Abstract
The mechanistic target of rapamycin (mTOR) signaling integrates diverse environmental cues, including growth factors, nutrients and immunological signals. Activation of mTOR signaling stimulates protein synthesis and anabolic metabolism and coordinates cell growth, proliferation and fate decisions. In recent years, mTOR signaling has been linked to the entire spectrum of T cell biology, ranging from T cell development and activation to lineage specification and memory formation. Mechanistically, mTOR activation profoundly affects the expression and activity of many immunologically relevant transcription factors to propagate immune signaling and mediate effector functions. These transcription factors orchestrate cell metabolism (MYC, SREBPs and HIF1), lineage differentiation (T-bet, GATA3, RORγt, FOXP3 and Eomesodermin) and immune activation and functions (NF-κB, FOXOs, IRF4, STATs and GFI-1). This review discusses how mTOR signaling, through impinging upon transcriptional factors, regulates T cell development, activation, and effector and memory differentiation.
Keywords: Eukaryotic transcription, transcription factors and co-activators
Introduction
The mechanistic target of rapamycin (mTOR) pathway is an evolutionarily conserved mechanism that primarily controls cell growth and metabolism. It consists of two protein complexes, mTOR complex 1 (mTORC1) and complex 2 (mTORC2). They share the kinase catalytic subunit mTOR, but are distinguished by two scaffolding subunits, RAPTOR (regulatory associated protein of mTOR) and RICTOR (RAPTOR-independent companion of mTOR), respectively. mTORC1, which is better studied, is activated by growth factors and nutrients mainly through phosphoinositide 3-kinase (PI3K)-AKT pathway.1 After activation, mTORC1 phosphorylates the translational initiation factor 4E (eIF4E) binding protein 1 (4E-BP1) and S6 kinase (S6K), which promote protein synthesis primarily from transcripts with 5′ terminal oligopyrimidine (TOP) motifs, 5′ TOP-like motifs or pyrimidine-rich translational elements (PRTE).2,3 Activation of mTORC1 is sensitive to rapamycin, an immunosuppressive drug. In contrast, how mTORC2 is activated is poorly understood, although mTORC2 is well known to phosphorylate AKT at Ser 473 and contributes to AKT activation. Whereas mTORC2 is insensitive to rapamycin in short-term treatment, long-term or high dose of rapamycin treatment can block mTORC2 activity in a variety of cells4,5 and CD4+ T cells,6,7 but not in effector CD8+ T cells.8
Recent studies have demonstrated that mTOR signaling is extensively involved in lymphocyte biology. Numerous immune signals, including T cell receptor (TCR), co-stimulatory signals and various cytokines, activate mTOR, which in turn regulates lymphocyte metabolism and dictates lineage differentiation and immune functions.9-11 Interestingly, many of these physiological effects are mediated by transcription factors whose expression or activity is dependent upon mTOR activation. Here, we review the impacts of mTOR signaling on transcription factors in different aspects of T cell biology, including lymphocyte development, activation and lineage differentiation.
mTORC2 Promotes Thymocyte Development Through Concerted Actions of NF-κB and FOXO Pathways
The NOTCH signaling pathway commits BM-derived progenitor cells to T-lineage fate and extinguishes non-T cell potentials. After engaging NOTCH ligands expressed on thymic epithelial cells, NOTCH receptors are cleaved to release the intracellular domain of NOTCH (ICN), which enters nucleus and regulates various target genes.12 PI3K-AKT pathway has been shown to act downstream of NOTCH and support thymocyte metabolism, proliferation and generation of double-positive (DP) cells. Lee et al. showed that loss of RICTOR in thymocytes impairs NOTCH-mediated NF-κB activation and increases activity of transcription factors FOXO1/3. Expression of constitutive active AKT, or combining constitutive active IKK2 and silencing of FOXO1/3, restores the defective NOTCH-induced differentiation in RICTOR-deficient thymocytes.13 Therefore, mTORC2, via activating AKT, relays signals from NOTCH receptors to NF-κB and FOXO pathways, and in turn promote DN to DP transition and normal T cell development. The mechanism through which mTORC2 activates NF-κB may involve the interaction between mTOR and IKK,14 or direct phosphorylation of PKC isoforms,15,16 which are critical for NF-κB activation in lymphocytes.17,18 Whether mTORC1 affects transcriptional programs in lymphocyte development remains to be addressed.
mTORC1 Promotes T cell Activation Through MYC and SREBPs
Naïve lymphocytes maintain a quiescent status in which they primarily rely on catabolism, especially fatty acid β-oxidation, to maintain homeostasis. Aberrant activation of mTORC1 upon deletion of its negative regulator Tsc1 disrupts quiescence of T cells, which predisposes them to apoptotic cell death.19-21 The downstream transcription factors mediating such events are unclear. Upon immune challenges, lymphocyte activation and clonal expansion necessitate rapid metabolic reprogramming, namely from catabolism to anabolism including glycolysis and lipid synthesis.10 The metabolic reprogramming is preceded by upregulation of several key transcription factors. One of the earliest activated factors is the oncogene MYC, which is responsible for initiating glycolysis and glutaminolysis. Acute deletion of MYC impairs TCR-induced expression of multiple glycolytic and glutaminolytic enzymes. Consequently, MYC-deficient T cells fail to activate and proliferate in response to TCR ligation.22 MYC upregulation is impaired by rapamycin treatment22 or deletion of Raptor,23 suggesting that mTOR signaling controls MYC expression. Previous studies using rapamycin have demonstrated that mTOR signaling enhances expression of MYC by promoting its translation, but not transcription or protein stability.24,25 In glioblastoma cells, activation of mTORC2 reduces class IIa histone deacetylase activity, which leads to increased FOXO acetylation. This in turn increases MYC expression by relieving microRNA-34c-dependent suppression.26 It is unclear whether this mechanism operates in T cells.
Aside from promoting glycolysis, mTOR signaling also activates lipid biosynthesis.27 Rapid upregulation of sterol synthesis accompanies lymphocyte activation.28 Kidani et al. demonstrated that the master transcription factors for lipid biosynthesis, the sterol regulatory element-binding proteins SREBP1 and SREBP2, activate fatty acid and cholesterol synthesis in T cells upon mitogenic stimulation.29 T cells deficient in the SREBP chaperone molecule SCAP, which controls the processing and subsequent transcriptional activity of SREBPs,30 fail to undergo metabolic reprograming, including lipid biosynthesis, glycolysis and ATP production. These cells have severe defects in growth and proliferation after mitogenic activation. Since MYC is activated normally in SCAP-deficient T cells, these metabolic defects are likely to be independent of MYC. Notably, the activation defect in SCAP-deficient T cells can be rescued by exogenous cholesterol, highlighting the importance of cholesterol synthesis in T cell activation.29 Importantly, the induction and processing of SREBPs are inhibited by rapamycin and PI3K inhibitors,29 or by the loss of Raptor.23 However, the expression of SREBP1 and SREBP2 mRNA is not affected by Raptor deficiency, suggesting that mTOR controls the expression of SREBPs through posttranscriptional regulation.23 PI3K-AKT-mTORC1 axis, but not mTORC2, promotes processing of SREBP1 in fibroblasts and epithelial cells.31,32 A recent study has showed that mTORC1 promotes SREBP activation by inducing nuclear exclusion of Lipin 1; nuclear accumulation of Lipin 1 inactivates SREBPs because it promotes the association of SREBPs to the nuclear matrix.33 Whether this is the case in T cells remains to be determined. Thus, the PI3K-mTORC1 axis orchestrates lipid synthesis by promoting SREBP expression and processing in T cells.
mTOR Dictates CD4+ T Cell Lineage Differentiation Through Multiple Transcription Factors
After activation, CD4+ T cells differentiate into different effector lineages, or become induced regulatory T cells (iTreg) dependent on specific cytokine milieu. Different effector lineages are promoted by distinct cytokine-mediated JAK-STAT transcriptional programs and defined by lineage-specific transcription factors.34 Delgoffe et al. first demonstrated that mTOR-deficient CD4+ T cells fail to differentiate into TH1, TH2 or TH17 effector lineages. This impairment of effector cell differentiation is due to failure of proper activation of specific STAT transcription factors, namely, STAT4 for TH1, STAT6 for TH2 and STAT3 for TH17.35 mTORC1 and mTORC2 have discrete functions in promoting effector lineage differentiation and transcription factor activation. Deficiency of Ras homolog enriched in brain (RHEB, an important activator of mTORC1) leads to normal TH2 but impaired TH1 and TH17 differentiation, because of reduced STAT4 and STAT3 activation, respectively. This is also correlated with diminished expression of T-bet, a TH1 master transcription factor, and RORγt, a TH17 master transcription factor.7 However, RAPTOR-deficient T cells showed relatively normal TH1, but defective TH17 differentiation.36 Recently, Yang et al. found that RAPTOR deficiency impairs TCR-induced metabolic reprogramming and IL-4 receptor expression, leading to a severe defect in TH2 differentiation. Such defect is associated with impaired STAT activation and GATA3 induction.23 The discrepancy between RHEB and RAPTOR deficiencies likely results from RHEB-independent activation of mTORC1.37 Indeed, RHEB is not required for sustained mTORC1 activation in CD4+ T cells.23
In contrast, loss of RICTOR preserves TH1 and TH17, but impairs TH2 differentiation, which is associated with reduced STAT6 activation and defective expression of GATA3.7 A separate report found that mTORC2 promotes TH2 and TH1 differentiation through PKCθ-NF-κB and AKT signaling, respectively, without affecting STAT activation.6 These discrepancies remain to be resolved. The mechanistic basis underlying mTOR-mediated activation of various transcription factors also warrants further investigation.
We have limited understanding as to how mTOR signaling regulates T cell lineage transcription factors. It is unclear whether mTOR signaling directly regulates their activation or through other intermediates. Nonetheless, the PI3K-mTOR axis has been shown to promote GATA3 translation, which could partially explain the defective TH2 differentiation in RAPTOR and RICTOR-deficient T cells.38 Furthermore, several mechanisms could underlie mTORC1-mediated transcriptional control of TH17 differentiation. First, mTOR has been shown to directly phosphorylate STAT3 and promotes its activation in various systems,39,40 which could potentially explain the defective STAT3 phosphorylation in mTOR-deficient T cells.35 Second, PI3K-mTORC1-S6K1 axis suppresses GFI-1, a transcription repressor that inhibits TH17 differentiation.41,42 mTORC1-S6K1 activates EGR2 (Early growth response 2), a transcription factor that inhibits GFI-1 expression by directly binding to its promoter. Accordingly, transduction of constitutive active form of S6K1 partially rescues TH17 differentiation of rapamycin-treated T cells.36 Third, PI3K-mTORC1-S6K2 axis promotes RORγt nuclear translocation.36 S6K2 is the nuclear-localized counterpart of S6K1, and its expression is enhanced by mTORC1 activation likely through posttranscriptional mechanisms. S6K2 interacts with RORγt and facilitates its nucleus translocation.36 Lastly, mTOR signaling induces HIF1 expression and HIF1-dependent glycolytic program in differentiating TH17 cells, which promotes TH17 cell generation and at the same time, suppresses iTreg differentiation.43 mTOR signaling enhances HIF1 mRNA transcription and protein translation, as well as HIF1 transcriptional activity that is likely mediated by interaction between RAPTOR and HIF1.31,44-46 Thus, the mTORC1-HIF1 axis reciprocally controls TH17 and iTreg differentiation.
Although mTOR deficiency disables effector lineage differentiation, it diverts CD4+ T cells into iTreg cells even without exogenous iTreg-polarizing cytokines.35 Activation of mTOR by PI3K-AKT pathway suppresses the expression of FOXP3, the master transcription factor for regulatory T cells.47-51 Conversely, inhibition of mTOR with rapamycin or by limiting essential amino acids promotes FOXP3 expression and differentiation of iTreg cells.47,48,52 Both mTORC1 and mTORC2 contribute to the suppression of FOXP3 expression, as deletion of either RHEB or RICTOR fails to spontaneously generate iTreg cells.7,35 However, despite mounting evidence linking mTOR to FOXP3, little is known regarding the detailed molecular mechanism underlying mTOR-mediated FOXP3 suppression. It is also important to note that deletion of RAPTOR or RICTOR impairs the function or generation of thymus-derived FOXP3+ Treg cells, respectively.53
mTORC1 Promotes CD8+ T Cell Effector Differentiation Through HIF1 and IRF4
After TCR activation and initial clonal expansion, CD8+ T cells differentiate into effector cytotoxic effector cells by expressing an array of effector molecules, including perforin, interferon-γ (IFN-γ) and granzymes. The transcription factor hypoxia-inducible factor 1 (HIF1) is induced early during T cell activation in an mTOR-dependent manner.8,22,46 Although its deficiency does not affect initial T cell activation, proliferation or metabolic reprogramming,8,22 HIF1 is required for sustained glycolysis and pyruvate metabolism in effector CD8+ T cells. Furthermore, HIF1-deficient effector CD8+ T cells have defective expression of perforin and certain granzymes, but not other effector molecules, such as interferon-γ, suggesting that the mTORC1-HIF1 axis regulates a specific transcriptional program in effector CD8+ T cells.8 TCR-induced mTOR signaling promotes HIF1 protein synthesis, likely through 4E-BP1-eIF4 mediated cap-dependent translation.31,46
Interferon regulatory factor 4 (IRF4) is a transcription factor critical for many immune cell differentiation and function.54 IRF4 expression is induced by TCR engagement and correlates with TCR strength.55,56 Rapamycin treatment impairs TCR-mediated IRF4 induction, indicating that mTOR signaling regulates IRF4 expression.55 Similar to HIF1, IRF4 is dispensable for early T cell activation and proliferation. However, IRF4 maintains CD8+ T cell survival and is required for ongoing clonal expansion and effector function in a dose-dependent manner.55,56 Further, IRF4 sustains the high aerobic glycolysis in activated CD8+ by promoting the expression of multiple glycolytic enzymes.56 Consequently, IRF4-deficient CD8+ T cells fail to generate a productive effector response upon viral challenge.55,56 The molecular mechanism whereby mTOR signaling promotes TCR-induced IRF4 expression remains unclear.
mTOR Controls Memory CD8+ Generation Through T-bet and Eomesodermin
After effector cell generation, a small subset of T cells differentiates into long-lived memory T cells. A metabolic transition from anabolism to catabolism accompanies memory T cell differentiation. Several studies have demonstrated that whereas mTORC1 promotes effector CD8+ T cell generation, it negatively regulates memory T cell development. Rapamycin treatment or silencing of RAPTOR enhances memory cell generation and function.57-59 Importantly, inhibition of mTORC1 reduces IL-12-induced expression of T-bet, a transcription factor essential for effector differentiation, and activates Eomesodermin (Eomes), a transcription factor that enhances memory cell formation.59,60 Thus, mTOR dictates effector and memory differentiation in part through differentially regulating the corresponding transcription factors. Interestingly, a recent study indicated that mTOR promotes IgG1 memory B cells differentiation into plasma cells by suppressing the transcription repressor BACH2,61 indicating different roles of mTOR signaling in memory T and B cell differentiation and function.
Concluding Remarks
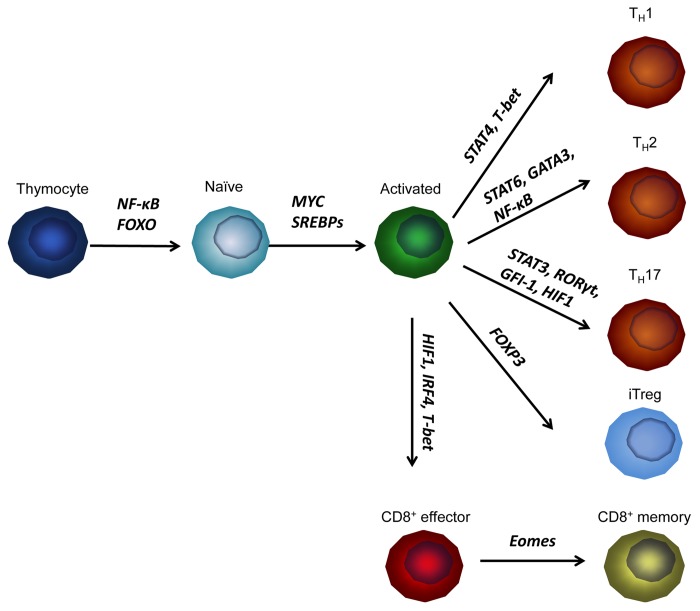
The physiological importance of mTOR signaling is underscored by the fact that dysregualtion of mTOR is associated with many human diseases. Although mTOR signaling was traditionally linked to protein translation, recent studies have demonstrated that mTOR also controls gene transcription in T cells as well as other experimental systems.62 mTOR signaling modulates T cell development, activation, differentiation, memory generation, and function through a diverse set of transcription factors (Fig. 1),10 highlighting the complexity of the involvement of mTOR in lymphocyte biology. These transcription factors orchestrate cell metabolism (MYC, SREBPs and HIF1), lineage differentiation (T-bet, GATA3, RORγt, FOXP3 and Eomesodermin), and immune activation and functions (NF-κB, FOXOs, IRF4, STATs and GFI-1). Despite these exciting findings, we have insufficient understanding in regard to the molecular mechanisms that link mTOR to these transcription factors, especially in the context of T cell biology. How mTOR signaling regulates specific transcription factors in different T cell lineages is an important question awaiting future investigation. Another fascinating, yet poorly studied area is how transcription factors may influence mTOR signaling. Recent studies have revealed an intricate crosstalk between MYC and mTOR signaling. MYC cooperates with AKT-mTOR signaling to promote ribosome biogenesis.63 Pourdehnad et al. found that mTOR-dependent phosphorylation of 4EBP1 is required for cancer cell survival in MYC-driven tumor initiation and maintenance.64 4EBP1, but not S6K, is highly phosphorylated in cancer cells induced by MYC overexpression. New class of mTOR active site inhibitors, which potently inhibit 4EBP1-eIF4E axis, induce apoptosis in MYC-driven cancer cells, delay tumorigenesis, and prolong animal survival.64 Furthermore, MYC-deficient T cells exhibit impaired mTOR activity.22 These findings further highlight the complexity of the interplay between mTOR signaling and transcription factors. We anticipate that more mechanistic studies will unravel the detailed connections between mTOR and gene transcription, which may contribute to the development of novel diagnostics and treatments of immune-related diseases.
Figure 1. Schematics of T cell development, activation, lineage differentiation and memory formation. mTOR-controlled transcription factors that regulate different stages are highlighted in bold.
References
- 1.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 5.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–43. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–53. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, Cantrell DA. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209:2441–53. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–38. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng H, Chi H. mTOR and lymphocyte metabolism. Curr Opin Immunol. 2013;25:347–55. doi: 10.1016/j.coi.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng H, Chi H. The interplay between regulatory T cells and metabolism in immune regulation. Oncoimmunology. 2013;2:e26586. doi: 10.4161/onci.26586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson PK, Zúñiga-Pflücker JC. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin Immunol. 2011;23:350–9. doi: 10.1016/j.smim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Lee K, Nam KT, Cho SH, Gudapati P, Hwang Y, Park DS, Potter R, Chen J, Volanakis E, Boothby M. Vital roles of mTOR complex 2 in Notch-driven thymocyte differentiation and leukemia. J Exp Med. 2012;209:713–28. doi: 10.1084/jem.20111470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. Akt-dependent regulation of NF-kappaB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22:1490–500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao Y, Miao RQ, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–43. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–31. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su TT, Guo B, Kawakami Y, Sommer K, Chae K, Humphries LA, Kato RM, Kang S, Patrone L, Wall R, et al. PKC-beta controls I kappa B kinase lipid raft recruitment and activation in response to BCR signaling. Nat Immunol. 2002;3:780–6. doi: 10.1038/ni823. [DOI] [PubMed] [Google Scholar]
- 18.Sun Z, Arendt CW, Ellmeier W, Schaeffer EM, Sunshine MJ, Gandhi L, Annes J, Petrzilka D, Kupfer A, Schwartzberg PL, et al. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature. 2000;404:402–7. doi: 10.1038/35006090. [DOI] [PubMed] [Google Scholar]
- 19.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–97. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q, Liu Y, Chen C, Ikenoue T, Qiao Y, Li CS, Li W, Guan KL, Liu Y, Zheng P. The tuberous sclerosis complex-mammalian target of rapamycin pathway maintains the quiescence and survival of naive T cells. J Immunol. 2011;187:1106–12. doi: 10.4049/jimmunol.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien TF, Gorentla BK, Xie D, Srivatsan S, McLeod IX, He YW, Zhong XP. Regulation of T-cell survival and mitochondrial homeostasis by TSC1. Eur J Immunol. 2011;41:3361–70. doi: 10.1002/eji.201141411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, Guertin DA, Lamb RF, Chi H. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–56. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gera JF, Mellinghoff IK, Shi Y, Rettig MB, Tran C, Hsu JH, Sawyers CL, Lichtenstein AK. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem. 2004;279:2737–46. doi: 10.1074/jbc.M309999200. [DOI] [PubMed] [Google Scholar]
- 25.Wall M, Poortinga G, Hannan KM, Pearson RB, Hannan RD, McArthur GA. Translational control of c-MYC by rapamycin promotes terminal myeloid differentiation. Blood. 2008;112:2305–17. doi: 10.1182/blood-2007-09-111856. [DOI] [PubMed] [Google Scholar]
- 26.Masui K, Tanaka K, Akhavan D, Babic I, Gini B, Matsutani T, Iwanami A, Liu F, Villa GR, Gu Y, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–39. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamming DW, Sabatini DM. A Central role for mTOR in lipid homeostasis. Cell Metab. 2013;18:465–9. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen HW, Heiniger HJ, Kandutsch AA. Relationship between sterol synthesis and DNA synthesis in phytohemagglutinin-stimulated mouse lymphocytes. Proc Natl Acad Sci U S A. 1975;72:1950–4. doi: 10.1073/pnas.72.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–99. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–31. doi: 10.1172/JCI0215593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–36. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–20. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–44. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, et al. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ. Cell Rep. 2012;1:360–73. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cook KD, Miller J. TCR-dependent translational control of GATA-3 enhances Th2 differentiation. J Immunol. 2010;185:3209–16. doi: 10.4049/jimmunol.0902544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol. 2000;10:47–50. doi: 10.1016/S0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 40.Kim JH, Yoon MS, Chen J. Signal transducer and activator of transcription 3 (STAT3) mediates amino acid inhibition of insulin signaling through serine 727 phosphorylation. J Biol Chem. 2009;284:35425–32. doi: 10.1074/jbc.M109.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichiyama K, Hashimoto M, Sekiya T, Nakagawa R, Wakabayashi Y, Sugiyama Y, Komai K, Saba I, Möröy T, Yoshimura A. Gfi1 negatively regulates T(h)17 differentiation by inhibiting RORgammat activity. Int Immunol. 2009;21:881–9. doi: 10.1093/intimm/dxp054. [DOI] [PubMed] [Google Scholar]
- 42.Zhu J, Davidson TS, Wei G, Jankovic D, Cui K, Schones DE, Guo L, Zhao K, Shevach EM, Paul WE. Down-regulation of Gfi-1 expression by TGF-beta is important for differentiation of Th17 and CD103+ inducible regulatory T cells. J Exp Med. 2009;206:329–41. doi: 10.1084/jem.20081666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–76. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Land SC, Tee AR. Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. J Biol Chem. 2007;282:20534–43. doi: 10.1074/jbc.M611782200. [DOI] [PubMed] [Google Scholar]
- 45.Treins C, Giorgetti-Peraldi S, Murdaca J, Semenza GL, Van Obberghen E. Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J Biol Chem. 2002;277:27975–81. doi: 10.1074/jbc.M204152200. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura H, Makino Y, Okamoto K, Poellinger L, Ohnuma K, Morimoto C, Tanaka H. TCR engagement increases hypoxia-inducible factor-1 alpha protein synthesis via rapamycin-sensitive pathway under hypoxic conditions in human peripheral T cells. J Immunol. 2005;174:7592–9. doi: 10.4049/jimmunol.174.12.7592. [DOI] [PubMed] [Google Scholar]
- 47.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O’Connor E, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008;105:7797–802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–74. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu G, Burns S, Huang G, Boyd K, Proia RL, Flavell RA, Chi H. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol. 2009;10:769–77. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat Immunol. 2010;11:1047–56. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park Y, Jin HS, Lopez J, Elly C, Kim G, Murai M, Kronenberg M, Liu YC. TSC1 regulates the balance between effector and regulatory T cells. J Clin Invest. 2013;123:5165–78. doi: 10.1172/JCI69751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cobbold SP, Adams E, Farquhar CA, Nolan KF, Howie D, Lui KO, Fairchild PJ, Mellor AL, Ron D, Waldmann H. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc Natl Acad Sci U S A. 2009;106:12055–60. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng H, et al. mTORC1 couples immune signals and metabolic programming to establish T-cell function. Nature. 2013 doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu WD, Pan HF, Ye DQ, Xu Y. Targeting IRF4 in autoimmune diseases. Autoimmun Rev. 2012;11:918–24. doi: 10.1016/j.autrev.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 55.Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity. 2013;39:833–45. doi: 10.1016/j.immuni.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–65. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 57.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–12. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–7. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Q, Rao RR, Araki K, Pollizzi K, Odunsi K, Powell JD, Shrikant PA. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity. 2011;34:541–53. doi: 10.1016/j.immuni.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kometani K, Nakagawa R, Shinnakasu R, Kaji T, Rybouchkin A, Moriyama S, Furukawa K, Koseki H, Takemori T, Kurosaki T. Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity. 2013;39:136–47. doi: 10.1016/j.immuni.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 62.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–9. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS, Hung S, Astle MV, Bywater M, Wall M, et al. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci Signal. 2011;4:ra56. doi: 10.1126/scisignal.2001754. [DOI] [PubMed] [Google Scholar]
- 64.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci U S A. 2013;110:11988–93. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]