thought to provide an important scaffold to recruit transcriptional proteins.148 The interaction acetyl-lysine binding pocket of these proteins is considered “draggable” and already identified as potential therapeutic target in some cancers and inflammatory disease.148 Some of the bromodomain containing proteins, including BRD1, are differentially regulated in cerebral cortex and hippocampus after electronconvulsive seizures149 and could provide important drug targets to treat conditions such as SCZ that are associated with transcriptional dysregulation in the cerebral cortex and other forebrain areas.
Conclusion
An increasing number of genes encoding chromatin regulators are linked to mono- and polygenic forms of neurodevelopmental disease, including some cases with schizophrenia. Furthermore, it is generally accepted that a significant portion of the genetic risk architecture of schizophrenia is positioned outside of coding sequences and in regulatory elements for gene expression (promoter, enhancers, repressors etc). Such types of regulatory elements are commonly defined by virtue of specific types of histone modifications and other types of epigenetic decorations, and therefore the combined epigenomic/genomic analyses in specific disease cases is expected to provide deep insights into the underlying molecular mechanisms of disease. Early findings from postmortem brain studies link some of the gene expression alterations in schizophrenia to changes in promoterbound DNA methylation and post-translational histone modifications. However, the field lacks larger-scale studies that map on a comprehensive and genome-wide scale the various epigenetic markings in diseased tissue. Such studies are urgently needed, particularly because a number of animal studies link developmental risk factors, including maternal immune activation during pregnancy and rearing conditions in the early postnatal period, and certain drugs and toxins, to lasting epigenetic alterations in offspring brain. Finally, many chromatin regulators are considered “draggable” and bear potential promise for novel treatments of schizophrenia and other neuropsychiatric diseases.
Acknowledgments
Work in the author's laboratory is supported by the National Institutes of Health and the Brain Behavior Research Foundation (BBRF/NARSAD).
REFERENCES
- 1.Hennekens CH., Hennekens AR., Hollar D., Casey DE. Schizophrenia and increased risks of cardiovascular disease. Am Heart J. 2005;150:1115–1121. doi: 10.1016/j.ahj.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Laursen TM., Munk-Olsen T., Vestergaard M. Life expectancy and cardiovascular mortality in persons with schizophrenia. Curr Opin Psychiatry. 2012;25:83–88. doi: 10.1097/YCO.0b013e32835035ca. [DOI] [PubMed] [Google Scholar]
- 3.Saha S., Chant D., McGrath J. A systematic review of mortality in schizophrenia: is the differential mortality gap worsening over time? Arch Gen Psychiatry. 2007;64:1123–1131. doi: 10.1001/archpsyc.64.10.1123. [DOI] [PubMed] [Google Scholar]
- 4.Taly A. Novel approaches to drug design for the treatment of schizophrenia. Exp Opin Drug Discovery. 2013;8:1285–1296. doi: 10.1517/17460441.2013.821108. [DOI] [PubMed] [Google Scholar]
- 5.Kim DH., Stahl SM. Antipsychotic drug development. Curr Topics Behav Neurosci. 2010;4:123–139. doi: 10.1007/7854_2010_47. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman JA., Stroup TS., McEvoy JP., et al Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med. 2005;353:1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 7.Swartz MS., Perkins DO., Stroup TS., et al Effects of antipsychotic medications on psychosocial functioning in patients with chronic schizophrenia: findings from the NIMH CATIE study. Am J Psychiatry. 2007;164:428–436. doi: 10.1176/ajp.2007.164.3.428. [DOI] [PubMed] [Google Scholar]
- 8.Ibrahim HM. amminga CA. Schizophrenia: treatment targets beyond monoamine systems. Annu Rev Pharmacol Toxicol. 2011;51:189–209. doi: 10.1146/annurev.pharmtox.010909.105851. [DOI] [PubMed] [Google Scholar]
- 9.Catts VS., Fung SJ., Long LE., et al Rethinking schizophrenia in the context of normal neurodevelopment. Front Cell Neurosci. 2013;7:60. doi: 10.3389/fncel.2013.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorph-Petersen KA., Lewis DA. Stereological approaches to identifying neuropathology in psychosis. Biol Psychiatry. 2011;69:113–126. doi: 10.1016/j.biopsych.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreassen OA., Thompson WK., Dale AM. Boosting the power of schizophrenia genetics by leveraging new statistical tools. Schizophr Bull. 2014;40:13–17. doi: 10.1093/schbul/sbt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Murillo L., Gogos JA., Karayiorgou M. The genetic architecture of schizophrenia: new mutations and emerging paradigms. Annu Rev Med. 2012;63:63–80. doi: 10.1146/annurev-med-072010-091100. [DOI] [PubMed] [Google Scholar]
- 13.Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akbarian S., Nestler EJ. Epigenetic mechanisms in psychiatry. Neuropsychopharmacology. 2013;38:1–2. doi: 10.1038/npp.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sweatt JD. Epigenetic Regulation in the Nervous System: Basic Mechanisms and Clinical Impact. London, UK Waltham, MA Academic Press; 2013 [Google Scholar]
- 16.Petronis A., Mill J. Brain, Behavior, and Epigenetics. Berlin, Heidelberg, Germany: Springer; 2011 [Google Scholar]
- 17.Appasani K. Epigenomics, from Chromatin Biology to Therapeutics. Cambridge, UK New York, NY Cambridge University Press; 2012 [Google Scholar]
- 18.Schroeder FA., Chonde DB., Riley MM., et al FDG-PET imaging reveals local brain glucose utilization is altered by class I histone deacetylase inhibitors. Neurosci Lett. 2013;550:119–124. doi: 10.1016/j.neulet.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurita M., Holloway T., Garcia-Bea A., et al HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat Neurosci. 2012;15:1245–1254. doi: 10.1038/nn.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abel T., Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vecsey CG., Hawk JD., Lattal KM., et al Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakovcevski M., Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohacek J., Gapp K., Saab BJ., Mansuy IM. Transgenerational epigenetic effects on brain functions. Biol Psychiatry. 2013;73:313–320. doi: 10.1016/j.biopsych.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Paredes M., Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 25.Li G., Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou VW., Goren A., Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 27.Ederveen TH., Mandemaker IK., Logie C. The human histone H3 complement anno 2011. Biochim Biophys Acta. 2011;1809:577–586. doi: 10.1016/j.bbagrm.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Kinney SM., Chin HG., Vaisvila R., et al Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J Biol Chem. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin SG., Wu X., Li AX., Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song CX., Szulwach KE., Fu Y., et al Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maunakea AK., Nagarajan RP., Bilenky M., al Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma RP., Grayson DR., Guidotti A., Costa E. Chromatin, DNA methylation and neuron gene regulation-the purpose of the package. J Psychiatry Neurosci. 2005;30:257–263. [PMC free article] [PubMed] [Google Scholar]
- 34.Tan M., Luo H., Lee S., J et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Taverna SD., Li H., Ruthenburg AJ., Allis CD., Patel DJ. How chromatinbinding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosammaparast N., Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 38.Justin N., De Marco V., Aasland R., Gamblin SJ. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Curr Opin Struct Biol. 2010:730–738. doi: 10.1016/j.sbi.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 39.Jin C., Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin F., Li Y., Dixon JR., et al A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martins-de-Souza D., Gattaz WF., Schmitt A., et al Alterations in oligodendrocyte proteins, calcium homeostasis and new potential markers in schizophrenia anterior temporal lobe are revealed by shotgun proteome analysis. J Neural Transm. 2009;116:275–289. doi: 10.1007/s00702-008-0156-y. [DOI] [PubMed] [Google Scholar]
- 42.Regenold WT., Phatak P., Marano CM., Gearhart L., Viens CH., Hisley KC. Myelin staining of deep white matter in the dorsolateral prefrontal cortex in schizophrenia, bipolar disorder, and unipolar major depression. Psychiatry Res. 2007;151:179–188. doi: 10.1016/j.psychres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 43.Katsel P., Davis KL., Haroutunian V. Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: a gene ontology study. Schizophr Res. 2005;79:157–173. doi: 10.1016/j.schres.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Aston C., Jiang L., Sokolov BP. Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res. 2004;77:858–866. doi: 10.1002/jnr.20208. [DOI] [PubMed] [Google Scholar]
- 45.Hakak Y., Walker JR., Li C., et al Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98:4746–4751 . doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tkachev D., Mimmack ML., Ryan MM., et al Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- 47.Duncan CE., Webster MJ., Rothmond DA., Bahn S., Elashoff M., Shannon Weickert C. Prefrontal GABA(A) receptor alpha-subunit expression in normal postnatal human development and schizophrenia. J Psychiatr Res. 2010;44:673–681. doi: 10.1016/j.jpsychires.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Charych El., Liu F., Moss SJ., Brandon NJ. GABA(A) receptors and their associated proteins: implications in the etiology and treatment of schizophrenia and related disorders. Neuropharmacology. 2009;57:481–495. doi: 10.1016/j.neuropharm.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo TU., Kim AM., Viscidi E. Disease-specific alterations in glutamatergic neurotransmission on inhibitory interneurons in the prefrontal cortex in schizophrenia. Brain Res. 2008;1218:267–277. doi: 10.1016/j.brainres.2008.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akbarian S., Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Rev. 2006;52:293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Guidotti A., Auta J., Davis JM., et al GABAergic dysfunction in schizophrenia: new treatment strategies on the horizon. Psychopharmacology. 2005;180:191–205. doi: 10.1007/s00213-005-2212-8. [DOI] [PubMed] [Google Scholar]
- 52.Dracheva S., Elhakem SL., McGurk SR., Davis KL., Haroutunian V. GAD67 and GAD65 rnRNA and protein expression in cerebrocortical regions of elderly patients with schizophrenia. J Neurosci Res. 2004;76:581–592. doi: 10.1002/jnr.20122. [DOI] [PubMed] [Google Scholar]
- 53.Hashimoto T., Bazmi HH., Mirnics K., Wu Q., Sampson AR., Lewis DA. Conserved regional patterns of GABA-related transcript expression in the neocortex of subjects with schizophrenia. Am J Psychiatry. 2008;165:479–489. doi: 10.1176/appi.ajp.2007.07081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benes FM. Amygdalocortical circuitry in schizophrenia: from circuits to molecules. Neuropsychopharmacology . 2010;35:239–257. doi: 10.1038/npp.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beneyto M., Kristiansen LV., Oni-Orisan A., McCullumsmith RE., MeadorWoodruff JH. Abnormal glutamate receptor expression in the medial temporal lobe in schizophrenia and mood disorders. Neuropsychopharmacology. 2007;32:1888–1902. doi: 10.1038/sj.npp.1301312. [DOI] [PubMed] [Google Scholar]
- 56.Meador-Woodruff JH., Healy DJ. Glutamate receptor expression in schizophrenic brain. Brain Res Rev. 2000;31:288–294. doi: 10.1016/s0165-0173(99)00044-2. [DOI] [PubMed] [Google Scholar]
- 57.Hemby SE., Ginsberg SD., Brunk B., Arnold SE., Trojanowski JQ., Eberwine JH. Gene expression profile for schizophrenia: discrete neuron transcription patterns in the entorhinal cortex. Arch Gen Psychiatry. 2002;59:631–640. doi: 10.1001/archpsyc.59.7.631. [DOI] [PubMed] [Google Scholar]
- 58.Mirnics K., Middleton FA., Marquez A., Lewis DA., Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 59.Middleton FA., Mirnics K., Pierri JN., Lewis DA., Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogel-Ciernia A., Wood MA. Neuron-specific chromatin remodeling: A missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology. 2014;80:18–27. doi: 10.1016/j.neuropharm.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adachi M., Monteggia LM. Decoding transcriptional repressor complexes in the adult central nervous system. Neuropharmacology. 2014;80:45–52. doi: 10.1016/j.neuropharm.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdolmaleky HM., Cheng KH., Russo A., et al Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 63.Grayson DR., Jia X., Chen Y., et al Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iwamoto K., Bundo M., Yamada K., et al DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25:5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang B., Dean B., Thomas EA. Disease- and age-related changes in histone acetylation at gene promoters in psychiatric disorders. Trans! Psychiatry. 2011;1:e64. doi: 10.1038/tp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang HS., Matevossian A., Whittle C., et al Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aberg KA., McClay JL., Nerella S., et al Methylome-Wide Association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gavin DP., Kartan S., Chase K., Jayaraman S., Sharma RP. Histone deacetylase inhibitors and candidate gene expression: An in vivo and in vitro approach to studying chromatin remodeling in a clinical population. J Psych Res. 2009;43:870–876. doi: 10.1016/j.jpsychires.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 69.Auta J., Smith RC., Dong E., et al DNA-methylation gene network dysregulation in peripheral blood lymphocytes of schizophrenia patients. Schizophr Res. 2013;150:312–318. doi: 10.1016/j.schres.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ikegame T., Bundo M., Murata Y., Kasai K., Kato T., Iwamoto K. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Hum Genet. 2013;58:434–438. doi: 10.1038/jhg.2013.65. [DOI] [PubMed] [Google Scholar]
- 71.Kano S., Colantuoni C., Han F., et al Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Mol Psychiatry. 2013;18:740–742. doi: 10.1038/mp.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dempster EL., Pidsley R., Schalkwyk LC., et al Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mill J., Tang T., Kaminsky Z., Khare T., et al Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wockner LF., Noble EP., Lawford BR., et al Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry. 2014;4:e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rivera CM., Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zazzu V., Regierer B., Kuhn A., Sudbrak R., Lehrach H. IT Future of medicine: from molecular analysis to clinical diagnosis and improved treatment. New Biotechnol. 2013;30:362–365. doi: 10.1016/j.nbt.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 77.Elangovan Rl., Disanto G., Berlanga-Taylor AJ., Ramagopalan SV., Handunnetthi L. Regulatory genomic regions active in immune cell types explain a large proportion of the genetic risk of multiple sclerosis. J Hum Genet. 2014;59:211–215. doi: 10.1038/jhg.2014.3. [DOI] [PubMed] [Google Scholar]
- 78.Numata S., Ye T., Hyde TM., et al DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang D., Cheng L., Badner JA., et al Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–419. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwab SG., Wildenauer DB. Genetics of psychiatric disorders in the GWAS era: an update on schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2013;263(suppl 2):S147–S154. doi: 10.1007/s00406-013-0450-z. [DOI] [PubMed] [Google Scholar]
- 81.Ripke S., O'Dushlaine C., Chambert K., et al Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Volman V., Behrens MM., Sejnowski TJ. Downregulation of paralbumin at cortical GABA synapses reduces network gamma oscillatory activity. J Neurosci. 2011;31:18137–18148. doi: 10.1523/JNEUROSCI.3041-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Curley AA., Lewis DA. Cortical basket cell dysfunction in schizophrenia. J. Physiol. 201 2;590(Pt 4):71 5–724. doi: 10.1113/jphysiol.2011.224659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benes FM., Lim B., Matzilevich D., Walsh JP., Subburaju S., Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guidotti A., Auta J., Davis JM., et al Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 86.Blatt GJ., Fatemi SH. Alterations in GABAergic biomarkers in the autism brain: research findings and clinical implications. Anat Rec. 2011;294:1646–1652. doi: 10.1002/ar.21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Akbarian S., Kim JJ., Potkin SG., Hagman JO., Tafazzoli A., Bunney WE., Jr, et al Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- 88.Addington AM., Gornick M., Duckworth J., Sporn A., Gogtay N., Bobb A., et al GAD1 (2q31.1), which encodes glutamic acid decarboxylase (GAD67), is associated with childhood-onset schizophrenia and cortical gray matter volume loss. Mol Psychiatry. 2005;10:581–588. doi: 10.1038/sj.mp.4001599. [DOI] [PubMed] [Google Scholar]
- 89.Straub RE., Lipska BK., Egan MF., Goldberg TE., Kleinman JE., Weinberger DR. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- 90.Marenco S., Savostyanova AA., van der Veen JW., et al Genetic modulation of GABA levels in the anterior cingulate cortex by GAD1 and COMT. Neuropsychopharmacology. 2010;35:1708–1717. doi: 10.1038/npp.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bharadwaj R., Jiang Y., Mao W., et al Conserved chromosome 2q31 conformations are associated with transcriptional regulation of GAD1 GABA synthesis enzyme and altered in prefrontal cortex of subjects with schizophrenia. J Neurosci. 2013;33:11839–11851. doi: 10.1523/JNEUROSCI.1252-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heck A., Fastenrath M., Ackermann S., et al Converging genetic and functional brain imaging evidence links neuronal excitability to working memory, psychiatric disease, and brain activity. Neuron. 2014;81:1203–1213. doi: 10.1016/j.neuron.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meaney MJ. Epigenetics and the biological definition of gene x environment interactions. Child Dev. 2010;81:41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- 94.Brown AS., Patterson PH. Maternal infection and schizophrenia: implications for prevention. Schizophr Bull. 2011;37:284–290. doi: 10.1093/schbul/sbq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schmitt A., Malchow B., Hasan A., Falkai P. The impact of environmental factors in severe psychiatric disorders. Front Neurosci. 2014;8:19. doi: 10.3389/fnins.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Siegmund KD., Connor CM., Campan M., et al DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PloS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shulha HP., Cheung I., Guo Y., Akbarian S., Weng Z. Coordinated cell type-specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS Genetics. 2013;9:e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheung I., Shulha HP., Jiang Y., Matevossian A., Wang J., Weng Z., et al Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci USA. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lister R., Mukamel EA., Nery JR., et al Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McGowan PO., Sasaki A., D'Alessio AC., Dymov S., Labonte B., Szyf M., et al Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Labonte B., Suderman M., Maussion G., et al Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Labonte B., Yerko V., Gross J., et al Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry. 2012;72:41–48. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 103.McGowan PO., Sasaki A., Huang TC., et al Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PloS One. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Curley AA., Arion D., Volk DW., et al Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: clinical, protein, and cell type-specific features. Am J Psychiatry. 2011;168:921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hyde TM., Lipska BK., Ali T., Mathew SV., Law AJ., Metitiri OE., et al Expression of GABA signaling molecules KCC2, NKCC1, and GAD1 in cortical development and schizophrenia. J Neurosci. 2011;31:11088–11095. doi: 10.1523/JNEUROSCI.1234-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang TY., Hellstrom IC., Bagot RC., Wen X., Diorio J., Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J Neurosci. 2010;30:13130–13137. doi: 10.1523/JNEUROSCI.1039-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.ackowiak M., Bator E., Latusz J., Mordalska P., Wedzony K. Prenatal MAM administration affects histone H3 methylation in postnatal life in the rat medial prefrontal cortex. Eur Neuropsychopharmacol. 2014;24:271289. doi: 10.1016/j.euroneuro.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 108.Huang HS., Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PloS One. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Richetto J., Calabrese F., Riva MA., Meyer U. Prenatal immune activation induces maturation-dependent alterations in the prefrontal GABAergic transcriptome. Schizophr Bull. 2014;40:351–361. doi: 10.1093/schbul/sbs195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tang B., Jia H., Kast RJ., Thomas EA. Epigenetic changes at gene promoters in response to immune activation in utero. Brain Behav immun. 2013;30:168–175. doi: 10.1016/j.bbi.2013.01.086. [DOI] [PubMed] [Google Scholar]
- 111.Connor CM., Dincer A., Straubhaar J., Galler JR., Houston IB., Akbarian S. Maternal immune activation alters behavior in adult offspring, with subtle changes in the cortical transcriptome and epigenome. Schizophr Res. 2012;140:175–184. doi: 10.1016/j.schres.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Niwa M., Jaaro-Peled H., Tankou S., et al Adolescent stress-induced epigenetic control of dopaminergic neurons via glucocorticoids. Science. 2013;339:335–339. doi: 10.1126/science.1226931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Satta R., Maloku E., Zhubi A., et al Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc Natl Acad Sci U S A. 2008;105:16356–16361. doi: 10.1073/pnas.0808699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maloku E., Kadriu B., Zhubi A., et al Selective alpha4beta2 nicotinic acetylcholine receptor agonists target epigenetic mechanisms in cortical GABAergic neurons. Neuropsychopharmacology. 2011;36:1366–1374. doi: 10.1038/npp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ronan JL., Wu W., Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genetics. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kleln CJ., Botuyan MV., Wu Y., et al Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Winkelmann J., Lin L., Schormair B., et al Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;2:2205–2210. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Piton A., Gauthier J., Hamdan FF., Lafreniere RG., Yang Y., Henrion E., et al Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2011;16:867–880. doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cohen D., Lazar G., Couvert P., et al MECP2 mutation in a boy with language disorder and schizophrenia. Am J Psychiatry. 2002;159:148–149. doi: 10.1176/appi.ajp.159.1.148-a. [DOI] [PubMed] [Google Scholar]
- 120.Kirov G., Pocklington AJ., Holmans P., et al De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee Y., Mattai A., Long R., Rapoport JL., Gogtay N., Addington AM. Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatr Genet. 2012;22:206–209. doi: 10.1097/YPG.0b013e328353ae3d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Luo X., Huang L., Jia P., Li M., Su B., Zhao Z., et al Protein-protein interaction and pathway analyses of top schizophrenia genes reveal schizophrenia susceptibility genes converge on common molecular networks and enrichment of nucleosome (chromatin) assembly genes in schizophrenia susceptibility loci. Schizophr Bull. 2014;40:39–49. doi: 10.1093/schbul/sbt066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gilman SR., Chang J., Xu B., et al Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cazzullo CL., Smeraldi E., Penati G. The leucocyte antigenic system HL-A as a possible genetic marker of schizophrenia. Br J Psychiatry .1974;125:25–27. doi: 10.1192/bjp.125.1.25. [DOI] [PubMed] [Google Scholar]
- 125.Corvin A., Morris DW. Genome-wide Association studies: findings at the major histocompatibility complex locus in psychosis. Biol Psychiatry. 2014;75:276–283. doi: 10.1016/j.biopsych.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 126.Sanders AR., Goring HH., Duan J., et al Transcriptome study of differential expression in schizophrenia. Hum Mol Genet. 2013;22:5001–5014. doi: 10.1093/hmg/ddt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mitchell AC., Bharadwaj R., Whittle C., et al The genome in three dimensions: a new frontier in human brain research. Biol Psychiatry. 2014;75:961–969. doi: 10.1016/j.biopsych.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weinberger D., Harrison P. eds. Schizophrenia. Third ed. Oxford, UK: Wiley-Blackwell; 2011 [Google Scholar]
- 129.Ibrahim HM. amminga CA. Schizophrenia: treatment targets beyond monoamine systems. Annu Rev Pharmacol Toxicol. 2011;51:189–209. doi: 10.1146/annurev.pharmtox.010909.105851. [DOI] [PubMed] [Google Scholar]
- 130.Akbarian S. Epigenetics of schizophrenia. Curr Topics Behav Neurosci. 2010;4:611–628. doi: 10.1007/7854_2010_38. [DOI] [PubMed] [Google Scholar]
- 131.Li J., Guo Y., Schroeder FA., et al Dopamine D2-like antagonists induce chromatin remodeling in striatal neurons through cyclic AMP-protein kinase A and NMDA receptor signaling. J Neurochem. 2004;90:1117–1131. doi: 10.1111/j.1471-4159.2004.02569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dong E., Nelson M., Grayson DR., Costa E., Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A. 2008;105:13614–13649. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schwarzenbacher R., McMullan D., Krishna SS., et al Crystal structure of a glycerate kinase (TM1585) from Thermotoga maritima at 2.70 A resolution reveals a new fold. Proteins. 2006;65:243–248. doi: 10.1002/prot.21058. [DOI] [PubMed] [Google Scholar]
- 134.Dokmanovic M., Clarke C., Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 135.Hasan A., Mitchell A., Schneider A., Halene T., Akbarian S. Epigenetic dysregulation in schizophrenia: molecular and clinical aspects of histone deacetylase inhibitors. Eur Arch Psychiatry Clin Neurosci. 2013;263:273–284. doi: 10.1007/s00406-013-0395-2. [DOI] [PubMed] [Google Scholar]
- 136.Sharma RP., Grayson DR., Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: analysis of the National Brain Databank microarray collection. Schizophr Res. 2008;98:111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Narayan S., Tang B., Head SR., et al Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jakovcevski M., Bharadwaj R., Straubhaar J., et al Prefrontal cortical dysfunction after overexpression of histone deacetylase 1. Biol Psychiatry. 2013;74:696–705. doi: 10.1016/j.biopsych.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Morris MJ., Mahgoub M., Na ES., Pranav H., Monteggia LM. Loss of histone deacetylase 2 improves working memory and accelerates extinction learning. J Neurosci. 2013;33:6401–6411. doi: 10.1523/JNEUROSCI.1001-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cao P., Liang Y., Gao X., Zhao MG., Liang GB. Administration of MS-275 improves cognitive performance and reduces cell death following traumatic brain injury in rats. CNS Neurosci Ther. 2013;19:337–345. doi: 10.1111/cns.12082. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 141.Zhang ZY., Schluesener HJ. Oral administration of histone deacetylase inhibitor MS-275 ameliorates neuroinflammation and cerebral amyloidosis and improves behavior in a mouse model. J Neuropathol Exp Neurol. 2013;72:178–185. doi: 10.1097/NEN.0b013e318283114a. [DOI] [PubMed] [Google Scholar]
- 142.Grayson DR., Kundakovic M., Sharma RP. Is there a future for histone deacetylase inhibitors in the pharmacotherapy of psychiatric disorders? Mol Pharmacol. 2010;77:126–135. doi: 10.1124/mol.109.061333. [DOI] [PubMed] [Google Scholar]
- 143.Schroeder FA., Lin CL., Crusio WE., Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry. 2007;62:55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 144.Zhu H., Huang Q., Xu H., Niu L., Zhou JN. Antidepressant-like effects of sodium butyrate in combination with estrogen in rat forced swimming test: involvement of 5-HT(1A) receptors. Behav Brain Res. 2009;196:200–206. doi: 10.1016/j.bbr.2008.08.039. [DOI] [PubMed] [Google Scholar]
- 145.Covington HE. 3rd, Vialou VF., LaPlant Q., Ohnishi YN., Nestler EJ. Hippocampal-dependent antidepressant-like activity of histone deacetylase inhibition. Neurosci Lett. 2011;493:122–126. doi: 10.1016/j.neulet.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lin H., Geng X., Dang W., et al Molecular mechanisms associated with the antidepressant effects of the class I histone deacetylase inhibitor MS275 in the rat ventrolateral orbital cortex. Brain Res. 2012;1447:119–125. doi: 10.1016/j.brainres.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 147.Filippakopoulos P., Picaud S., Mangos M., et al Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Helin K., Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 149.Fryland T., Elfving B., Christensen JH., Mors O., Wegener G., Borglum AD. Electroconvulsive seizures regulates the Brd1 gene in the frontal cortex and hippocampus of the adult rat. Neurosci Lett. 2012;516:110–113. doi: 10.1016/j.neulet.2012.03.069. [DOI] [PubMed] [Google Scholar]