Abstract
By influencing the number of RNA molecules repeatedly synthesized from the same gene, the control of transcription reinitiation has the potential to shape the transcriptome. Transcription reinitiation mechanisms have been mainly addressed in vitro, through approaches based on both crude and reconstituted systems. These studies support the notion that transcription reinitiation and its regulation rely on dedicated networks of molecular interactions within transcription machineries. At the same time, comparison with in vivo transcription rates suggests that additional mechanisms, factors and conditions must exist in the nucleus, whose biochemical elucidation is a fascinating challenge for future in vitro transcription studies.
Keywords: RNA polymerase, reinitiation, termination, in vitro transcription, template competition, chromatin template
Introductory Remarks
In vitro reconstituted transcription systems, by allowing manipulation of individual components and analysis of individual transcription steps, has proven to be invaluable to understand core transcription mechanisms and to identify transcription cycle intermediates as regulatory targets. In particular, in vitro studies in the past 30 y have revealed that productive gene activation and the completion of a first transcription cycle mark transcribed genes in such a way that the rate of transcription for the subsequent cycles is modified (most frequently increased) with respect to the “opening” transcription event. Being exclusively based on in vitro analysis, the distinction between first and subsequent transcription cycles could appear as somehow artificial. In particular, for constitutively expressed genes, the “first” transcription round, as distinct from subsequent, reinitiation-dependent cycles, tends to appear as an abstraction. However, no gene, even if “housekeeping,” is continuously transcribed under all conditions. For example, highly efficient transcription of rRNA and tRNA genes by RNA polymerase (RNAP) I and III, as well as ribosomal protein gene transcription by RNAP II, are known to be dramatically up- or downregulated in function of nutrient availability, cell proliferation and other cellular and environmental states, including disease.1-4 And there is general evidence that, both in regulated transcription of protein-coding genes by RNAP II and in RNAP III transcriptional response to nutrient availability, reinitiation is a specific target of regulation.5,6 Regulatory changes in gene transcription, in particular the induction and maintenance of activated states, may thus be viewed as processes involving a transition between different modes of recruitment of the transcription machinery to target genes. And such recruitment modes can best be characterized through in vitro mechanistic analysis.
A few review articles published in the past 15 y provide comprehensive accounts of the different reinitiation strategies appeared over the course of evolution to meet different types of requirements, ranging from the need for fast and abundant transcript production in support of cell growth, to fine-tuned regulation involving gene looping and RNAP II recycling.5-8 This review will instead focus on the wide range of in vitro strategies developed to unearth specific features and requirements of the different transcription systems, with the aim of favoring improved experimental approaches for the elucidation of reinitiation mechanisms.
In Vitro Dissection of Transcription Cycles
As pointed out above, the key aspect of gene transcription addressed by reinitiation studies is the possibility that molecular events occurring during a transcription cycle (ideally the first one) can influence the rate of successive transcription rounds by modifying the availability for reinitiation of the template and/or transcription proteins.7 Early evidence of this phenomenon came from studies of RNAP I and III systems. Later studies evidenced a similar behavior for RNAP II and other transcription systems, and unveiled the existence of different molecular strategies to reach similar goals.
Studies of RNA polymerase III and I transcription
The first of such strategies discovered in eukaryotes is the initial establishment of a stable protein-DNA intermediate on RNAP III-transcribed genes, that was shown to resist to dilution and to competitor DNA for several rounds of transcription per template.9,10 Such an intermediate was later shown to consist of basal transcription factors that form a stable promoter complex in a sequence-directed manner. In these and other pioneering in vitro studies, conducted on both RNAP I and RNAP III systems, the occurrence of multiple transcription rounds was inferred on the basis of quantification of the number of transcript molecules produced from a known number of template molecules. Since the total amount of transcription observed was greater than one transcript for every promoter present in the reaction, it could be concluded that reinitiation had to take place.10-12
Mechanistic analysis of the reinitiation process was strongly improved by the introduction of inhibitory molecules that, by selectively blocking individual steps of the transcription cycle, allowed to separately characterize the kinetic parameters of initiation and reinitiation reactions.
In the yeast RNAP III system this was achieved by the use of the polyanion heparin. This molecule selectively inactivates polymerase molecules not engaged in elongation (either because unassembled or because they underwent termination), excluding them from participation in new transcription cycle. Heparin-resistant elongation complexes are obtained by forcing RNAP III to stall as a ternary complex containing a short nascent RNA at an early position on the template. Upon addition of the missing nucleotide in the presence of heparin, stalled RNAP III molecules resume elongation and eventually terminate, but cannot reinitiate. Under these conditions it is reasonable to assume an unambiguous correspondence between the transcription output observed and the number of active elongation complexes assembled before inhibitor addition. The ratios between the outputs of inhibited and non-inhibited reactions provide a direct estimate of the number of transcription rounds having occurred in a given time period.6,13 First initiation and reinitiation by human RNAP III could similarly be distinguished by the use of the detergent Sarkosyl, thus allowing to generalize the notion that all the factors required for initiation by RNAP III are sequestered into a promoter complex that is stable for multiple transcription cycles in reactions that can be entirely reproduced and characterized in vitro.14 Sarkosyl was also successfully used as a selective inhibitor of reinitiation in RNAP I-dependent in vitro systems, thus consolidating the notion that a template-committed complex render RNAP I promoters competent for multiple rounds of transcription.15-17
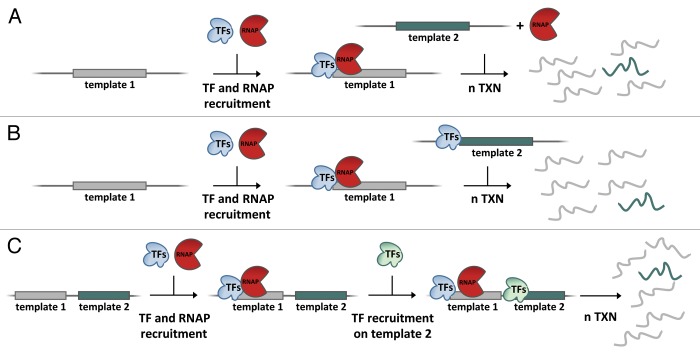
A further improvement of RNAP III in vitro analyses arose from the identification and cloning of part of the yeast RNAP III machinery, that allowed the reconstitution of particularly active in vitro transcription reactions from both native and recombinant transcription factor preparations.18,19 These systems facilitated the study of the kinetic parameters of the first initiation event and those of subsequent cycles, and led to realize that RNAP III reinitiation on the promoter complex is remarkably faster after the completion of the first cycle. This observation, together with template competition assays showing that, unexpectedly, recycling RNAP III was retained on the same template for multiple transcription rounds, led to propose a facilitated reinitiation pathway for RNAP III involving recapture of terminating RNAP III by promoter-bound transcription factors.20-22 An important feature of these studies was the use, for template competition assays, of sub-stoichiometric polymerase concentrations with respect to promoter complexes. Under such conditions, only polymerase molecules having completed the first transcription cycle have the possibility of being sequestered by a promoter complex for a new cycle. In the most stringent competition protocol, the two competing promoter complexes were juxtaposed on the same plasmid DNA, to warrant them the same a priori probability of being met by polymerase after termination of the first round.21 Figure 1 comparatively illustrates the use of template competition assays in reinitiation studies.
Figure 1. Template competition assays for the analysis of transcription reinitiation. This classical in vitro strategy allows revealing the retention of transcription factors (TFs) or RNA polymerase (RNAP) to a first transcribed, test template by challenging it with a competitor template. (A) Testing for TF retention through multiple transcription cycles. A test gene (template 1) is pre-incubated with TFs and RNAP. Upon starting the first transcription cycle, a competitor gene (template 2) is added together with RNAP, and multiple transcription cycles are permitted (n TXN). Persistence of TF on template 1, and thus exclusion of template 2 from transcription, is revealed by template 1/template 2 transcript ratio. Retention can be tested for individual TFs or combinations of them. (B) Testing for RNAP retention through multiple transcription cycles. Test gene (template 1) is pre-incubated with TFs and RNAP. Upon starting the first transcription cycle, competitor template 2, pre-assembled with TFs, is added and multiple transcription cycles are permitted (n TXN). Persistence of RNAP on template 1 is revealed by template 1/template 2 transcript ratio. (C) Testing for RNAP retention through competition between physically linked templates. RNAP is specifically sequestered by template 1 through a template-specific TF set. A promoter complex is then allowed to assemble on template 2 carried by the same DNA molecule. Upon completing the first transcription cycle on template 1, RNAP will be available for multiple cycles to the two juxtaposed templates without any bias in its local concentration (as it can be the case when the competitor template is provided on a different DNA molecule). The actual occurrence of in cis competition requires a high occupancy of competitor template by promoter complexes. Exclusion of template 2 because of RNAP retention on template 1 is revealed as above. In all the protocols, the component (TF or RNAP) whose stable sequestration is under analysis is provided in sub-stoichiometric amounts.
The availability of highly purified (and partly recombinant) in vitro RNAP III and RNAP I transcription systems, together with the ability to compare single and multiple rounds of transcription, were also essential for the identification of novel transcription stimulatory activities, some of which turned out to affect reinitiation.23-27
Studies of RNA polymerase II transcription
Despite recent progress toward in vitro reconstitution of coupled RNAP II transcription and 3′ end processing28 and dissection of the different steps of RNAP II termination,29 a complete in vitro reproduction of series of transcription cycles, including termination, is still lacking for RNAP II. As a consequence, in vitro analysis does not allow focusing on the reinitiation properties of RNAP II molecules having undergone termination. It does, however, allow monitoring the re-utilization by RNAP II of promoter complexes after the first transcription cycle. This can be directly tested by the so-called “colliding polymerase assay,” in which a first elongating polymerase is stalled at the end of a G-less cassette due to GTP omission. Polymerases having initiated new transcription cycles on the same promoter complex collide with stalled polymerase molecules in front of them, and are thus visualized through the appearance of RNA products progressively shorter by about 30 nucleotides. By exploiting such a protocol, it was possible to uncover a requirement for reinitiation of basal transcription factors, and to directly show that Sarkosyl blocks RNAP II reinitiation.30,31 Recently, this approach has been exploited to reveal a role of hnRNP R, in conjunction with Mediator, in transcription reinitiation on the human c-fos gene.32
Other studies of RNAP II reinitiation exploited simpler in vitro kinetic analyses with model promoters in the presence or absence of inhibitors. Such studies have consolidated the notion that subsequent cycles of RNAP II transcription occur faster than first round initiation, due to the fact that one or more factors remain committed to promoters after initiation33,34 (reviewed in5). Interestingly, core promoter elements, besides their basic role in preinitiation complex assembly, may also exert a significant influence on template reutilization for multiple cycles, as first shown for the TATA element.34
As a further insight coming from in vitro RNAP II reinitiation studies, analyses using naked (non-chromatin assembled) templates generally failed to reveal effects of activators on reinitiation (the main effect being an increase in the number of functional promoter complexes),35 while activators were found to dramatically stimulate reinitiation on chromatin templates, as assessed by comparison of transcriptional stimulation under multiple and single round transcription conditions (based on the use of Sarkosyl).36-39 Through a similar approach, chromatin remodeling and histone acetylation were also shown to favor transcription reinitiation on a chromatin template in vitro.40 It is thus plausible that, in the context of chromatin, activators favor the reutilization of promoter complexes for multiple transcription cycles. The importance of chromatin context in regulated transcription reinitiation, however, should not lead to underappreciate the role of DNA-encoded core promoter architecture in the same process. As a particularly significant illustration of this possibility, the intrinsic (chromatin-independent) features of diverse p53-responsive core promoters have been shown (through in vitro experiments employing Sarkosyl as a reinitiation inhibitor) to dictate differences in both the rate of gene induction and the duration of reinitiation-dependent transcriptional response.41
As reminded above, a common limitation of RNAP II studies is the use of specifically devised templates lacking native termination signals and only supporting a run-off mode of transcription termination. Under these conditions, reutilization of the same promoter complex for multiple cycles can be studied, but reinitiation mechanisms involving termination-coupled RNA polymerase recycling cannot be put in evidence. RNAP II reinitiation models attributing a key role to gene looping in polymerase recycling from terminator to promoter are based on in vivo detection of protein-protein, protein-DNA and DNA-DNA contacts, mainly through chromatin immunoprecipitation and chromosome conformation capture techniques.42 Experimental support to RNAP II gene looping from in vitro studies is still lacking, probably because gene loop conformations may be dependent on general nuclear architecture in a subtle manner,43 and their functional significance is likely to be found more in complex chromatin-level transactions, such as transcriptional memory or directionality, than in the control of basal or activated transcription output.44,45
Studies of other RNA polymerases
Despite the importance of bacterial transcription studies for our general knowledge of transcriptional mechanisms and their control,46 reinitiation in bacterial systems could not be easily addressed by in vitro transcription approaches. This is mainly due to the peculiarity of bacterial transcription control, which does not rely on stable promoter complexes, and whose central mechanism is the dissociation of σ factor after each round of RNA synthesis, followed by competition among different σ factors for binding to RNA polymerase to redirect it to promoters for a new cycle.47 This might explain the very limited number of transcription cycles typical of purified bacterial systems, usually based on Escherichia coli RNA polymerase.48 By showing that transcription-dependent loss of σ can be negatively modulated, previous studies have suggested the possibility that regulating the extent and time of σ release during elongation can influence not only elongation, but also RNA polymerase recycling and thus reinitiation.49,50 Other in vitro studies have also shown that proteins as diverse as RapA, a bacterial homolog of eukaryotic SWI/SNF proteins, and the ribosomal protein S1 can enhance transcription reinitiation, probably by counteracting inhibitory RNA polymerase-nucleic acid interactions, thus favoring polymerase recycling.48,51
By contrast, transcription reinitiation could be thoroughly addressed in Archaea thanks to the availability of an in vitro system, reconstituted from purified Pyrococcus furiosus RNA polymerase and recombinant TBP and TFB, able to support multiple rounds of specifically initiated and terminated transcription. Template competition experiments with limiting RNA polymerase concentration highlighted that polymerase is retained on the same template for multiple transcription cycles, through a RNAP III-like mechanism.52
Transcription reinitiation by monomeric bacteriophage T7 RNA polymerase has also been investigated through in vitro kinetic analysis employing heparin as a reinitiation inhibitor. Even in this simple system, in which neither transcription factors nor dissociable subunits can uncouple the rates of the first and subsequent transcription cycles, RNA polymerase was found to undergo a transcription-dependent transition to an initiation-incompetent state, affecting transcription rate at sub-saturating polymerase concentrations.53
Monitoring Transcription-Dependent Changes in the Transcription Machinery
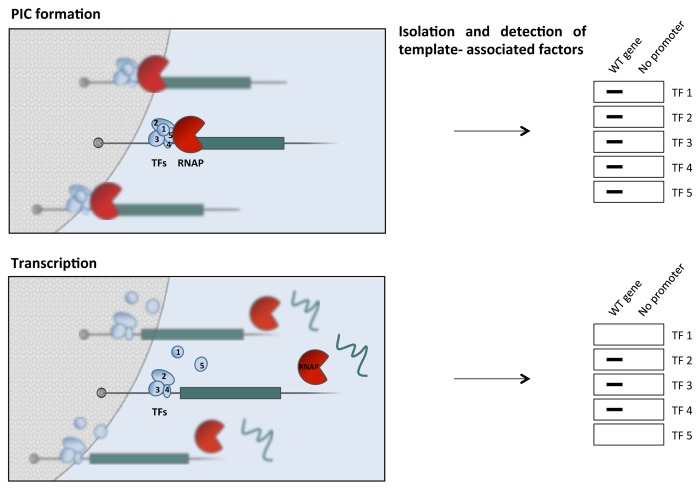
Given the remarkable number of basal factors participating in RNAP II transcription complexes, and their specific contributions to different steps of the transcription cycle,54 the fate of these factors during transcription is a relevant issue, strictly related to the one of reinitiation. This issue has been successfully addressed through the use of immobilized templates in combination with immunoblotting55,56 (Fig. 2). After completion of the first round of transcription, or after transition from initiation to elongation, the components of the transcription machinery that remain bound to the promoter are easily isolated and detected. Through this approach, a RNAP II reinitiation intermediate was identified that includes transcription factors TFIID, TFIIA, TFIIH, TFIIE and Mediator. Reinitiation by RNAP II is facilitated by recognition of such a scaffold complex, whose stabilization by transcriptional activators could be a key mechanism in gene activation. Specialized functions of Mediator in reinitiation could be revealed by the same approach.57,58
Figure 2. Immobilized template assays for the analysis of transcription-dependent changes in transcription complexes. Templates are immobilized on magnetic beads and incubated with nuclear extract to allow for promoter complex (PIC) formation and transcription. Templates can be isolated, and the associated factors analyzed by immunoblot (shown on the right) or other techniques at any step of the transcription cycle. A promoter-mutated template can provide a control for non-specifically associated proteins. A similar approach can also allow monitoring the actual occurrence of RNA and/or RNAP release at termination.
Immobilized templates, allowing the controlled step-wise assembly of transcription intermediates, have also facilitated the in vitro analysis of reinitiation by human RNAP III and its regulation by the general repressor Maf1.59 In principle, immobilized template assays offer the possibility (still underexploited) to monitor how transcription cycling affects the composition of transcription complexes, as well as the covalent modification state of involved proteins, in any transcription system.
Disentangling Transcription Termination and Reinitiation
Recycling of an RNA polymerase molecule for a new transcription cycle obviously requires proper termination. Therefore, any factor affecting transcript and/or polymerase release at the termination step can affect transcription reinitiation. Several in vitro studies, mainly in the RNAP III and RNAP I systems, indeed provided evidence for a role in reinitiation of factors variously affecting termination.7 Conformational or chemical modifications of RNA polymerase and/or the transcription machinery, occurring during the termination process, might also affect RNA polymerase recycling and thus reinitiation. This possibility is exemplified by the RNAP II C-terminal domain, which must undergo tightly controlled dephosphorylation before or during the termination phase in order for RNAP II to be recruited to the promoter complex in an unphosphorylated state for a new cycle.60,61
In the case of RNAP III, a first in vitro evidence for a non-trivial requirement of termination for facilitated recycling came from comparison of reinitiation rates on templates supporting either natural or run-off termination. After run-off termination, RNAP III reinitiation rate was found to be low, reflecting mere reiteration of the first cycle. This suggested that a RNAP III conformational change occurring during termination might render it competent for facilitated reinitiation.20 In support to this hypothesis, a peptide nucleic acid roadblock placed a few bp downstream of a RNAP III terminator was found to selectively interfere with transcription reinitiation, possibly by hampering a conformational change required for productive polymerase recycling.62 Further in vitro characterization of RNAP III reinitiation mechanism exploited an incomplete RNAP III form defective in termination and/or reinitiation, and the ability to reconstitute a fully functional enzyme by adding the missing subunits. This study revealed a specific role of the Rpc11 subunit in RNAP III recycling in concomitance with the termination reaction.63 Further supporting the link between termination and RNAP III reloading for reinitiation, a recent genome-wide location analysis of human RNAP III showed that the strength of the terminator sequence can strongly affect RNAP III occupancy of tRNA genes.64 Facilitated RNAP III reinitiation has also been shown to occur on unusually long transcription units in in vitro reconstituted systems,21,65 a fact which could explain why termination-deficient RNAP III mutants, reading through average-length terminators and terminating at a downstream located failsafe terminator, are not affected in their overall transcription efficiency (and thus in reinitiation) in vivo.66
Recently, in vitro assembled RNAP III elongation complexes immobilized on beads have been exploited for a deep mechanistic analysis of RNAP III termination. The structure of RNAP III-synthesized transcripts was found to dictate the release of elongation complexes at the end of transcription units, with the poly(dT) termination signal inducing backtracking of RNAP III to the nearest RNA hairpin.67 The centrality of RNA structure in termination re-launches the hypothesis of a functional role of the transcript in reinitiation,68 which should be amenable to further mechanistic investigation thanks to the use of immobilized transcription complexes.
Bridging the Gap Between In Vitro and In Vivo Transcription Rates
Whenever possible, faithful in vitro recapitulation of eukaryotic gene transcription should not overlook the role of chromatin, whose protein components have the potential to influence the transcription process at any step. Regulated RNAP II transcription, and the stimulation of reinitiation by activators, could more easily be revealed in vitro by using templates assembled into chromatin,36,39,69 thus pointing to chromatin organization as an important factor contributing to activator-dependent reinitiation in eukaryotic nuclei. In addition to chromatin, the native cellular context of transcription also includes less well characterized factors unavoidably lost in vitro, such as three-dimensional organization of the nuclear genome70 and macromolecular crowding, which can dramatically influence transcription reaction kinetics via volume exclusion effects.71,72 Since reinitiation might involve, at least in some cases, RNA polymerase recapture after termination, crowding could strongly contribute, together with specific recapture mechanisms, to reinitiation rate. More generally, setting up in vitro conditions that mimic the macromolecular crowding typical of eukaryotic nuclei will be required in order to fully understand the mechanisms by which amazingly high transcription frequencies (on the order of 1 transcript/gene/sec6) can be attained in vivo.
Concluding Remarks
A large number of in vitro transcription studies in bacterial, archaeal and eukaryotic systems have consolidated the notion that reinitiation is a key aspect of the transcription process and may represent an important, yet still underappreciated target of transcriptional regulation. The recycling of RNA polymerase after each transcription round can impact on reinitiation rate in all transcription systems, yet it could only be evaluated in mechanistic detail in those systems (RNAP III in particular) in which the transcription cycle can be recapitulated in vitro in all its steps, including termination, on natural templates. Given the importance of RNAP II transcription in gene regulatory networks, an in vitro system capable of supporting multiple RNAP II transcription cycles is thus certainly among the desiderata. More generally, in vitro systems faithfully supporting reinitiation will require not only the use of chromatin templates, but also the reproduction of the macromolecular crowding effects thought to occur in eukaryotic nuclei. Progress in biochemical studies of transcription reinitiation will not only deepen our understanding of a fundamental aspect of gene expression, but it will also help in increasing the robustness of gene transcription and directed RNA production for both biotechnological and nanotechnological applications.71
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Work in authors’ laboratory was supported by a grant from the Italian Ministry of Education, University and Research (PRIN 2009 Program, 200973ST5Y). M.C.B. is presently supported by a fellowship from Associazione Italiana Ricerca sul Cancro (AIRC) to the University of Milano.
References
- 1.Moir RD, Willis IM. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim Biophys Acta. 2013;1829:361–75. doi: 10.1016/j.bbagrm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB. Dysregulation of RNA polymerase I transcription during disease. Biochim Biophys Acta. 2013;1829:342–60. doi: 10.1016/j.bbagrm.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laferté A, Favry E, Sentenac A, Riva M, Carles C, Chédin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–40. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–40. doi: 10.1016/S0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 5.Hahn S. Activation and the role of reinitiation in the control of transcription by RNA polymerase II. Cold Spring Harb Symp Quant Biol. 1998;63:181–8. doi: 10.1101/sqb.1998.63.181. [DOI] [PubMed] [Google Scholar]
- 6.Dieci G, Bosio MC, Fermi B, Ferrari R. Transcription reinitiation by RNA polymerase III. Biochim Biophys Acta. 2013;1829:331–41. doi: 10.1016/j.bbagrm.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Dieci G, Sentenac A. Detours and shortcuts to transcription reinitiation. Trends Biochem Sci. 2003;28:202–9. doi: 10.1016/S0968-0004(03)00054-9. [DOI] [PubMed] [Google Scholar]
- 8.Shandilya J, Roberts SG. The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling. Biochim Biophys Acta. 2012;1819:391–400. doi: 10.1016/j.bbagrm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Lassar AB, Martin PL, Roeder RG. Transcription of class III genes: formation of preinitiation complexes. Science. 1983;222:740–8. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- 10.Bogenhagen DF, Wormington WM, Brown DD. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982;28:413–21. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- 11.Wandelt C, Grummt I. Formation of stable preinitiation complexes is a prerequisite for ribosomal DNA transcription in vitro. Nucleic Acids Res. 1983;11:3795–809. doi: 10.1093/nar/11.11.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cizewski V, Sollner-Webb B. A stable transcription complex directs mouse ribosomal RNA synthesis by RNA polymerase I. Nucleic Acids Res. 1983;11:7043–56. doi: 10.1093/nar/11.20.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kassavetis GA, Riggs DL, Negri R, Nguyen LH, Geiduschek EP. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–66. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovelman R, Roeder RG. Sarkosyl defines three intermediate steps in transcription initiation by RNA polymerase III: application to stimulation of transcription by E1A. Genes Dev. 1990;4:646–58. doi: 10.1101/gad.4.4.646. [DOI] [PubMed] [Google Scholar]
- 15.Kato H, Nagamine M, Kominami R, Muramatsu M. Formation of the transcription initiation complex on mammalian rDNA. Mol Cell Biol. 1986;6:3418–27. doi: 10.1128/mcb.6.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto RT, Nogi Y, Dodd JA, Nomura M. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 1996;15:3964–73. [PMC free article] [PubMed] [Google Scholar]
- 17.Schnapp A, Grummt I. Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J Biol Chem. 1991;266:24588–95. [PubMed] [Google Scholar]
- 18.Kassavetis GA, Joazeiro CA, Pisano M, Geiduschek EP, Colbert T, Hahn S, Blanco JA. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–64. doi: 10.1016/0092-8674(92)90399-W. [DOI] [PubMed] [Google Scholar]
- 19.Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 1996;273:249–67. doi: 10.1016/S0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
- 20.Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–52. doi: 10.1016/S0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 21.Ferrari R, Rivetti C, Acker J, Dieci G. Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc Natl Acad Sci U S A. 2004;101:13442–7. doi: 10.1073/pnas.0403851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrari R, Dieci G. The transcription reinitiation properties of RNA polymerase III in the absence of transcription factors. Cell Mol Biol Lett. 2008;13:112–8. doi: 10.2478/s11658-007-0041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Roeder RG. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–57. doi: 10.1016/S1097-2765(00)80074-X. [DOI] [PubMed] [Google Scholar]
- 24.Tavenet A, Suleau A, Dubreuil G, Ferrari R, Ducrot C, Michaut M, Aude JC, Dieci G, Lefebvre O, Conesa C, et al. Genome-wide location analysis reveals a role for Sub1 in RNA polymerase III transcription. Proc Natl Acad Sci U S A. 2009;106:14265–70. doi: 10.1073/pnas.0900162106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dieci G, Ruotolo R, Braglia P, Carles C, Carpentieri A, Amoresano A, Ottonello S. Positive modulation of RNA polymerase III transcription by ribosomal proteins. Biochem Biophys Res Commun. 2009;379:489–93. doi: 10.1016/j.bbrc.2008.12.097. [DOI] [PubMed] [Google Scholar]
- 26.Braglia P, Dugas SL, Donze D, Dieci G. Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:1545–57. doi: 10.1128/MCB.00773-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panova TB, Panov KI, Russell J, Zomerdijk JC. Casein kinase 2 associates with initiation-competent RNA polymerase I and has multiple roles in ribosomal DNA transcription. Mol Cell Biol. 2006;26:5957–68. doi: 10.1128/MCB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mariconti L, Loll B, Schlinkmann K, Wengi A, Meinhart A, Dichtl B. Coupled RNA polymerase II transcription and 3′ end formation with yeast whole-cell extracts. RNA. 2010;16:2205–17. doi: 10.1261/rna.2172510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porrua O, Libri D. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol. 2013;20:884–91. doi: 10.1038/nsmb.2592. [DOI] [PubMed] [Google Scholar]
- 30.Szentirmay MN, Sawadogo M. Sarkosyl block of transcription reinitiation by RNA polymerase II as visualized by the colliding polymerases reinitiation assay. Nucleic Acids Res. 1994;22:5341–6. doi: 10.1093/nar/22.24.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szentirmay MN, Sawadogo M. Transcription factor requirement for multiple rounds of initiation by human RNA polymerase II. Proc Natl Acad Sci U S A. 1991;88:10691–5. doi: 10.1073/pnas.88.23.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuda A, Shimada M, Nakadai T, Nishimura K, Hisatake K. Heterogeneous nuclear ribonucleoprotein R cooperates with mediator to facilitate transcription reinitiation on the c-Fos gene. PLoS One. 2013;8:e72496. doi: 10.1371/journal.pone.0072496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawley DK, Roeder RG. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987;262:3452–61. [PubMed] [Google Scholar]
- 34.Yean D, Gralla J. Transcription reinitiation rate: a special role for the TATA box. Mol Cell Biol. 1997;17:3809–16. doi: 10.1128/mcb.17.7.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992;11:2229–40. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraus WL, Kadonaga JT. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–42. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheridan PL, Mayall TP, Verdin E, Jones KA. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 1997;11:3327–40. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandaltzopoulos R, Becker PB. Analysis of activator-dependent transcription reinitiation in vitro. Methods Enzymol. 2003;370:487–501. doi: 10.1016/S0076-6879(03)70042-1. [DOI] [PubMed] [Google Scholar]
- 39.Sandaltzopoulos R, Becker PB. Heat shock factor increases the reinitiation rate from potentiated chromatin templates. Mol Cell Biol. 1998;18:361–7. doi: 10.1128/mcb.18.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuguchi G, Vassilev A, Tsukiyama T, Nakatani Y, Wu C. ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J Biol Chem. 2001;276:14773–83. doi: 10.1074/jbc.M100125200. [DOI] [PubMed] [Google Scholar]
- 41.Morachis JM, Murawsky CM, Emerson BM. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 2010;24:135–47. doi: 10.1101/gad.1856710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hampsey M, Singh BN, Ansari A, Lainé JP, Krishnamurthy S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv Enzyme Regul. 2011;51:118–25. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–24. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. Gene loops enhance transcriptional directionality. Science. 2012;338:671–5. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lainé JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–9. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Hippel PH. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–5. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 47.Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell. 2005;20:335–45. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 48.Sukhodolets MV, Garges S, Adhya S. Ribosomal protein S1 promotes transcriptional cycling. RNA. 2006;12:1505–13. doi: 10.1261/rna.2321606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bar-Nahum G, Nudler E. Isolation and characterization of sigma(70)-retaining transcription elongation complexes from Escherichia coli. Cell. 2001;106:443–51. doi: 10.1016/S0092-8674(01)00461-5. [DOI] [PubMed] [Google Scholar]
- 50.Deighan P, Pukhrambam C, Nickels BE, Hochschild A. Initial transcribed region sequences influence the composition and functional properties of the bacterial elongation complex. Genes Dev. 2011;25:77–88. doi: 10.1101/gad.1991811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sukhodolets MV, Cabrera JE, Zhi H, Jin DJ. RapA, a bacterial homolog of SWI2/SNF2, stimulates RNA polymerase recycling in transcription. Genes Dev. 2001;15:3330–41. doi: 10.1101/gad.936701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spitalny P, Thomm M. A polymerase III-like reinitiation mechanism is operating in regulation of histone expression in archaea. Mol Microbiol. 2008;67:958–70. doi: 10.1111/j.1365-2958.2007.06084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferrari R, Rivetti C, Dieci G. Transcription reinitiation properties of bacteriophage T7 RNA polymerase. Biochem Biophys Res Commun. 2004;315:376–80. doi: 10.1016/j.bbrc.2004.01.071. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol. 2009;21:344–51. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–90. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 56.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–9. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 57.Rani PG, Ranish JA, Hahn S. RNA polymerase II (Pol II)-TFIIF and Pol II-mediator complexes: the major stable Pol II complexes and their activity in transcription initiation and reinitiation. Mol Cell Biol. 2004;24:1709–20. doi: 10.1128/MCB.24.4.1709-1720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reeves WM, Hahn S. Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol Cell Biol. 2003;23:349–58. doi: 10.1128/MCB.23.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cabart P, Lee J, Willis IM. Facilitated recycling protects human RNA polymerase III from repression by Maf1 in vitro. J Biol Chem. 2008;283:36108–17. doi: 10.1074/jbc.M807538200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 1999;13:1540–52. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bataille AR, Jeronimo C, Jacques PE, Laramée L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–70. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 62.Guffanti E, Corradini R, Ottonello S, Dieci G. Functional dissection of RNA polymerase III termination using a peptide nucleic acid as a transcriptional roadblock. J Biol Chem. 2004;279:20708–16. doi: 10.1074/jbc.M311295200. [DOI] [PubMed] [Google Scholar]
- 63.Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–28. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Canella D, Bernasconi D, Gilardi F, LeMartelot G, Migliavacca E, Praz V, Cousin P, Delorenzi M, Hernandez N, Cycli XC, CycliX Consortium A multiplicity of factors contributes to selective RNA polymerase III occupancy of a subset of RNA polymerase III genes in mouse liver. Genome Res. 2012;22:666–80. doi: 10.1101/gr.130286.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dieci G, Giuliodori S, Catellani M, Percudani R, Ottonello S. Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae. J Biol Chem. 2002;277:6903–14. doi: 10.1074/jbc.M105036200. [DOI] [PubMed] [Google Scholar]
- 66.Rijal K, Maraia RJ. RNA polymerase III mutants in TFIIFα-like C37 that cause terminator readthrough with no decrease in transcription output. Nucleic Acids Res. 2013;41:139–55. doi: 10.1093/nar/gks985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–80. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emde G, Frontzek A, Benecke BJ. Secondary structure of the nascent 7S L RNA mediates efficient transcription by RNA polymerase III. RNA. 1997;3:538–49. [PMC free article] [PubMed] [Google Scholar]
- 69.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–51. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–9. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan C, Saurabh S, Bruchez MP, Schwartz R, Leduc P. Molecular crowding shapes gene expression in synthetic cellular nanosystems. Nat Nanotechnol. 2013;8:602–8. doi: 10.1038/nnano.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sokolova E, Spruijt E, Hansen MM, Dubuc E, Groen J, Chokkalingam V, Piruska A, Heus HA, Huck WT. Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proc Natl Acad Sci U S A. 2013;110:11692–7. doi: 10.1073/pnas.1222321110. [DOI] [PMC free article] [PubMed] [Google Scholar]