Abstract
Clinical studies find that childhood adversity and stress-ful life events in adulthood increase the risk for major depression and for suicide. The predispositions to either major depression or suicide are thought to depend on genetic risk factors or epigenetic effects. We investigated DNA methylation signatures postmortem in brains of suicides with diagnosis of major depressive disorder. DNA methylation levels were determined at single C-phosphate-G (CpG) resolution sites within ventral prefrontal cortex of 53 suicides and nonpsychiatric controls, aged 16 to 89 years. We found that DNA methylation increases throughout the lifespan. Suicides showed an 8-fold greater number of methylated CpG sites relative to controls (P<2.2x10-16), with greater DNA methylation changes over and above the increased methylation observed in normal aging. This increased DNA methylation may be a significant contributor to the neuropathology and psychopathology underlying the risk of suicide in depression.
Keywords: aging, depression, DNA methylation, epigenetics, mood disorder, suicide
Abstract
Los estudíos clínicos han encontrado que la niñez adversa y los acontecimientos vitales estresantes en la adultez aumentan el riesgo para la depresión mayor y para el suícídío. Se piensa que la predísposición tanto para la depresión como para el suicidio depende de factores de ríesgo genético o de efectos epígenéticos. Se investigaron los signos de metilación del ADN en cerebros postmortem de pacíentes suícídas con díagnóstíco de depresíón mayor. Los níveles de metílacíón del ADN se determínaron en resolución única de Cítosinafosfato-Guanina (CfG) en la corteza prefrontal ventral de 53 pacíentes suicidas y de controles no psiquíátricos con edades entre 16 y 89 años. Se encontró que la metílación del ADN aumenta a lo largo de la vída. Los pacientes suícidas mostraron un número 8 veces mayor de sitios CfG metílados en relacíón con los controles (p<2,2x10-16), con cambíos mayores en la metilación del ADN, por sobre lo observado en el envejecímíento normal. Este aumento en la metílación del ADN puede contribuir de manera signifícativa a la neuropatología y a la psicopatología que están a la base del riesgo de suicidio en la depresión.
Abstract
D'après des études cliniques, le malheur dans l'enfance et les événements de vie stressant à l'âge adulte augmentent le risque de dépression caractérisée (majeure) et de suicide. Les prédispositions à la dépression caractérisée ou au suicide dépendent probablement de facteurs de risque génétiques ou d'effets épigénétiques. Nous avons analysé des signatures postmortem de la méthylation de l'ADN dans des cerveaux de suicidés diagnostiqués comme ayant eu un épisode dépressif caractérisé. Nous avons déterminé les taux de méthylation de l'ADN avec une résolution CpG (C-phosphate-G) à site unique dans le cortex préfrontal ventral de 53 suicidés et témoins non psychiatriques âgés de 16 à 89 ans. Nous avons trouvé que la méthylation de l'ADN augmente tout au long de la vie. Le nombre de sites CpG méthylés étaient multipliés par 8 chez les suicidés par rapport aux témoins (p<2,2x10-16), avec des changements de méthylation de l'ADN plus importants, allant au-delà de l'accroissement de la méthylation observé dans le vieillissement normal. Cette augmentation de la méthylation de l'ADN peut participer significativement à la neuropathologie et à la psychopathologie qui sous-tend le risque de suicide dans la dépression.
Introduction
The World Health Organization estimates that nearly 1 million people die from suicide yearly (World Health Organization figures, 2012). Psychological autopsies find that about 90% of suicides in Western nations have a psychiatric illness at the time of suicide.1 Of those, about 60% are suffering from a mood disorder. Suicide risk is influenced by a wide range of factors. Although some genetic risk factors have been identified, environmental events such as childhood adversity or repeated exposure to life-threatening situations typical in war and active combat have also been reported to contribute to the increased risk of suicidal behavior.2 Exposure to stress can confer lasting biological and behavioral effects, such as altered responses to stress, throughout the lifespan, and as such these effects may be mediated by epigenetic factors like DNA methylation (through addition of a methyl group to cytosine residues). Studies indicate that epigenetic alterations may contribute to the risk of suicide.3 For example, DNA methylation levels at the NGFI-A binding site of the human glucocorticoid receptor (hGR) promoter and at the rRNA promoter are increased in suicides with a history of childhood adversity compared with either controls and suicides without history of adversity. In both cases, hypermethylation was associated with decreased gene expression in hippocampus.4,5 History of childhood abuse in suicides has also been associated with site-specific hypermethylation in hippocampal hGR1B and 1C promoters.6 Moreover, serotonergic and γ-aminobutyric acid (GABA)ergic systems have been extensively studied in the context of suicide.7 The C allele of the 5-HT2A receptor gene polymorphism is hypermethylated in leukocytes of suicide attempters though not in prefrontal cortex (PFC) of suicides.8 Increased DNA methylation was detected in the GABAα receptor α1 gene in prefrontal cortex of suicides, together with increased DNA methyltransferase 3B (DNMT3B) in various brain regions such as the amygdala and brain stem.9 In the prefrontal cortex of suicides, promoter, hypermethylation, and lower gene expression were found for tyrosine kinase B (TrKB1).10 Hypermethylation of the brain-derived neurotrophic factor (BDNF) promoter IV has been reported in Wernicke's area in suicides versus controls.11 Less methylation of specific CpGs in the promoter region of the S-adenosylmethionine decarboxylase (AMD1) and arginase (ARG2) genes was correlated with increased gene expression in Brodmann Area 44 in suicides compared with nonsuicide controls.12
While the majority of these studies involved candidate genes previously implicated in suicide-related neuropathologies, Labonté et al utilized a genome-wide technique to examine DNA methylation changes in the hippocampi of suicide completers versus nonpsychiatric, sudden-death comparison subjects. A total of 366 differentially methylated promoters (273 hypermethylated and 93 hypomethylated) were found, including genes involved in learning, memory, and behavior.13 In the present study, we performed DNA methylation profiling on the orbital prefrontal cortex of 53 suicides and controls with a diagnosis of major depressive disorder (N=25) and in nonpsychiatric controls (N=28) focusing on DNA methylation perturbations associated with aging. Although a number of studies have reported directional epigenetic perturbations associated with aging in a multitude of tissue and organ systems including the human brain, to date no study has examined the dynamics of age-related epigenetic perturbations in suicide.14-20 These data represent a genome-scale study of DNA methylation dynamics associated with aging and suicide in postmortem human brain.
Materials and methods
Samples and subjects
Human tissue specimens were obtained according to an institutional approved protocol. Tissue was coded and samples had no person identifiers. The brains were collected at forensic autopsies. With informed consent, family members were interviewed as part of a psychological autopsy to obtain demographic information and any evidence of psychiatric or somatic disease. At least one family member per sample, for both cases and controls, agreed to be interviewed for the purpose of a psychological autopsy, which generated an Axis I and Axis II diagnosis.21 Depressed-suicide cases (N=25) were selected according to the following criteria: death by suicide, diagnosis of major depressive disorder (DSM-IV),22 absence of psychotropic or illegal drugs on toxicological screens, death without prolonged agonal state or protracted medical illness. Controls (N=28) did not have an Axis I psychiatric disorder, were drug-free and died suddenly without a prolonged agonal period from causes other than suicide. Due to the potentially confounding effect of pharmacological treatments in epigenetic studies, only brain samples with toxicological screens that ruled out. recent, consumption of medications (including illicit and psychotropic drugs) were investigated.
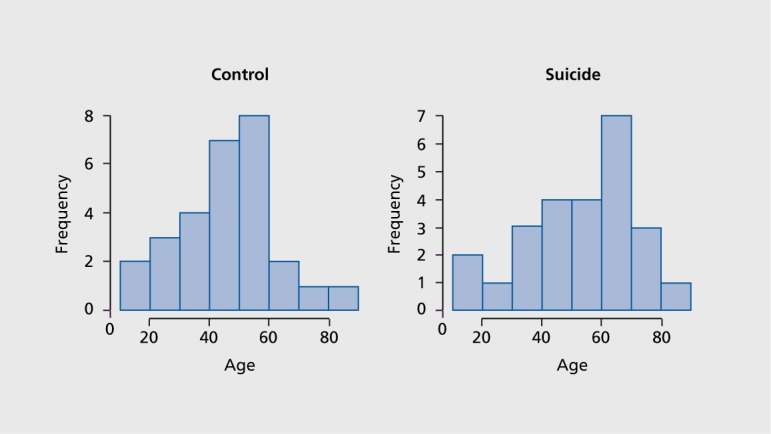
Distribution of age at death in suicides and nonpsychiatric, sudden death controls were comparable Figure 1. averaging 47±17 and 52±17 years among suicides and controls, respectively. All suicides also had a diagnosis of major depressive disorder. Brain pH ranged from 5.4 to 6.9 and the average postmortem interval (PMI) was 15±17 hours. Methylation patterns did not differ significantly by sex, pH, and PMI within specimens examined. Toxicological screens of body fluids and, in the majority of cases, brain tissue were used to rule out. exposure to medications within the last 3 months antemortem that might alter DNA methylation.23
Figure 1. Histogram of age at death among 28 nonpsychiatric control and 25 depressed suicide samples.
Sample dissection
A total of 53 cortical specimens from ventral prefrontal cortex were examined (Table I). The right cerebral hemisphere was sliced coronally at intervals of 2 to 5 cm, and the slices were rapidly frozen in Freon (dichlorodifluoromethane or 1,1,1,2 tetrafluoroe thane). Frozen specimens were then stored at -80°C until use. Prefrontal cortex Brodmann area 47 (BA47) was dissected from frozen brain sections that had been transferred from -80 to -20°C for 2 h prior to gray matter sampling.
Table I. Sample description with gender and diagnosis distributions.
| Diagnosis | Male | Female |
| Suicide cases | 15 | 10 |
| Nonpsychiatric controls | 19 | 9 |
Illumina Infinium HumanMethylation27 BeadChip
CpG methylation of 27 578 CPG sites was determined using Illumina Infinium HumanMethylation27 BeadChip, described previously.24 In accordance with the manufacturer's protocol, 1 ug of genomic DNA was bisulfite converted with the DNA Methylation Bisulfite Kit (Zymo). Samples were then subject to whole-genome amplification (WGA) and enzymatic fragmentation. Each sample was then hybridized to the locus-specific oligos on the BeadChip: T for the unmethylated state and C for the methylated state. Single base extension was then performed with labeled didcoxynuclcotides (ddNTPs) that can be measured and quantified. Methylation β values were quantified, by the intensity of the C allele over the combined intensities of the C and T alleles. To avoid confusion between the β value and parameters in the linear models used in our statistical analyses, hereafter we refer to the β value as methylation value. The Infinium assay is an extension of the GoldenGate assay.24 Methylation values for all samples were exported from GenomeStudio and samples with call rates below 95% were eliminated from subsequent analyses.
Illumina Infinium HumanMethylation QC and analysis
In addition to call-rate filtering, further data filtering was applied based on the following criteria: (i) probes that overlap with potential SNPs (annotated by dbSNP130) to eliminate methylation signal variability due to interindividual allelic variability; (ii) probes on sex chromosomes to avoid methylation bias in gender; and (iii) probes with detection P-values=;0.05 in over 10% of the depressed suicide cases and control samples, respectively. The total number of target CpG sites after filtering was 20 493. CpG sites with coefficient of variation (CV) >0.1 (15 249 CpGs) were considered as informative and used for all subsequent analyses.
A linear regression model using centered age (xi1) and phenotype (xi2)(xE) as the predictor variables and methylation value (xi1) as the response variable was fit by the R function aov:
yi = β0+β1xi1+β2xi2+β3xi1xi2+εi, Eq. 1
where i denotes target CpGs. In order to eliminate potential age bias, we transformed standard age data to centered age (as xj =aj-ā where j corresponds to a specific sample). The sum of squares for the three variables in the model (xi1 xi2, and xi1xi2) was computed by aov, and the ratio of each variable was calculated. The contribution of each variable to DNA is demonstrated by the distribution of the ratio of all target CpGs.
Significance of correlation between age and increase in DNA methylation among suicide cases compared with controls was computed using a t-statistic of β1 and the corresponding degrees of freedom. We also defined a null linear model
yi = β0+β1xi1+εi Eq. 2
We conducted an Analysis of Variance (ANOVA) F-test of the two models and computed P-values for a combined age and phenotype effect on DNA methylation for each CpG site. Since the filtered CpG sites from the HumanMethylation27 BeadChip were 15 249, the P-values generated by the ANOVA F-test were corrected for multiple testing using the Benjamini-Hochberg linear step-up procedure.25
DNA methylation changes with respect to factors like age and gender were evaluated with a simple linear model yi = β0+β1xi1+εi (where x1 refers to age or gender). P-values for a specific variable were estimated with a one-sided t-test. For each factor, we tested two null hypotheses: (i) DNA methylation does not increase with the confounding variable; and (ii) DNA methylation does not decrease with the confounding variable.
Gene ontology and pathway analysis
Genes with significant target CpGs were imported into Ingenuity Pathway Analysis Software (Ingenuity Systems, http://www.ingenuity.com/). Gene ontology analysis was performed and enriched function categories were produced using Ingenuity Knowledge Base Genes as the reference set. Enrichment P-values for biological functions were corrected for multiple testing using the Benjamini-Hochberg linear step-up procedure, with the threshold for significant, functions set a priori at P<0.05.
Validation of Illumina methylation assay at selected CpG sites by bisulfite pyrosequencing
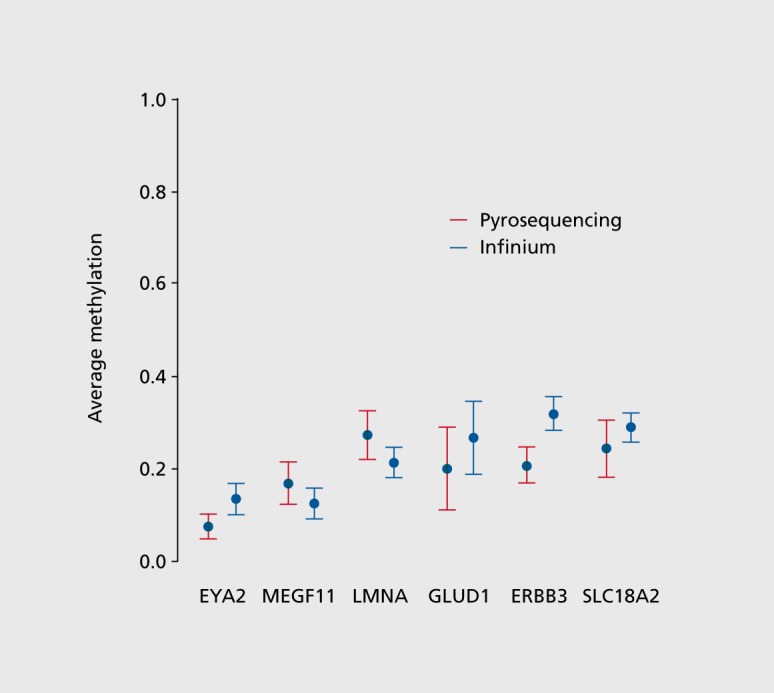
Methylation data from the Illumina assay was validated by quantifying methylation levels at randomly selected CpG sites by bisulfite pyrosequencing. The % CpG methylation at selected sites proximal to the genes Eya2 (eyes absent homolog 2), Megf11 (multiple EGFlike-domains 11), Lmna (lamin A/C),Gludl (glutamate dehydrogenase 1), Erbb3 (v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3) and Slc18a2 (solute carrier family 18 member 2) were quantified by bisulfite conversion of genomic DNA followed by PCR amplification and pyrosequencing Figure 2. 26 500 ng of genomic DNA from the same brain region and the batch of DNA used in the Illumina methylation assay was bisulfite-treated using the EpiTect Bisulfite kit (Qiagen) and stored at -20C until further analysis. The bisulfite converted DNA was amplified by PCR using the PyroMark PCR kit. (Qiagen) according to the manufacturer's instructions and PCR primers provided in Table II. PCR and sequencing primers were selected using the PyroMark Assay Design software (Version 2.0; Qiagen) and tested for linearity using standards with known methylation levels. PCR products were sequenced using a PyroMark Q96 MD system (Qiagen) following the manufacturer's suggested protocol. Percent methylation at each CpG site was quantified using the PyroMark CpG software (Version 1.0.10, Qiagen).
Figure 2. Validation of CpG methylation data using methylation pyrosequencing in 6 randomly selected genomic regions including 19 CpG dinucleotides. CpG methylation levels were mapped between Illumina Human Methylation BeadChip and methylation pyrosequencing platform.
Results
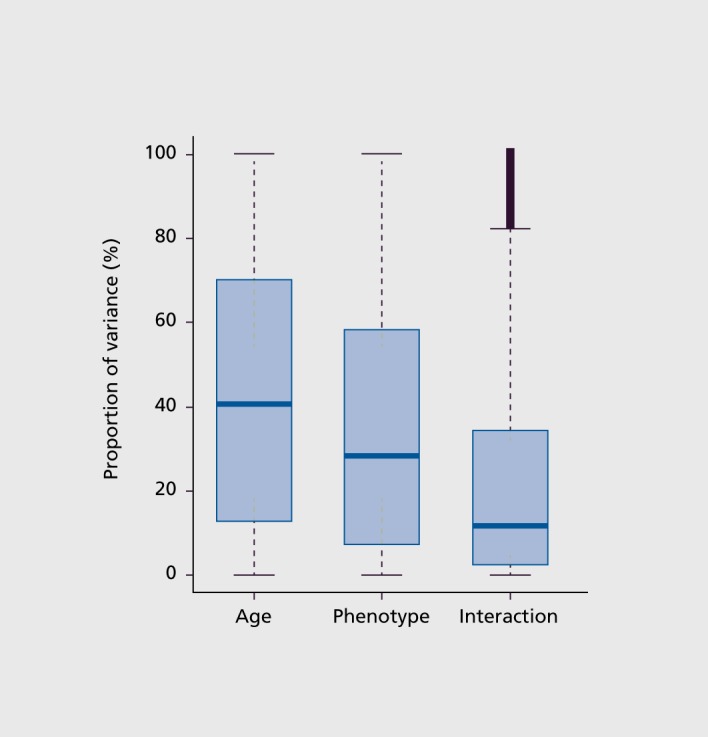
DNA methylation data analysis which included age and diagnosis, as well as interaction of these factors, revealed that CpG methylation variability in major depression suicides and controls is predominantly attributable to aging (Figure 3). These analyses are based on examination of 15 249 CpG sites in common across all 53 samples (referred to as “target” CpG sites through application of probe filtering, described in Methods). While diagnostic state (or phenotype) also accounted for some of the methylation variability in the data, age was the dominant factor (Figure 3).
Figure 3. DNA methylation patterns are significantly impacted by ageing. Among the samples, the variance component attributed to age is greater than that of diagnosis (phenotype) and age x phenotype interaction. Box plot shows the contribution of age, phenotype, and interaction of agexphenotype variance components, that account for the total methylation variability.
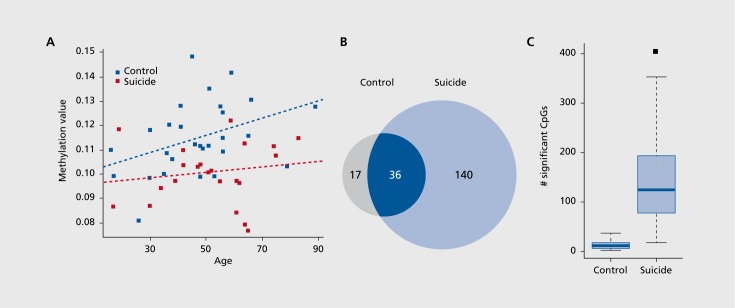
Given these observations, we next investigated age-related DNA methylation patterns in major depression suicide and control groups separately. In each diagnostic group (ie, suicide or control) a simple linear model with age as the covariate was applied to all target CpG sites. All significant CpG sites showed an increase in DNA methylation with increasing age Figure 4A. In contrast, no CpG site showed a decrease in DNA methylation relative to chronological age in either the suicide or control groups. Aging is positively correlated with CpG methylation (Figure 4A). Remarkably, the number of CpG sites with an increase in DNA methylation in the suicide group was 8-fold greater than in the control brains (Figure 4B, Fisher exact P-value<2.2x10-6). A total of 140 CpG sites showed increased CpG methylation, whereas only 17 CpG sites in controls exhibited an increase. There were 36 CpG sites common to both groups that showed increased CpG methylation (Figures 4A and B). To determine whether these findings are due to possible differences in age distribution of major depression suicide and control samples (Figure 1), we performed permutation tests where we randomly selected suicide and control samples within 10-year age intervals while maintaining the same number of suicide cases and controls in each age interval. This analysis eliminates potential biases in our results due to differences in age distributions among the suicide and control samples. Indeed, application of the permutation test also revealed that the major depression suicide group had substantially more CpG sites that undergo increased CpG methylation compared with controls (Figure 4C). These results indicate that there is much greater DNA methylation change across the lifespan in the prefrontal cortex in major depression suicides.
Figure 4. Aging and methylation in suicide brains. (A) Scatter plot showing DNA methylation is correlated with chronological age. (B) Venn diagram of significant target CpG sites with positive correlation between methylation and aging (P<0.05 corrected for multiple testing using Benjamini-Hochberg linear step-up procedure). (C) Box plot showing that the increase in the number of CpG sites in suicide brains is not attributed to slight differences in age distribution of suicide cases vs controls. The plot represents results of permutation test via resampling of suicide and control samples in a series of 10-year age intervals (exhausting all 63 possible suicide sample combinations and 1000 random sampling of age-matched controls), confirming significantly greater number of methylation changes in suicide brains (P<0.001).
Table II. Primers used in validation of Illumina methylation data via bisulfite pyrosequencing. The F, R, and S in the region column correspond to the forward, reverse, and sequencing primers.
| Gene | Region | Primer sequence | Biotinylated strand | Amplicon size | #CpG analyzed |
| F | GAGTTTTTTAGGAGTGGTATTTGGTATAGT | ||||
| EYA2 | R | TCCTCCCTACAACCCAAACAT | Forward | 154 | 3 |
| S | CCAAACATAACCCCTAT | ||||
| F | GGGTAGGGTAGGAGATAGGGATTTAAGAT | ||||
| MEGF11 | R | ACCCTCTTCTCTTTAAATCTCAATTTC | Reverse | 156 | 3 |
| S | GGGATTTAAGATAGGGG | ||||
| F | ATTAAAAGGGTTTTTGGTGAGGTTTGA | ||||
| LMNA | R | CCTCTCTAACCAAATCCATTCTACA | Reverse | 142 | 1 |
| S | GGGGATTTGAGTGAT | ||||
| F | ATGGGGAAGGAGAGATTTAGT | ||||
| GLUD1 | R | AACCACCCCAACTTCTTCAAAATAAT | Reverse | 182 | 5 |
| S | AGTATATGGTTGTAGGG | ||||
| F | AGGTTTGAAGTTTTGGAGAAAATAATTAGG | ||||
| ERBB3 | R | AATTACTACCCAAAACCCTACT | Reverse | 166 | 3 |
| S | TTTTGGAGAAAATAATTAGGTT | ||||
| F | GGTGTAAAGGGTGGTTTTTTTAGGAA | ||||
| SLC18A2 | R | AACACCAACCAAAACTTTCTTATAACTCC | Forward | 200 | 4 |
| S | CTTTAAACCTCAATATCCCTA |
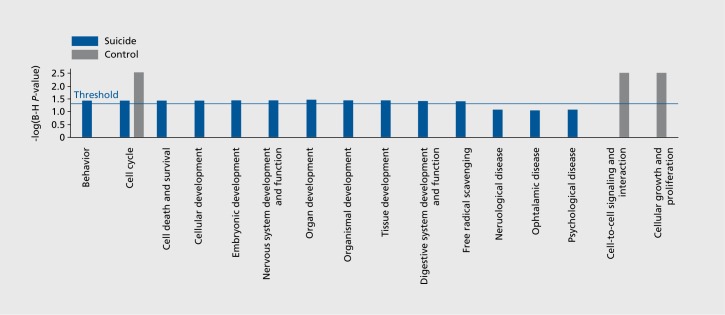
CpG sites that exhibited age-dependent DNA methylation were mapped to the human reference genome to find the associated genes. This identified a total of 17 and 136 genes uniquely to the control and major depression suicide groups, respectively, with 34 genes in common in the two groups. To distinguish the biological functions in major depression suicide and controls, genes that were uniquely mapped to either the suicide or control groups, a comparison of biological functions via Ingenuity Pathway Analysis software was employed. Genes showing increased methylation in major depression suicide were significantly associated with behavior, cell cycle, cell death and survival, cellular and embryonic development Figure 5. and correspondingly genes from the controls were significantly associated with cell cycle, cell-to-cell signaling and interaction, and cellular growth and proliferation.
Figure 5. Bar chart showing comparison of suicide and control diseases and functions via Ingeuity Pathway analysis.
Discussion
The present study shows increased age-related DNA methylation perturbations in prefrontal cortex in major depression suicide compared with nonpsychiatric controls. These perturbations involve a gain of methylation during aging, indicated by an 8-fold greater degree of methylation across the lifespan in major depression suicides, pointing to a mechanistic link between aberrant hypermethylation in suicide and related psycho- and neuropathologies, aging, and cellular senescence. Although this study cannot establish a causal relationship between increased DNA methylation and risk for suicide, it does show a remarkable increase in the numbers of cytosine sites in suicide brains as compared with ageand sex-matched controls across the lifespan. It should be noted that, the suicides in the present study also had a diagnosis of mood disorder, which in general is found in about 60% of suicides. Thus, these findings may underlie the neuropathology related to mood disorder and/or suicide. Accumulating evidence from postmortem studies reveals differences in morphology and neurotransmitter and trophic indices in the brains of suicides and controls, such as a reduction in neuronal density.27-29 Similarly, there is much evidence from postmortem studies indicating loss of neurons or glia in mood disorders.23,30-32 There are both structural and functional imaging data showing abnormalities in the brain associated with suicidal behavior and with mood disorders. The prefrontal cortex is both thinner and hypoactive in major depression and in association with suicidal behavior, independently of mood disorder.32 Future studies need to examine the effects of major depression and suicide separately.
Our findings suggest that some of the same molecular and cellular factors associated with aging may also contribute to the neuropathology of suicide, mood disorders, and related neuropathologies. The etiological basis of mood disorders appears to be more environmental and less genetic and familial as patients get older. The influence of epigenetic factors on suicide or depression may be greater as patients age relative to the impact of genetic factors. Aging is associated with cellular senescence involving telomere shortening, oxidative stress, and accumulation of genetic and epigenetic modifications.34-36 In particular, epigenetic modifications, such as DNA methylation and post-translational modifications of histone proteins, are central to cellular functioning, and those associated with aging can, in a subtle and progressive fashion, negatively influence proper cellular functioning. Such perturbations will likely have a cumulative effect, on neuronal functioning and have the potential to lead to neuropathology, severe mental illness or even suicide.
Acknowledgments
This work was supported by NIMH grants: MH40210, MH62185, MH082041, MH094774, MH090964, and the American Foundation for Suicide Prevention.
Contributor Information
Fatemeh Haghighi, James J. Peters Veterans Affairs Medical Center; Fishberg Department of Neuroscience, Mount Sinai School of Medicine; Department of Psychiatry, Division of Molecular Imaging and Neuropathology, New York State Psychiatric Institute, Columbia University; New York, USA.
Yurong Xin, Department of Psychiatry, Division of Molecular Imaging and Neuropathology, New York State Psychiatric Institute, Columbia University; New York, USA.
Benjamin Chanrion, Department of Psychiatry, Division of Molecular Imaging and Neuropathology, New York State Psychiatric Institute, Columbia University; New York, USA.
Anne H. O'Donnell, Department of Psychiatry, Division of Molecular Imaging and Neuropathology, New York State Psychiatric Institute, Columbia University; New York, USA.
Yongchao Ge, Department of Neurology, Icahn School of Medicine at Mount Sinai; New York, USA.
Andrew J. Dwork, Department of Psychiatry, Division of Molecular Imaging and Neuropathology, New York State Psychiatric Institute, Columbia University; New York, USA.
Victoria Arango, Department of Psychiatry, Division of Molecular Imaging and Neuropathology, New York State Psychiatric Institute, Columbia University; New York, USA.
J. John Mann, Department of Psychiatry, Division of Molecular Imaging and Neuropathology, New York State Psychiatric Institute, Columbia University; New York, USA.
REFERENCES
- 1.Harris EC., Barraclough B. Suicide as an outcome for mental disorders. A meta-analysis. Br J Psychiatry. 1997;170:205–228. doi: 10.1192/bjp.170.3.205. [DOI] [PubMed] [Google Scholar]
- 2.Mann JJ., Currier D. Relationships of genes and early-life experience to the neurobiology of suicidal behaviour. In: O'Connor RC, Platt S, Gordon J, eds. International Handbook of Suicide Prevention Research, Policy and Practice. Chichester, UK; Malden, MA: John Wiley & Sons. 2011:133–150. [Google Scholar]
- 3.Labonte B., Turecki G. The epigenetics of suicide: explaining the biological effects of early life environmental adversity. Arch Suicide Res. 2010;14:291–310. doi: 10.1080/13811118.2010.524025. [DOI] [PubMed] [Google Scholar]
- 4.McGowan PO., Sasaki A., D'Alessio AC., et al Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGowan PO., Sasaki A., Huang TC., et al Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labonte B., Yerko V., Gross J., et al Differential glucocorticoid receptor exon 1 (B), 1 (C), and 1 (H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry. 20121;72:41–48. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 7.Boldrini M., Mann JJ. Depression and suicide. In: Zigmond MJ, Rowland LP, Coyle JT, eds. Neurobiology of Brain Disorders. Elsevier Publishing. In press. [Google Scholar]
- 8.De Luca V., Viggiano E., Dhoot R., Kennedy JL., Wong AH. Methylation and QTDT analysis of the 5-HT2A receptor 102C allele: analysis of suicidally in major psychosis. J Psychiatr Res. 2009;43:532–537. doi: 10.1016/j.jpsychires.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Poulter MO., Du L., Weaver IC., et al GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry. 2008;64:645–652. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Ernst C., Deleva V., Deng X., et al Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch Gen Psychiatry. 2009;66:22–32. doi: 10.1001/archpsyc.66.1.22. [DOI] [PubMed] [Google Scholar]
- 11.Keller S., Sarchiapone M., Zarrilli F., et al TrkB gene expression and DNA methylation state in Wernicke area does not associate with suicidal behavior. J Affect Disord. 2011;135:400–404. doi: 10.1016/j.jad.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Gross JA., Fiori LM., Labonte B., Lopez JP., Turecki G. Effects of promoter methylation on increased expression of polyamine biosynthetic genes in suicide. J Psychiatr Res. 2013;47:513–519. doi: 10.1016/j.jpsychires.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labonte B., Suderman M., Maussion G., et al Genome-wide methylation changes in the brains of suicide completers. Am J Psychiatry. 2013;170:511–520. doi: 10.1176/appi.ajp.2012.12050627. [DOI] [PubMed] [Google Scholar]
- 14.Bjornsson HT., Sigurdsson Ml., Fallin MD., et al Intra-individual change over time in DNA methylation with familial clustering. JAMA. [Research Support, N.I.H., Extramural Research Support, Non-U. S. Gov't]. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boks MP., Derks EM., Weisenberger DJ., et al The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Twin Study]. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen BC., Houseman EA., Marsit CJ., et al Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't]. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakyan VK., Down TA., Maslau S., et al Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. [Research Support, Non-U.S. Gov't]. 2010;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraga MF., Ballestar E., Paz MF., et al Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez DG., Nails MA., Gibbs JR., et al Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. [Research Support, N.I.H., Extramura Research Support, N.I.H., Intramural]. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell JT., Tsai P-C., Yang T-P., Pidsley R., Nisbet J., Glass D. Epigenomewide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly TM., Mann JJ. Validity of DSM-III-R diagnosis by psychological autopsy: a comparison with clinician ante-mortem diagnosis. Acta Psychiatrica Scandinavica. 1996;94:337–343. doi: 10.1111/j.1600-0447.1996.tb09869.x. [DOI] [PubMed] [Google Scholar]
- 22.Frances A., First MB., Pincus H. . DSM-IV Guidebook. Washington, DC: American Psychiatric Association. 1995 [Google Scholar]
- 23.Underwood MD., Kassir SA., Bakalian MJ., Galfalvy H., Mann JJ., Arango V. Neuron density and serotonin receptor binding in prefrontal cortex in suicide. Int J Neuropsychopharmacol. 2012;15:435–447. doi: 10.1017/S1461145711000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bibikova M., Lin Z., Zhou L., et al High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Series B (Methodological). 1995;57:289–300. [Google Scholar]
- 26.Dupont JM., Tost J., Jammes H., Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Arango V., Underwood MD., Mann JJ. Fewer pigmented locus coeruleus neurons in suicide victims: preliminary results. Biol Psychiatry. 1996;39:112–120. doi: 10.1016/0006-3223(95)00107-7. [DOI] [PubMed] [Google Scholar]
- 28.Rajkowska G. Morphometric methods for studying the prefrontal cortex in suicide victims and psychiatric patients. Ann N Y Acad Sci. 1997;836:253–268. doi: 10.1111/j.1749-6632.1997.tb52364.x. [DOI] [PubMed] [Google Scholar]
- 29.Hercher C., Turecki G., Mechawar N. Through the looking glass: examining neuroanatomical evidence for cellular alterations in major depression. J Psychiatr Res. 2009;43:947–961. doi: 10.1016/j.jpsychires.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Arango V., Mann JJ. Abnormalities of brain structure and function in mood disorders. In: Charney DS, Nestler EJ, editors. The Neurobiology of Mental Illness. 3rd ed. New York, NY: Oxford University Press. 2009:515–529. [Google Scholar]
- 31.Boldrini M., Santiago AN., Hen R., et al Hippocampal granule neuron number and dentate gyrus volume in antidepressant-treated and untreated major depression. Neuropsychopharmacology. 2013;38:1068–1077. doi: 10.1038/npp.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajkowska G., Miguel-Hidalgo JJ., Wei J., et al Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol Psychiatry. 1999;45:1085–1098. doi: 10.1016/s0006-3223(99)00041-4. [DOI] [PubMed] [Google Scholar]
- 33.Van Heeringen K., Mann JJ. The neurobiology of suicide. Lancet Psychiatry. In press. 2014 doi: 10.1016/S2215-0366(14)70220-2. [DOI] [PubMed] [Google Scholar]
- 34.Engelfriet PM., Jansen EH., Picavet HS., Dolle ME. Biochemical markers of aging for longitudinal studies in humans. Epidemiol Rev. In press. doi: 10.1093/epirev/mxs011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gladyshev VN. The origin of aging: imperfectness-driven non-random damage defines the aging process and control of lifespan. Trends Genet. 2013;29:506–512. doi: 10.1016/j.tig.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Otin C., Blasco MA., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 20136;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]