Abstract
Regulation of transcription elongation via pausing of RNA polymerase has multiple physiological roles. The pausing mechanism depends on the sequence heterogeneity of the DNA being transcribed, as well as on certain interactions of polymerase with specific DNA sequences. In order to describe the mechanism of regulation, we introduce the concept of heterogeneity into the previously proposed alternative models of elongation, power stroke and Brownian ratchet. We also discuss molecular origins and physiological significances of the heterogeneity.
Keywords: RNA polymerase, translocation, transcription elongation factors, transcription pausing, sequence heterogeneity of DNA, Brownian ratchet, power stroke
Introduction
For over 50 years, the mechanism of transcription by DNA-dependent RNA polymerase (RNAP) has been intensively investigated. Based on the accumulated knowledge, transcription is divided into three major stages, namely, initiation, chain elongation and chain termination. Initiation consists of RNAP binding to a specific promoter DNA and strand separation of the DNA that allows NTP binding to the template strand at the enzyme active site. Elongation is a repetitive but temporally discontinuous formation of phosphodiester bonds. Termination describes the complete dissociation of the RNA strand from the RNA-DNA hybrid retained in RNAP in response to specific signals.1 Different from initiation or termination, which occurs at specific DNA sites, elongation involves a gene-scale DNA tracking system, accompanied by bond formation at every base pair. Hence, specific DNA conformations/flexibilities, DNA lesions, and DNA-binding proteins are of specific concern in the regulation of elongation.
One of the major mechanisms that regulates elongation in all kingdoms of life is sequence-dependent pausing.2,3 In bacteria, pausing is important for the coupling of transcription with translation,4 and for providing opportunities for regulatory factors to bind the elongation complex.5,6 In eukaryotes, slow elongation, achieved by pausing around an exon-intron junction, allows spliceosome assembly, which increases the efficiency of alternative splicing.7 One of the main consequences of pausing is backtracking of RNAP relative to the RNA-DNA hybrid, which extrudes the 3′ end of the RNA from the active center.8,9 A deep sequencing study detected backtracking in a large fraction of paused polymerases in the yeast genome relative to nucleosome positioning.10 Backtracking plays a role to retain polymerases in promoter-proximal regions in order to maintain association with σ70 in Escherichia coli11 or NELF in eukaryotes,12,13 as a way to control specific gene transcription. Backtracking is also a mechanism to increase fidelity by providing a chance for proofreading.14 Contrary to such positive roles for backtracking, an irreversibly and stably backtracked complex forms a roadblock to replication of genomic DNA,15 and is highly toxic to the cell.16-18 Bacterial GreA/B, archeal TFS, and eukaryotic TFIIS rescue irreversibly backtracked complexes by promoting endonucleolytic cleavage and removal of the extruded 3′ end of transcripts.19-22 Bacterial NusG increases the elongation rate by inhibiting backtracking,23,24 which, for example, is critical for uninterrupted and rapid elongation of rRNA.25 Archaea and eukaryotes have Spt5, the counterpart of NusG.26-28 Therefore, polymerase pausing and protein factors that suppress pausing, especially related to backtracking, are essential for a broad variety of transcriptional regulatory steps in all kingdoms of life. So far, polymerase elongation and pausing have been discussed as parts of a homogeneous model that considers DNA as a monotonous polymer similar to a protein filament. However, pausing is a consequence of a heterogeneous effect, namely polymerase recognition of specific DNA sequences during elongation. In this review, we summarize previously proposed models of elongation and add the concept of this sequence-specific heterogeneity into the models to explain pausing. We also discuss the Brownian ratchet mechanism, which does not accurately depict polymerase elongation, as was originally pointed out by Shimamoto.29
Power stroke and Brownian ratchet mechanisms of elongation including pausing
The movement of the RNA-DNA hybrid across the surface of the catalytic cleft in the enzyme is essential for elongation and for the generation of an empty site at the active center, where NTP binds. This movement is a one base pair forward translocation of RNAP along the DNA under conditions that prevent backward translocation or backtracking. Alternative mechanisms for the forward translocation have been proposed: the power-stroke model and the Brownian ratchet model. Their core concepts were taken from studies on unidirectional movement of motor proteins, such as myosin and kinesin, that are accompanied by ATP hydrolysis (reviewed in Refs30-34). In the power-stroke model, the conversion of chemical energy into mechanical work by a motor protein is coupled in the same elementary step. Chemical energy is directly generated by ATP hydrolysis in the protein or indirectly via conformational changes due to the release of inorganic phosphate (Pi). In the case of transcription elongation, the energy is supplied during the transfer of an NMP moiety from pyrophosphate (PPi) to the 3′ end of RNA. Hence, in the power-stroke model for elongation, the forward translocation is synchronized to either phosphodiester-bond formation or PPi release, with a transition state existing between the pre- and post-translocated states. Because the chemical energy is required for driving a forward translocation, the activation energy must be much higher than 0.5 kBT, the averaged value of thermal energy per degree of freedom, where kB is the Boltzmann constant and T is the absolute temperature. This condition is sometimes mistakenly neglected, but necessary for the description with rate equations. If this mechanism is the case for elongation by E. coli RNAP, the transition state involves PPi release, because it has been shown that a nucleotide addition precedes translocation and PPi release.35 Below, we also discuss the case for pausing in the context of the power-stroke model.
In the Brownian ratchet model, there is no transition state with significant activation energy, and there are no energy barriers larger than the order of kBT. Hence, the transition state theory and rate equations cannot be applied to describe translocation (see Ref29 for more fundamentals). The forward translocation is not synchronized to the chemical steps. A net forward bias in the translocation would be generated by a cognate NTP binding to the active site of the elongation complex, which prevents backward translocation and simultaneously progresses the elongation to the next cycle by condensation of the NTP with the transcript.36,37 In an analysis for force-distance relationship of elongation by E. coli RNAP, the effective distance over which force acts is a single base pair during elongation within a pause-free sequence, which is equal to the moving distance of polymerase to complete translocation.38 This was interpreted as the absence of the transition state between the pre- and post-translocated states, and consistent with Brownian ratchet translocation during smooth elongation.
Interestingly, the forward translocation of RNAP is smooth or restricted, depending upon the sequence of the DNA being transcribed. We are calling this heterogeneous tracking. This is distinguished from the mechanism of homogenous tracking by the motor proteins.30-34 Therefore, RNAP pauses at specific sequences during elongation, as exemplified by a biochemical study using yeast RNAP II and a transcription factor TFIIS mutant. TFIIS is known to induce endonucleolytic transcript cleavage near the 3′ RNA end by interacting with the active center of RNAP II.20-22 The study revealed that during RNAP II elongation, the cleavage-deficient TFIIS mutant carrying alanine substitutions in the catalytic loop, TFIISAA, specifically binds to the RNAP II that transiently pauses on timescales of 100 ms to 1 s and promotes its backtracking.39 RNAP II encountered such sequences every 10–100 bps, where the forward translocation was restricted.39 This was interpreted as forward translocation being the slowest process in the single nucleotide addition and limits elongation in the position sensitive to TFIISAA. It is noteworthy that TFIISAA has dominant lethal effect on yeast cells, suggesting a physiological significance for control of the sequence-dependent pausing.39,40
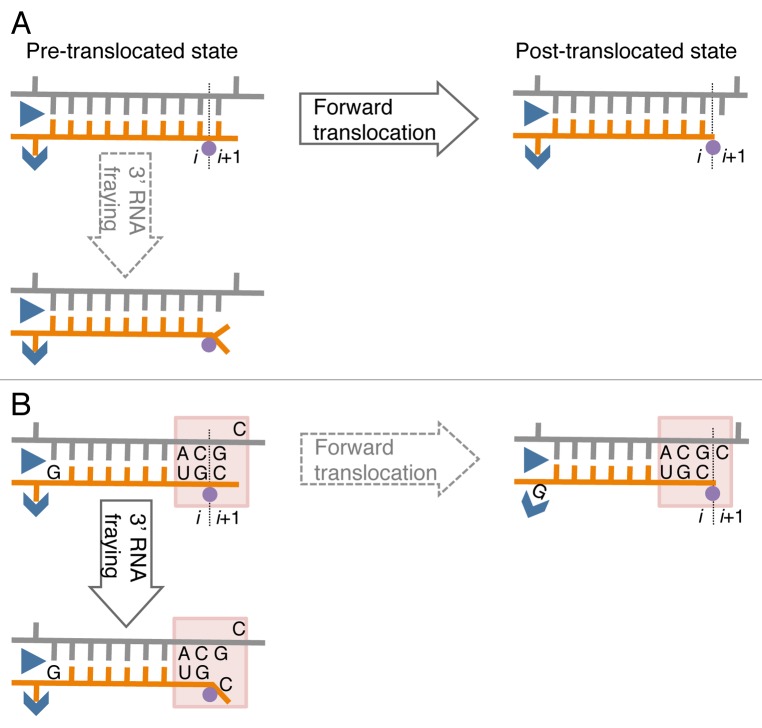
Until now, pausing and pause-free elongation have been described in terms of homogeneous tracking according to the two pawl-ratchet (Brownian ratchet) model.36 This model proposes two rapid Brownian fluctuations during elongation: (1) translocation fluctuation of RNA-DNA hybrid and (2) conformational fluctuation of the RNAP active site including the bridge helix and the trigger loop, elements also involved in catalysis and substrate binding.36 The fluctuations are supposed to occur much more frequently than formation of a phosphodiester bond during pause-free elongation. If the same rapid fluctuations are independent of the transcribed sequence, they cannot be a source of sequence-specific pausing. In contrast to pause-free sequences, when RNAP encounters RNA-DNA hybrids of an unusual conformation or flexibility, the hybrid movement through the catalytic cleft may be restrained, hindering forward translocation. At these pause sites, the movement of the hybrid may become synchronized to phosphodiester bond formation or PPi release, followed by a return to the movement uncoupled to the chemical step in pause-free sites. In other words, at pre-translocation pause sites, the energy released from PPi dissociation is utilized for the forward translocation representing a transient switch to a power-stroke translocation mechanism. The DNA sites coding for pauses may have two consecutive pyrimidine/purine duplets in the non-coding DNA strand, where the downstream pyrimidine residue corresponds to the 3′ end of the RNA, and a purine residue corresponds to the RNA residue in the upstream end of the RNA-DNA hybrid in the elongation complex (Fig. 1). These sequences have been identified as pause sites in a number of bulk biochemical and single molecule transcription studies for E. coli RNAP and yeast/human RNAP II.39,41-43 Interestingly, recent NMR studies revealed that pyrimidine/purine steps within dsDNA increase mobility of the sugar-moiety of the pyrimidine nucleotide,44,45 suggesting that the pyrimidine/purine neighbor spanning the junction between the RNA-DNA hybrid and the downstream DNA increases mobility of the sugar-phosphate backbones, possibly via sugar pucker rearrangements.44 This unique property may promote melting or fraying of the 3′ RNA end in the hybrid from the template DNA strand and prevent forward translocation (Fig. 1B). Furthermore, in other studies, a frayed 3′ RNA end was shown to interfere with phosphoryl transfer and promote backtracking.46-48 The purine residue at the 5′ end of the RNA strand in the RNA-DNA hybrid may hinder forward translocation by a steric clash with the catalytic cleft while in the single-stranded form or stabilization in the double-stranded form (Fig. 1B). An X-ray crystal structure of the bacterial elongation complex suggested that an RNAP Switch-3 domain that is involved in the RNA separation from the hybrid tightly interacts with the 5′ RNA end of the hybrid. Thus, bulky purines, contrary to pyrimidines, could interfere with the function of the Switch-3 domain.49 These sequence-specific effects may explain the pauses occurring every 10–100 bps, i.e., what we call heterogeneous tracking of RNAP during elongation.
Figure 1. A model of sequence-specific pausing. (A) Pause-free elongation. RNA (orange), template DNA strand (gray), catalytic Mg2+ (magenta circle), and two RNAP domains (blue) involving 5′ RNA separation from the hybrid, i.e., Switch 3 (arrow head) and lid (triangle) domains are shown. The 3′ RNA-binding site (i) and the NTP binding site (i+1) are also indicated. (B) Elongation at the pausing site. The two sequence elements involved in transcription pausing are shown: (1) 3′ ACGC 5′ sequence in the transcribed DNA strand (grey) corresponding to the junction between the RNA-DNA hybrid and the downstream dsDNA in the elongation complex (indicated by shaded box); this sequence increases mobility/flexibility of the RNA/DNA backbones, which promotes fraying of the 3’ RNA end. (2) G residue in the RNA at the upstream end of the hybrid contributes to immobilization of the hybrid in the catalytic cleft of RNAP by interacting with the Switch 3 domain in the post-translocated state, or by interacting with the lid domain in the pre-translocated state.
In order to explain pausing caused by Brownian motion, one has to assume Brownian motion that is as slow as or slower than the process of single nucleotide addition. Such slow Brownian motion will be observed with very rare structural configurations of the elongation complex, which occur at low energy ~kBT. An example is a combination of the limited orientation and position of DNA duplex, RNA-DNA hybrid, bridge helix and trigger loop. Pausing can be explained by the Brownian ratchet mechanism only if these structural configurations and the process of phosphodiester-bond formation have comparable frequencies. A new optical method that directly measures changes in the protein and the hybrid motions coupled to water in a broad time domain is required for verifying the Brownian ratchet mechanism.
Endogenous and exogenous mechanisms for translocation control
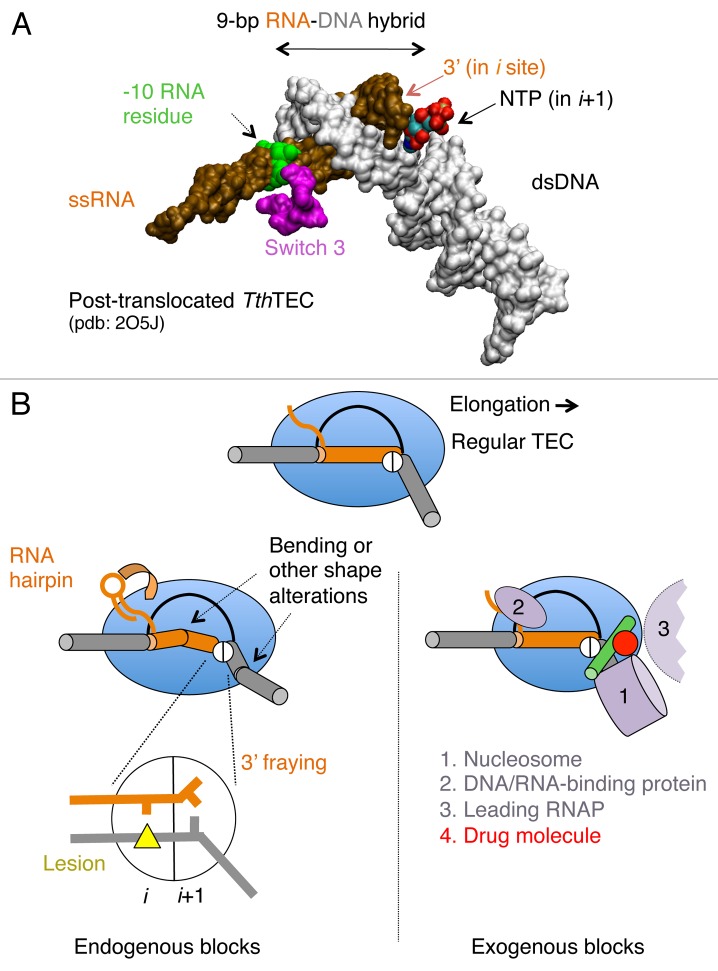
During elongation, RNAP frequently encounters alterations of dsDNA, nascent RNA structure, DNA lesions, and misincorporation events at the 3′ end of the RNA.14,47,50,51 Polymerase also faces histones in nucleosomes and other DNA binding proteins in front of advancing polymerase.52,53 These different types of encounters can block forward translocation. Some of these blocks have been confirmed experimentally, whereas the others require further validation (Fig. 2).
Figure 2.Cis- and trans-acting factors affecting translocation. (A) Structure of RNA-DNA hybrid and dsDNA in TEC by T. thermophilus (Tth) RNAP. The structural targets for translocation regulators are indicated by arrows. (B) A schematic structure of TEC: RNAP (blue oval), upstream and downstream dsDNA (gray cylinders), RNA-DNA hybrid (brown cylinder), transcription bubble (black line) and the bridge helix (green) are shown; the active center in RNAP is represented by a circle with i and i+1 subsites. The inset displays the pre-translocated configuration of the active center with DNA lesion in i site (yellow triangle) and the 3′ RNA residue in a frayed configuration in i+1 site. The left side shows cis-acting translocation inhibitors: bending, or other structural alteration of the hybrid, the front-end DNA duplex and hairpin in the nascent RNA interacting with RNAP (shown by curved arrow). The right side displays the trans-acting inhibitors: a drug molecule bound to bridge helix reducing its mobility/bending (red dot), protein factors bound to dsDNA, nascent RNA or RNAP, and the second RNAP molecule in a head-to-tail configuration (all in magenta).
Elongating RNAP maintains a 9–10-bp RNA-DNA hybrid in its catalytic cleft (Fig. 2A), which is essential for high transcription processivity and maintenance of the correct DNA register. In bacterial RNAP, the hybrid is 9-bp and 10-bp long in the post-translocated and pre-translocated ternary elongation complexes (TECs), respectively.49,54 An expansion of the hybrid beyond this length is limited by the lid and rudder, which are small domains of RNAP located at the 5′ end of the hybrid and by the bridge helix at the 3′ end.49,55,56 Shortening of the hybrid below 7 base pairs leads to termination.1 In the yeast RNAP II, the hybrid appears to be 1 bp shorter than in the bacterial RNAP.48,55,56 However, this conclusion requires additional validation because all available structures of the yeast RNAP II were generated to contain mismatched RNA/DNA of the hybrid upstream from the -8 position, which prevents backtracking and maintains structure.48,55,57,58 In TECs that have been crystallized, the hybrid always adopts a conformation that is intermediate between the A and B forms of a double helix.55 However, this observed uniform structure may be misleading because TEC crystallization typically involves a laborious selection to find base pair composition and length of the hybrid that is structurally stable and inhibits RNAP translocation in the crystal lattice. Thus, the X-ray results on the length and structure of the hybrid in TEC being strongly biased by the technical limits imposed.
The X-ray structures of TECs poised at different steps during translocation and catalysis revealed multiple orientations of the 3′ RNA end, the incoming cognate and non-cognate NTPs, and the template DNA residue (i+1) near the active center of the enzyme.54,58-61 Apart from local protein changes in the flexible regions surrounding the RNA-DNA hybrid and the downstream dsDNA, which include the switches, fork loop, trigger loop, lid and rudder elements, the structure of RNAP showed minor variations in different translocation intermediates.49,57,58 One wall of the catalytic cleft is part of a mobile clamp domain from the N-terminal part of the largest subunit of RNAP.62 Binding of the hybrid to the catalytic cleft induces closure of the clamp, leading to encirclement of the RNA-DNA hybrid and partial clamping of the downstream DNA duplex. An inner surface of the cleft facing the hybrid is composed of the switch and fork domains from the two largest RNAP subunits (Fig. 2A). Notably, the catalytic cleft contains small flexible loops forming sequence specific contacts with the major grove of the hybrid and cavities that accommodate bulges and/or flipped out residues in the hybrid.49,55,56 Because of the helical structure of dsDNA, translocation includes the rotation of the RNA-DNA hybrid within the catalytic cleft (~12 degrees per 1-bp shift). During this process, all local protein contacts between the nascent RNA and DNA are rearranged until the enzyme reaches the post-translocated register. Below, we describe inhibitors that block hybrid movement and translocation.
Endogenous effects
Any local irregularity in the structure of the downstream DNA duplex or of the RNA-DNA hybrid (e. g., intrinsic bending or other non-canonical double-helical forms of the double helix) will interfere with the hybrid rotation and passage during translocation through the narrow catalytic cleft (Fig. 2B, left panel). Because the most extensive RNAP contacts with the hybrid are localized close to the active center, translocation is particularly sensitive to the chemistry and structure of the RNA-DNA base pairing at the 3′ end of the nascent RNA, as well as to DNA lesions in the template strand entering the active site. The 3′ residue RNA-binding site (i) and the NTP binding site (i+1) subdivide the active site, each having a different capacity to accommodate different base pairs as well as 3′ RNA-DNA mismatches and DNA lesions. For instance, RNAP appears to preferentially place pyrimidine and a mismatch at the 3′ RNA end (i site), and bulky DNA lesions at the i+1, which is less restrictive than the i site. This stabilizes the pre-translocated register. A lesion with photo-activated cyclobutane pyrimidine dimer (CPD) appears not to interfere with forward translocation before entering the i site.51 This tolerance is explained by the recent finding that translocation occurs without loading of this lesion to i+1 site, with the CPD being maintained on top of the bridge helix in a flipped-out configuration.51 A strong translocation block occurs after the CPD enters the i and i-1 sites. In contrast, the i site appears to be more tolerant than the i+1 site for other types of DNA lesions such as 8-oxoguanine (8-oxoG).63 Thus, translocation register is dictated by the ability of different parts of the catalytic cleft to accommodate DNA lesions and RNA-DNA base pairs of variable chemical structure. Finally, formation of a secondary structure in the nascent transcript immediately upstream from the RNA-DNA hybrid also interferes with translocation by anchoring the RNA in single-strand RNA binding site on RNAP located beneath the flap domain of β subunit.49,64 A direct interaction of the nascent 5′ RNA with the coil-coiled motif at the tip of the flap may also restrict the hybrid movement during translocation (Fig. 2B).65
Exogenous effects
In Figure 2B, we classify several exogenous inhibitors to translocation: (1) protein roadblocks imposed downstream of RNAP, (2) protein tethers that bind to both the nucleic acids and RNAP, restraining translocation, (3) a leading RNAP that stalls during elongation to impose a roadblock to the trailing RNAP, and (4) small drug molecules that bind to the oscillating elements of RNAP, such as trigger loop, bridge helix, and forks.49,54,57,58 The first class includes nucleosomes, DNA-bound proteins such as transcription repressors (LacI, GalR, etc.), and limited movement imposed by adjacent RNAP molecules located in tandem transcribing the same DNA.66,67 A translocation block has been confirmed experimentally for RNAP II encountering nucleosomes53 and for RNAPs transcribing in tandem.67
Another class includes transcription elongation factor Nun encoded by H022 bacteriophage, which simultaneously binds RNAP, DNA and RNA-DNA hybrid at a significant distance from the RNAP active site to physically interfere with forward translocation (Vitiello et al., submitted). Nun has been shown to strongly arrest RNAP elongation in vivo and in vitro by stabilizing the enzyme in a pre-translocation register.68 Another similar example includes elongation factor NusG as found in Bacillus subtilis, which induces pausing of RNAP by interacting with the non-transcribed DNA strand in the transcription bubble as well as the upstream DNA duplex.69,70 Streptolidigin and tagetitoxin drugs, which act on bacterial RNAP, and α-amanitin, which acts on eukaryotic RNAP II, exemplify the third class of translocation inhibitors.57,71,72 These drugs bind to the bridge helix or the trigger loop (Fig. 2B) and reduce their mobility, which is required for translocation.36
The RNA-DNA hybrid in TECs is a target for control of translocation
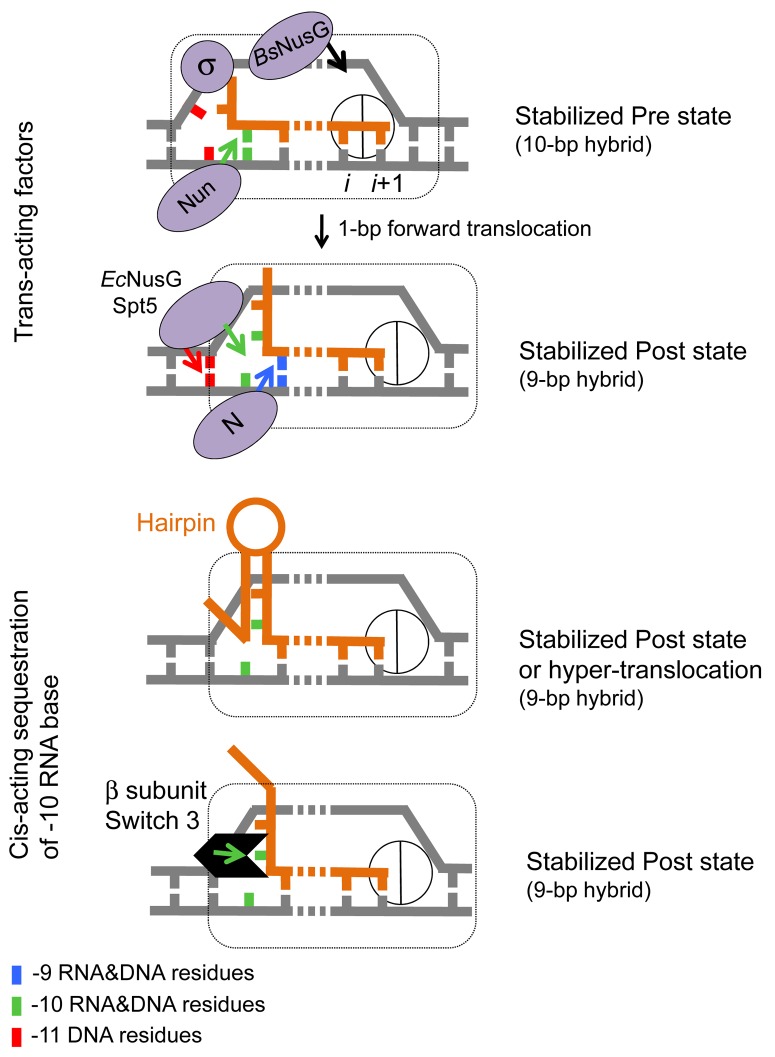
Structural studies of Thermus thermophilus TECs suggested that the hybrid length increases to 10-bp immediately after bond formation and is followed by restoration of the original 9-bp hybrid after translocation.49,54 In this view, the 10-bp and 9-bp hybrids are the signatures of the pre- and post-translocated state of TEC, respectively. This assumption is consistent with the reported biochemical properties of T. thermophilus TECs assembled on a synthetic RNA/DNA scaffold containing 8–11-bp hybrids.73 TECs made with a 10-bp hybrid exhibited a high rate of pyrophosphorolysis relative to those made with a 9-bp hybrid,73 indicating that removal of 1 base pair at the -10 position of the hybrid causes forward translocation. Transcription factors and cis-acting elements in the DNA and nascent RNA may also promote or inhibit translocation by targeting the upstream end of the hybrid (Fig. 3). For instance, Nun protein of bacteriophage H022, which causes pre-translocation arrest in vivo and in vitro, likely inhibits translocation by stabilizing the -10 base pair position of the hybrid (Vitiello et al., submitted).68 In contrast, N protein of bacteriophage λ, which binds to the RNAP catalytic cleft near the RNA exit channel,74,75 may inhibit base pairing at the -10 position of the hybrid favoring forward translocation. NusG protein from E. coli interacts with the DNA duplex at the upstream end of the transcription bubble.76 A similar interaction has been proposed by Murakami and colleagues for the yeast Spt5 protein based on their structural analysis of Spt4/5 dimer bound to the clamp domain of archaeal RNAP.77 This upstream interaction may aid translocation by promoting re-annealing of DNA strands immediately upstream from the 9-bp hybrid position. NusG and Spt5 have been shown, as is the case for N protein,78 to stimulate RNAP elongation in vitro.24,76 The cis-acting hairpin formed in the RNA upstream from the -9 position of the RNA-DNA hybrid likely sequesters the -10 nt residue into the stem removing it from the hybrid (Fig. 3) during transcription termination.1 Interestingly, structural data also indicate that, in the post-translocated TEC, the -10 RNA base is sequestered in a protein pocket made by the Switch-3 domain of the bacterial β subunit (Fig. 3),49 and mutations in this domain exhibit severe translocation defects in vitro.79 Proteins such as N, Nun and NusG, bound close to the hybrid end, may alter the Switch 3 domain to decrease or increase sequestration of the -10 residue of RNA. It is not clear how the B. subtilis NusG and H022 Nun proteins have evolved to recognize the pre-translocated state of RNAP, as opposed to their homologs in E. coli, NusG and λ N, which target the post-translocated state.68,80 However, there are several lines of evidence that indicate that NusG can switch between transcription stimulation and inhibition modes depending on sequence context and/or bacterial species.69,70,81 Nun can also switch to N-like anti-termination activity in vivo82 and in vitro, as it has been shown by engineering of “hybrid” proteins between N and Nun in domain swapping experiment.83
Figure 3. Translocation modulators target RNA-DNA hybrid and transcription bubble in bacterial TEC. Top panel: Nun protein of bacteriophage H022 interferes with translocation68 by stabilizing the -10 base pair of the hybrid and tethering RNAP to the hybrid. We proposed that the homologous N protein of bacteriophage λ stabilizes the -9 base pair of the hybrid and prevents the -10 base pair to favor translocation. E. coli NusG and its eukaryotic homolog Spt5 promote translocation by facilitating re-annealing of DNA immediately upstream from the 9-bp hybrid. B. subtilis NusG tethers RNAP to pre-translocated register by binding to the middle part of transcription bubble.70 σ70 subunit interferes with translocation by binding to the “-10-like” sequence at the upstream end of transcription bubble.96 Bottom panel: cis-acting RNA hairpin and Switch 3 domain in β subunit promote translocation by preventing expansion of the hybrid to 10-bp length.79
Physiological significances of pausing in bacteria and in eukaryotes
The physiological role of transcriptional regulation via RNAP II pausing has been extensively investigated in eukaryotic cells. Promoter-proximal pausing of RNAP II controls expression of heat shock genes and proto oncogenes in Drosophila and mammalian cells, respectively.2 ChIP-chip analysis indicated that these pauses are also detected at a large number of untranscribed genes in Drosophila and mammalian cells under variable experimental conditions.12,84-86 There are factors that induce promoter-proximal pausing of RNAP II, namely, DSIF (DRB sensitivity-inducing factor) and NELF (negative elongation factor).2,13,87 The involvement of these factors in the control of pausing implies that transcription pausing induced by translocation blocks followed by backtracking of RNAP may not be sufficient to explain all promoter-proximal pausing in vivo, although it has been proposed that pausing in Drosophila does involve backtracking of RNAP II.88 Below, we summarize recent findings on promoter-proximal pausing and discuss the pausing mechanism in the context of heterogeneous tracking of RNAP over specific sequences that can modulate the tracking.
A prominent role of promoter-proximal pausing was initially proposed to explain the rapid transcription response to environmental stimuli, such as heat-shock or cell differentiation, where the transcription initiation step is bypassed.84 However, recent studies indicate more divergent regulatory roles of promoter-proximal pausing: in Drosophila, pausing is employed to maintain a basal level of expression of genes coding for membrane receptors, transcriptional regulators involved in immune response, and factors affecting signal transduction pathways.89 Promoter-proximal pausing is a key step in the concerted activation of genes involved in embryogenesis,90 and for shutting off the heat shock genes by NELF-mediated pathway during recovery from heat shock.91 Promoter-proximal paused RNAP II may additionally contribute to gene repression by blocking promoter access by other RNAP II molecules.88
The recent development of high-throughput sequencing technology92 has allowed short RNA species retaining a 7-methylguanosine cap at their 5′ ends to be deep-sequenced.13 A large fraction of the 3′ ends of these short RNA species was mapped to Drosophila genomic positions that coincided with sites susceptible to permanganate, revealing that RNAP II often paused at positions located between +25 and +60 bps relative to transcriptional start sites throughout the genome.13 This result was consistent with the permanganate-ChIP-seq data, which indicated that promoter-proximal pausing occurs between +20 and +60 bps from transcriptional start sites at thousands of Drosophila promoters.93 The positions of the sequenced 3′ ends were shifted downstream in cells defective for TFIIS, indicating that the paused RNAP II may be backtracked.13 Interestingly, the estimated DNA melting temperature of the regions surrounding the 3′ end of these short RNA species was higher than those of most transcribed regions of the genome.13 The authors claimed that RNAP II pauses transiently within the downstream regions, where the RNA-DNA hybrid is less stable, and backtracks upstream from these regions to generate a more stable hybrid;13 however, the mechanical link to a translocation block of these sequences remains unclear.
In E. coli, promoter-proximal pausing has also been reported for rplN and ompX genes, as well as for the genes for λ and 82 phages.94,95 Gre factors are required for escape from these pause sites. A permanganate footprint analysis in E. coli revealed that RNAP associated with σ70 frequently pauses at promoter-proximal regions carrying a -10 like sequence.96,97 This pausing was detected at 10-20% of the E .coli promoters, and it was enhanced by deletion of the greA gene.95 This suggested that promoter-proximal pausing caused by RNAP backtracking in E. coli is a general phenomenon. ChIP-chip analysis also indicated that RNAPs undergo pausing near the transcription start sites in more than 20% of transcribed genes in E. coli.98 A fraction of paused RNAPs detected by ChIP-chip analysis likely includes “moribund complexes” that retain σ70 and short abortive transcripts.99,100 Formations of the moribund complexes have been shown to depend on promoter sequence.101,102 The abortive type of RNA synthesis by the moribund complex is much slower than RNA synthesis by a productive elongation complex.99 The release of abortive RNA transcripts may require backtracking.29 Because the RNA retained in the moribund complex is shorter than 20-nt and has a 5′ triphosphate,103 deep sequencing of nascent transcripts that are cleaved into short fragments and have either 5′ triphosphate or 5′ monophosphate ends could allow discrimination of the moribund complexes from any paused elongation complexes.
ChIP-chip analysis revealed that promoter-proximal pausing in S. cerevisiae and B. subtilis is not as robust as in Drosophila and E. coli.104,105 In yeast, these pauses are typically sensitive to transcript cleavage factor TFIIS: RNAP II pause sites identified by deep sequencing of nascent 3′ transcripts are shifted several bps downstream from their original location in cells lacking the dst1 gene coding for TFIIS.10 This result indicates that pausing in yeast involves backtracking of RNAP II followed by TFIIS-stimulated cleavage of the 3′ proximal transcript, which is required to resume elongation. ChIP-chip analysis also detected increase in promoter-proximal pausing caused by deletion of the greA gene in B. subtilis.105 These observations argue that promoter-proximal pausing due to RNAP backtracking appears to be a common theme in prokaryotes and eukaryotes and that the activity of TFIIS or Gre factors that rescue backtracked RNAP plays a key role in gene regulation at transcription pause sites. Future characterization of the variable cis-acting DNA signals and trans-acting protein factors that regulate elongation by RNAP in vivo will help to establish a firm link between transcription pausing and heterogeneous tracking of polymerase.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Donald Court for critical reading of the manuscript and Maria Kireeva for discussions. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research to M.K. M.I. is partially supported by a fellowship from JSPS.
Glossary
Abbreviations:
- bp
base pair
- ChIP-chip
chromatin immunoprecipitation followed by microarray
- CPD
cyclobutane pyrimidine dimer
- dsDNA
double-stranded DNA
- kB
the Boltzmann constant
- NTP
ribonucleoside triphosphate
- 8-oxoG
8-oxoguanine
- Pi
inorganic phosphate
- PPi
pyrophosphate
- RNAP
RNA polymerase
- T
absolute temperature
- TEC
ternary elongation complex
References
- 1.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10:1151–62. doi: 10.1016/S1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 2.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–31. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–6. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 4.Landick R, Carey J, Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci U S A. 1985;82:4663–7. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/S0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 6.Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–25. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- 7.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/S0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 9.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–38. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 10.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perdue SA, Roberts JW. A backtrack-inducing sequence is an essential component of Escherichia coli σ(70)-dependent promoter-proximal pausing. Mol Microbiol. 2010;78:636–50. doi: 10.1111/j.1365-2958.2010.07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–8. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imashimizu M, Oshima T, Lubkowska L, Kashlev M. Direct assessment of transcription fidelity by high-resolution RNA sequencing. Nucleic Acids Res. 2013;41:9090–104. doi: 10.1093/nar/gkt698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–43. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–45. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Helmrich A, Ballarino M, Nudler E, Tora L. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol. 2013;20:412–8. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- 19.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–66. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 20.Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′----5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–56. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 21.Reines D, Ghanouni P, Li QQ, Mote J., Jr. The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J Biol Chem. 1992;267:15516–22. [PMC free article] [PubMed] [Google Scholar]
- 22.Hausner W, Lange U, Musfeldt M. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J Biol Chem. 2000;275:12393–9. doi: 10.1074/jbc.275.17.12393. [DOI] [PubMed] [Google Scholar]
- 23.Burova E, Hung SC, Sagitov V, Stitt BL, Gottesman ME. Escherichia coli NusG protein stimulates transcription elongation rates in vivo and in vitro. J Bacteriol. 1995;177:1388–92. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SME. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres M, Balada JM, Zellars M, Squires C, Squires CL. In vivo effect of NusB and NusG on rRNA transcription antitermination. J Bacteriol. 2004;186:1304–10. doi: 10.1128/JB.186.5.1304-1310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Contreras X, Yamaguchi Y, Handa H, Peterlin BM, Guo S. Repression of RNA polymerase II elongation in vivo is critically dependent on the C-terminus of Spt5. PLoS One. 2009;4:e6918. doi: 10.1371/journal.pone.0006918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–51. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–56. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimamoto N. Nanobiology of RNA polymerase: biological consequence of inhomogeneity in reactant. Chem Rev. 2013;113:8400–22. doi: 10.1021/cr400006b. [DOI] [PubMed] [Google Scholar]
- 30.Oosawa F, Hayashi S. The loose coupling mechanism in molecular machines of living cells. Adv Biophys. 1986;22:151–83. doi: 10.1016/0065-227X(86)90005-5. [DOI] [PubMed] [Google Scholar]
- 31.Tyska MJ, Warshaw DM. The myosin power stroke. Cell Motil Cytoskeleton. 2002;51:1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- 32.Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–22. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- 33.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–95. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishii Y, Nishiyama M, Yanagida T. Mechano-chemical coupling of molecular motors revealed by single molecule measurements. Curr Protein Pept Sci. 2004;5:81–7. doi: 10.2174/1389203043486838. [DOI] [PubMed] [Google Scholar]
- 35.Malinen AM, Turtola M, Parthiban M, Vainonen L, Johnson MS, Belogurov GA. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res. 2012;40:7442–51. doi: 10.1093/nar/gks383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–93. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 37.Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 38.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–5. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imashimizu M, Kireeva ML, Lubkowska L, Gotte D, Parks AR, Strathern JN, Kashlev M. Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J Mol Biol. 2013;425:697–712. doi: 10.1016/j.jmb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell. 2010;38:202–10. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hein PP, Palangat M, Landick R. RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry. 2011;50:7002–14. doi: 10.1021/bi200437q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawryluk PJ, Ujvári A, Luse DS. Characterization of a novel RNA polymerase II arrest site which lacks a weak 3′ RNA-DNA hybrid. Nucleic Acids Res. 2004;32:1904–16. doi: 10.1093/nar/gkh505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–94. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikolova EN, Bascom GD, Andricioaei I, Al-Hashimi HM. Probing sequence-specific DNA flexibility in a-tracts and pyrimidine-purine steps by nuclear magnetic resonance (13)C relaxation and molecular dynamics simulations. Biochemistry. 2012;51:8654–64. doi: 10.1021/bi3009517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duchardt E, Nilsson L, Schleucher J. Cytosine ribose flexibility in DNA: a combined NMR 13C spin relaxation and molecular dynamics simulation study. Nucleic Acids Res. 2008;36:4211–9. doi: 10.1093/nar/gkn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–5. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–19. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Sydow JF, Brueckner F, Cheung AC, Damsma GE, Dengl S, Lehmann E, Vassylyev D, Cramer P. Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell. 2009;34:710–21. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–62. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 50.Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim Biophys Acta. 2010;1799:389–401. doi: 10.1016/j.bbagrm.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Walmacq C, Cheung AC, Kireeva ML, Lubkowska L, Ye C, Gotte D, Strathern JN, Carell T, Cramer P, Kashlev M. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol Cell. 2012;46:18–29. doi: 10.1016/j.molcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Epshtein V, Toulmé F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–27. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal elements that control the topography of the barrier to transcription. Cell. 2012;151:738–49. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–8. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 55.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–82. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 56.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–6. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 57.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–8. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 58.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–54. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–9. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 60.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–6. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–41. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–76. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 63.Damsma GE, Cramer P. Molecular basis of transcriptional mutagenesis at 8-oxoguanine. J Biol Chem. 2009;284:31658–63. doi: 10.1074/jbc.M109.022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toulmé F, Mosrin-Huaman C, Artsimovitch I, Rahmouni AR. Transcriptional pausing in vivo: a nascent RNA hairpin restricts lateral movements of RNA polymerase in both forward and reverse directions. J Mol Biol. 2005;351:39–51. doi: 10.1016/j.jmb.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 65.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–3. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 66.Mosrin-Huaman C, Turnbough CL, Jr., Rahmouni AR. Translocation of Escherichia coli RNA polymerase against a protein roadblock in vivo highlights a passive sliding mechanism for transcript elongation. Mol Microbiol. 2004;51:1471–81. doi: 10.1111/j.1365-2958.2003.03926.x. [DOI] [PubMed] [Google Scholar]
- 67.Saeki H, Svejstrup JQ. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol Cell. 2009;35:191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hung SC, Gottesman ME. The Nun protein of bacteriophage HK022 inhibits translocation of Escherichia coli RNA polymerase without abolishing its catalytic activities. Genes Dev. 1997;11:2670–8. doi: 10.1101/gad.11.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yakhnin AV, Babitzke P. Mechanism of NusG-stimulated pausing, hairpin-dependent pause site selection and intrinsic termination at overlapping pause and termination sites in the Bacillus subtilis trp leader. Mol Microbiol. 2010;76:690–705. doi: 10.1111/j.1365-2958.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yakhnin AV, Yakhnin H, Babitzke P. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc Natl Acad Sci U S A. 2008;105:16131–6. doi: 10.1073/pnas.0808842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuzenkova Y, Roghanian M, Bochkareva A, Zenkin N. Tagetitoxin inhibits transcription by stabilizing pre-translocated state of the elongation complex. Nucleic Acids Res. 2013;41:9257–65. doi: 10.1093/nar/gkt708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark AD, Jr., et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–52. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kashkina E, Anikin M, Tahirov TH, Kochetkov SN, Vassylyev DG, Temiakov D. Elongation complexes of Thermus thermophilus RNA polymerase that possess distinct translocation conformations. Nucleic Acids Res. 2006;34:4036–45. doi: 10.1093/nar/gkl559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mishra S, Mohan S, Godavarthi S, Sen R. The interaction surface of a bacterial transcription elongation factor required for complex formation with an antiterminator during transcription antitermination. J Biol Chem. 2013;288:28089–103. doi: 10.1074/jbc.M113.472209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–49. doi: 10.1016/S0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 76.Tomar SK, Artsimovitch I. NusG-Spt5 proteins-Universal tools for transcription modification and communication. Chem Rev. 2013;113:8604–19. doi: 10.1021/cr400064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A. 2011;108:546–50. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rees WA, Weitzel SE, Das A, von Hippel PH. Regulation of the elongation-termination decision at intrinsic terminators by antitermination protein N of phage lambda. J Mol Biol. 1997;273:797–813. doi: 10.1006/jmbi.1997.1327. [DOI] [PubMed] [Google Scholar]
- 79.Kent T, Kashkina E, Anikin M, Temiakov D. Maintenance of RNA-DNA hybrid length in bacterial RNA polymerases. J Biol Chem. 2009;284:13497–504. doi: 10.1074/jbc.M901898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SME. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sevostyanova A, Artsimovitch I. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 2010;38:7432–45. doi: 10.1093/nar/gkq623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robledo R, Gottesman ME, Weisberg RA. Lambda nutR mutations convert HK022 Nun protein from a transcription termination factor to a suppressor of termination. J Mol Biol. 1990;212:635–43. doi: 10.1016/0022-2836(90)90226-C. [DOI] [PubMed] [Google Scholar]
- 83.Henthorn KS, Friedman DI. Identification of functional regions of the Nun transcription termination protein of phage HK022 and the N antitermination protein of phage lambda using hybrid nun-N genes. J Mol Biol. 1996;257:9–20. doi: 10.1006/jmbi.1996.0142. [DOI] [PubMed] [Google Scholar]
- 84.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–11. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–12. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 89.Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26:933–44. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–3. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ghosh SK, Missra A, Gilmour DS. Negative elongation factor accelerates the rate at which heat shock genes are shut off by facilitating dissociation of heat shock factor. Mol Cell Biol. 2011;31:4232–43. doi: 10.1128/MCB.05930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 93.Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol Cell. 2013;50:711–22. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stepanova E, Wang M, Severinov K, Borukhov S. Early transcriptional arrest at Escherichia coli rplN and ompX promoters. J Biol Chem. 2009;284:35702–13. doi: 10.1074/jbc.M109.053983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–85. doi: 10.1016/S1097-2765(00)00126-X. [DOI] [PubMed] [Google Scholar]
- 96.Hatoum A, Roberts J. Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol Microbiol. 2008;68:17–28. doi: 10.1111/j.1365-2958.2008.06138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nickels BE, Mukhopadhyay J, Garrity SJ, Ebright RH, Hochschild A. The sigma 70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nat Struct Mol Biol. 2004;11:544–50. doi: 10.1038/nsmb757. [DOI] [PubMed] [Google Scholar]
- 98.Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–6. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kubori T, Shimamoto N. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1996;256:449–57. doi: 10.1006/jmbi.1996.0100. [DOI] [PubMed] [Google Scholar]
- 100.Sen R, Nagai H, Hernandez VJ, Shimamoto N. Reduction in abortive transcription from the lambdaPR promoter by mutations in region 3 of the sigma70 subunit of Escherichia coli RNA polymerase. J Biol Chem. 1998;273:9872–7. doi: 10.1074/jbc.273.16.9872. [DOI] [PubMed] [Google Scholar]
- 101.Susa M, Kubori T, Shimamoto N. A pathway branching in transcription initiation in Escherichia coli. Mol Microbiol. 2006;59:1807–17. doi: 10.1111/j.1365-2958.2006.05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Imashimizu M, Tanaka K, Shimamoto N. Comparative Study of Cyanobacterial and E. coli RNA Polymerases: Misincorporation, Abortive Transcription, and Dependence on Divalent Cations. Genet Res Int. 2011;2011:572689. doi: 10.4061/2011/572689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McClure WR. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 104.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–46. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 105.Kusuya Y, Kurokawa K, Ishikawa S, Ogasawara N, Oshima T. Transcription factor GreA contributes to resolving promoter-proximal pausing of RNA polymerase in Bacillus subtilis cells. J Bacteriol. 2011;193:3090–9. doi: 10.1128/JB.00086-11. [DOI] [PMC free article] [PubMed] [Google Scholar]