Abstract
Human RNA polymerase III transcribes small untranslated RNAs that contribute to the regulation of essential cellular processes, including transcription, RNA processing and translation. Analysis of this transcription system by in vitro transcription techniques has largely contributed to the discovery of its transcription factors and to the understanding of the regulation of human RNA polymerase III transcription. Here we review some of the key steps that led to the identification of transcription factors and to the definition of minimal promoter sequences for human RNA polymerase III transcription.
Keywords: Human RNA polymerase III, in vitro, basal transcription, activated transcription, TFIIIA, TFIIIB, TFIIIC, PTF, SNAPC, promoter, enhancer
The discovery of constituents of eukaryotic transcription systems was enabled by the development of in vivo and in vitro methods for their study. Depending on the organism to be studied, the relative importance of each of these two general approaches has been different. Our knowledge about transcription of human genes would be considerably smaller without the results that were obtained by employing in vitro methods. Here, we will mainly focus on in vitro studies of gene expression that contributed to discoveries in the human RNA polymerase (RNAP) III transcription system.
In contrast to unicellular yeast, it has until today been and will most likely remain for a many more years impossible to study human RNAP III transcription in cells that have been grown under conditions resembling at least approximately to an in vivo situation in a human being. For that reason, it has hitherto been difficult to address questions concerning the physiological regulation of human RNAP III transcription in its natural environment. However, the identification of the basal components of the RNAP III transcription system could be achieved without directly resorting to a human “in vivo model system.” Yet, the identification of human RNAP III subunits and of accessory transcription factors repeatedly took advantage of in vivo systems that were established in other species. Many discoveries of components of the human RNAP III transcription system were fostered by work performed in unicellular (for example, S. cerevisiae and S. pombe) and multicellular eukaryotes (among others, X. laevis or D. melanogaster). Often, results obtained in these model systems helped researchers identify proteins of the human RNAP III transcription system. For instance, the amino acid sequences of transcription factors that were identified and cloned by employing in vivo and in vitro methods in these organisms served as guide for comparing and identifying orthologous transcription factors in human cells (e.g., TFIIIA, TFIIIB [BDP1], TFIIIC35 [GTF3C6]). However, several of the human RNAP III transcription factors were not cloned by homology, but due to extensive purification from human cells (TFIIIB [BRF2], PTF/SNAPc, TFIIIC [GTF3C1–5]). Purification of these factors by employing biochemical methods was only possible, because functional assays were developed that allowed detecting their specific activities. These assays included in vitro transcription, electrophoretic mobility shift assays (EMSA) and footprinting techniques (descriptions of these techniques are found in1-3). Two techniques were essential for the biochemical purification and functional analysis of human transcription factors: (i) the elaboration of protocols that allowed deriving protein extracts from human cells and (ii) the development of cell-free in vitro transcription assays.4-6 Template DNA for in vitro transcription was provided by genes cloned into plasmid DNA. In the case of RNAP III transcription, usually complete and generally short transcribed sequences were included into the template DNA. The genes transcribed by RNAP III and the promoters recognized by its cognate transcription factors have been reviewed7,8 and are schematically depicted in Figures 1–3. From cellular extracts, individual transcription activities were purified and used for reconstitution of RNAP III transcription in vitro. In 1980, Segall and colleagues described the reconstitution of human tRNA and 5S RNA transcription in vitro by two (tRNA) or three (5S RNA) partially purified protein fractions, respectively. According to their elution profile from phosphocellulose (P11 or PC) chromatography, these fractions were referred to as P11 (or PC) fractions -0.1 (-A), -0.35 (-B) and -0.6 (-C), containing the transcription activities TFIIIA (P11–0.1), TFIIIB (P11–0.35) and TFIIIC (P11–0.6). RNAP III eluted in fractions P11–0.35 and P11–0.6.9 From these fractions, the individual transcription activities were further purified over several subsequent chromatographical steps. In the following, we will give a short and non-exhaustive overview over some key-steps in the identification and cloning of human RNAP III transcription factors that have been made possible by biochemical purification and assay of their activities by in vitro transcription.
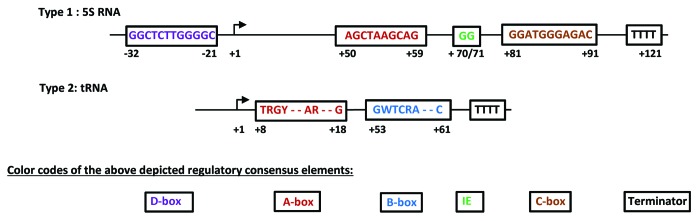
Figure 1. Schematic representation of the regulatory elements of human RNA polymerase III transcribed type 1 and type 2 genes. Nucleic acid nomenclature: R = G/A; Y = C/T; W = A/T. Designations of individual boxes are represented in appropriate color codes below type 1 and 2 gene schemes. Type 1 gene: 16 out of 17 human RNA5S genes retrieved at www.ncbi.nlm.nih.gov/nucleotide are identical; T84 within the C-box of RNA5S9 is substituted by a C. The sequence blocks of A- and C-boxes, as well as of the intermediate element (IE) in the human 5S gene are chosen by analogy to described sequence blocks in the X. laevis 5S RNA gene.129,130 The 5S gene A-box sequence lacks the conserved T in the first position of the tRNA consensus A box. The human D-box element sequence upstream of the transcription start site of the 5S gene was published.131 Type 2 genes: are representatively depicted by human tRNA consensus sequences that have been derived from published data.132-134 tRNA termination occurs at variable sites downstream of the B-box.
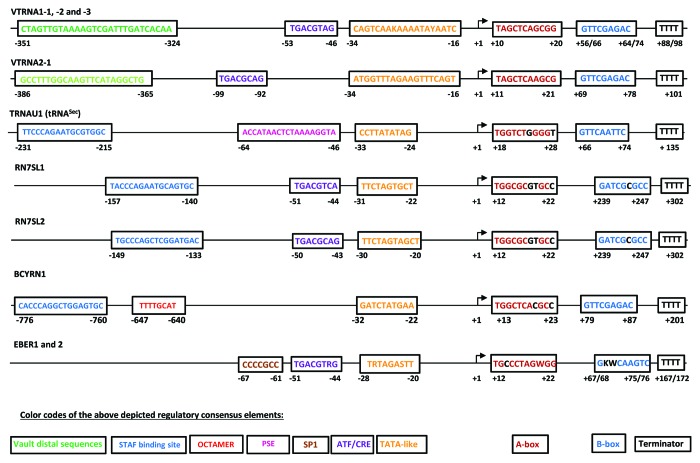
Figure 3. Schematic representation of human RNA polymerase III transcribed genes with regulatory elements 5′ and 3′ of the transcription start site (type 4 genes). Nucleic acid nomenclature: K = G/T; Y = C/T; R = G/A; S = G/C; W = A/T. Nucleotide sequences of individual presumably regulatory elements are shown for each gene. Designations of individual boxes are represented in appropriate color codes below type 4 gene schemes. Experimental proof for the functionality of these elements has been shown for the ATF/CRE element in the human 7SL and herpesvirus 4 EBER genes.89,90,121,138 Inter-species sequence conservation was shown for VTRNA1–1, -2, -3 and VTRNA2–1 regulatory elements. Vault distal elements comprise the shown sequences and two distinct additional further distal sequence blocks for each gene.139,140 PSE and STAF-binding sites in the human TRNAU1, RN7SL1 and 2, as well as the BCYRN1 gene were delineated by sequence homology with PSE and STAF-binding site consensus sequences (Fig. 2). TATA-like elements were proposed for the VTRNA1–1, -2, -3, the 7SL1 and 2 genes, EBER genes (ETAB)138 and the mouse analog of the BCYRN1 gene.121,122,140 Sequences at similar positions in VTRNA2–1 and TRNAU1 genes are shown. A stimulatory influence of a SP1 binding site on in vitro transcription of the EBER2 gene was shown.138 An octamer element as a potential enhancer element was identified by sequence homology upstream of the BCYRN1 gene. Gene internal A and B boxes are appropriately depicted within the transcribed regions. Bases that differ from the tRNA consensus sequences (Fig. 1) for these elements are shown in black letters.
The General Human RNA Polymerase III Transcription Machinery
TFIIIA
The first description of a human transcription activity being exclusively required for the expression of the 5S gene in vitro was published in 1980 by R.G. Roeder and colleagues (Segall et al., 1980). The authors showed that 5S gene transcription requires crude fractions containing transcription factors TFIIIA (PC-A), TFIIIB (PC-B), TFIIIC (PC-C) and RNAP III (PC-B and PC-C), whereas tRNA transcription was reconstituted by the fractions containing TFIIIB, TFIIIC and RNAP III only. Ordered association of TFIIIA, TFIIIC and TFIIIB with the 5S gene was established by in vitro studies using partially purified human factors.10,11 The identification of the amino acid sequence of human TFIIIA12 and molecular cloning of the corresponding cDNA13,14 demonstrated high homology (57% identity) to X.laevis TFIIIA, which had been identified and cloned earlier.15,16
TFIIIB
Starting with phosphocellulose fraction B (PC-B or P11–0.35), several research groups were involved in the purification of human TFIIIB. K.H. Seifart and colleagues published in 1988 the partial purification of human TFIIIB.17 After the molecular cloning of the TATA-binding protein (TBP) from human cells18,19 and the discovery that TBP participates in U6 transcription in yeast20 and human cells21-23 it became clear that it is also involved in the transcription of RNAP III genes with internal promoter elements.24 Subsequently, TBP and associated factors were purified that reconstituted TFIIIB activity at gene internal promoters.25-29 Further purification resulted in the identification of human TFIIB-related factor 1 (BRF1/TFIIIB90),30,31 showing extensive sequence homology in its N-terminal half to the orthologous protein from S. cerevisiae32-34 and to the paralogous RNAP II transcription factor TFIIB.35,36 Depletion of TBP from cellular extracts led to the loss of in vitro transcription of all genes expressed by human RNAP III. However, reconstitution of transcription from gene internal promoter elements required a complex of TBP and associated factors (TAFs), whereas transcription of gene regulated by external promoters could be reconstituted by purified recombinant human TBP only.25,29 These results suggested the existence of distinct TFIIIB activities that are specifically required for either in vitro transcription of gene internal promoter-containing genes or for that of genes with promoter elements in the 5′ non-transcribed region. In 1995, these two activities were physically separated by chromatography.37 The proteins constituting both TFIIIB-activities were identified and cloned.38-41 TFIIIBα, being active in transcription of genes with promoter elements 5′ of the transcription start site (TSS) is composed of TBP, TFIIIB double prime (BDP1) and BRF2. TFIIIBβ, required for transcription of genes with gene-internal promoter elements, is composed of TBP, BDP1 and BRF1.
TFIIIC
TFIIIC was initially described as primary DNA-binding protein at tRNA and adenoviral VA genes (A- and B-box-containing gene internal promoter elements) and as secondary DNA-binding factor of the 5S gene (A- and C-box-containing gene internal promoter). Later, human TFIIIC-activity was chromatographically separated into two fractions that either comprised the DNA-binding activity (TFIIIC2) or that contained an essential, yet not primarily DNA-binding activity (TFIIIC1).1,42 The six subunits constituting the DNA-binding complex were identified43-46 and the corresponding cDNAs were cloned.47-52 Reconstitution of in vitro transcription by partially purified fractions indicated that TFIIIC1 may be related to the BDP1 subunit of TFIIIB, a suspicion that was corroborated by the fact that a C-terminally truncated version of BDP1 (recombinant human TFIIIB150) was able to partially replace TFIIIC1 in in vitro transcription assays.53
PBP/PTF/SNAPc
As for TFIIIC, also the identification of an activator of U6 transcription was achieved by purification from P11–0.6 (PC-C) fractions and analysis of its activity by in vitro assays, including DNase I footprinting, EMSA and in vitro transcription. This transcription factor, interacting with the proximal sequence element (PSE) of U6 and 7SK gene regulatory sequences was discovered due to its ability to reconstitute transcription of these genes in vitro. This factor, described as PSE-binding protein (PBP),23 PSE-binding transcription factor (PTF)54 or small nuclear RNA-activating protein complex (SNAPc)55 binds to the PSE and is required, together with OCT-1 for full activation of U6 and 7SK genes. The subunits of this complex were purified and their corresponding cDNAs were cloned.56-62 The human RNAP III transcription factors and the cloning of their subunits are listed in Table 1.
Table 1. Listing of the human RNA polymerase III transcription factor subunits and the papers describing the cloning of the respective cDNAs.
| Transcription factor | Subunits | Cloning described in ref. |
|---|---|---|
| TFIIIA | TFIIIA/GTF3A | 13,14 |
| TFIIIB | TBP | 18,19 |
| BRF1 (TFIIIB90) | 30,31 | |
| BRF2 (TFIIIB50) | 40,41 | |
| BDP1 (TFNR; TFIIIB150) | 39,40 | |
| TFIIIC | TFIIIC220/GTF3C1 | 47,48 |
| TFIIIC110/GTF3C2 | 49 | |
| TFIIIC102/GTF3C3 | 50 | |
| TFIIIC90/GTF3C4 | 51 | |
| TFIIIC63/GTF3C5 | 50 | |
| TFIIIC35/GTF3C6 | 52 | |
| PBP/PTF/SNAPc | PTFα/SNAPC4 | 61 |
| PTFβ/SNAPC3 | 59,60 | |
| PTFγ/SNAPC1 | 56,58 | |
| PTFδ/SNAPC2 | 57,58 | |
| -/SNAPC5 | 62 |
RNA polymerase III
Human RNA polymerase III was purified from HeLa cells and the cDNAs encoding its subunits were cloned.63-66 It was shown that two isoforms of RNAP III are present in human cells that are both capable of transcribing the three distinct types of RNAP III promoters in vitro, but that differed in their cellular expression patterns and functions.67,68 Specific functions in transcription initiation could be assigned to a ternary subcomplex of RNAP III. From the complete 17 subunit-containing enzyme, three subunits (RPC32; RPC39; RPC62) could be separated either by treatment with 4M urea or by sucrose gradient centrifugation. It was shown by in vitro transcription of intact RNAP III genes or of “tailed templates” that the remaining 14 RNAP III subunits were catalytically active but no longer capable of specifically initiating transcription from RNAP III promoters. These results showed that the ternary subcomplex of human RNAP III fulfills important roles in transcription initiation.64
Transcriptional Regulators of Human RNA Polymerase III Transcription
Repressors
Several transcriptional activators, co-activators, but also repressors of human RNAP III transcription in vitro have been identified, indicating that transcription by RNAP III may be fine-tuned according to the needs of a cell. Tumor suppressor proteins (e.g., TP53; RB1; PTEN; CDKN2A (ARF)) have been implicated in the inhibition of RNAP III transcription.69,70 In addition, negative regulators of human RNAP III transcription with no described tumor suppressor protein function were identified. These negative regulators of RNAP III transcription may, among other functions, adapt transcription levels in response to cellular stresses. For instance, in response to cell growth and nutrient availability, and similar to what was demonstrated in the yeast S. cerevisiae,71 MAF1 was shown to be a general repressor of human RNAP III transcription.72-76 Other examples of RNAP III transcriptional inhibitors include ACR1 and YY1, which were shown to inhibit the expression of Alu elements.77,78 In the case of the ACR1 protein, it was furthermore shown that its inhibitory activity was specific for Alu genes and that it did not repress tRNA or RNAP II-driven transcription.77 For the YY1 protein, however, also positive roles in RNAP III transcription have been proposed, such as facilitating SNAPc/PTF-binding to the PSE of the U6 gene79 and a possible binding to the A-box of the tRNAGln gene.80 DR1 (also known as negative cofactor 2 β [NC2beta]) and its dimerization partner DRAP1 (NC2alpha) were shown to associate with and repress RNAP III transcription.81,82 In addition to these repressors that may act by regulating the binding of RNAP III transcription factors to their cognate DNA elements and/or by affecting their interaction with the components of the RNAP III transcription machinery, there are also reports of repressors that act on the level of DNA methylation or chromatin-related mechanisms. Besides the KRAB-domain containing factor KOX1 that represses RNAP III transcription in cellular transfection assays,83 a repressive function was shown for the DNA methyl transferases DNMT1 and DNMT3a in human U6 transcription in vivo. This repression was recapitulated in transcription of the U6 gene in vitro by methylating template DNA with the heterologous Spiroplasma DNA methyltransferase M.SssI.84
Activators
The first example of an enhancer element stimulating RNAP III transcription was the octamer binding site in the U6 and 7SK gene 5′ flanking sequences.85-87 Shortly after recognizing a role for the octamer element in RNAP III transcription, it was shown that RNAP II transcriptional activator proteins, OCT-1 and OCT-2, bound to this site and stimulated transcription by RNAP III.54,88 Thereafter a couple of other examples of activator proteins stimulating RNAP III transcription have been published, including ATF (in transcription of Epstein Barr virus EBER1 and 2 genes89and of the 7SL gene90), SP1 (EBER genes)89 and MYC (tRNALeu gene; VA1 gene).91 The identification of the SPH element as an enhancer of the tRNASec gene and of ZNF143 (STAF), the transcriptional activator binding to this gene, was achieved in X. laevis.92,93 Several years later the human gene encoding ZNF143 was cloned.94,95 These discoveries demonstrated that several distinct activators could interact with and stimulate RNAP III transcription. However, and probably due to the small number of genes transcribed by RNAP III that are directed by promoters with regulatory elements located 5′ of the transcription start site, also the number of transcriptional activators that have been described remains limited.
Chromatin-associated regulation of RNAP III transcription
Compared with RNAP II transcription, only few in vitro studies of human RNAP III transcription were conducted with template DNA that was reconstituted into chromatin. TFIIIA was shown to prevent the assembly of chromatin on the 5S gene, but that preassembled chromatin excluded subsequent binding by TFIIIA.96 TFIIIC was shown to possess intrinsic acetyltransferase activity (GTF3C4/TFIIIC90 and GTF3C3/TFIIIC102) and to alleviate chromatin-mediated repression in the presence of HeLa nuclear extract.51,97 Thereafter, expression of tRNA and VA1 genes was analyzed by employing an in vitro transcription system that was reconstituted with recombinant and highly purified transcription factors, RNAP III and a chromatin assembly system consisting of purified HeLa histones, ACF and NAP1. In this system, it was shown that the acetyltransferase P300 is recruited to RNAP III genes and activates transcription.98 The authors showed that P300 is recruited by TFIIIC, probably involving a direct interaction of P300 with the GTF3C2 (TFIIIC110) subunit. The interaction stabilized TFIIIC-binding to tDNA in an acetyltransferase-independent manner. Surprisingly, and in contrast to what was shown for RNAP II transcription, P300 also stimulated tRNA in vitro transcription from a naked DNA template.98 P300 could not be replaced by PCAF in this reconstituted in vitro chromatin transcription assay. In contrast, the PCAF-related GCN5 complex (STAGA/TFTC)99 was shown to stimulate transcription of 5S and tRNA genes in vivo.100 In the case of U6 transcription, it was suggested that a nucleosome positioned between the DSE and the PSE approaches these to elements, thereby activating in vitro transcription of this gene.101,102 This model was confirmed through analyzes of the 7SK gene by employing in vivo footprinting techniques.103 In addition, the U6 gene was shown to be regulated by the chromodomain helicase DNA-binding protein 8 (CHD8). This chromatin remodeling factor was demonstrated to associate with ZNF143 (STAF) and to contribute to efficient in vitro expression of the human U6 gene that was reconstituted into chromatin.104
Co-activators
Positive co-factor 4 (PC4) was initially shown to stimulate RNAP II transcription in vitro.105,106 Several years later it was shown that PC4 acts also as co-activator of RNAP III transcription.107 Comparable to PC4, also topoisomerase 1 could be co-immunoprecipitated with an affinity-purified holo-TFIIIC complex and was shown to act as a positive cofactor of RNAP III transcription. Both PC4 and Topo1 were shown to reinforce interactions of TFIIIC with downstream promoter sequences. In addition, it was demonstrated for PC4 that it stimulates multiple round, but not single round transcription of the VA1 gene in vitro.107 Analogous to PC4, it was reported that Sub1, the S. cerevisiae ortholog of PC4, associates with RNAP III transcribed genes and enhances their expression.108,109
Distinction of Components Required for Basal or Activated RNAP III Transcription
RNAP III genes with promoters 5′ of the transcription start site
Due to the close physical association and fixed distance of the PSE with the TATA-box and the transcription start site (Fig. 2) and probably also because of the functional similarity to TFIIIC in driving RNAP III transcription from promoters that are regulated by gene internal promoter sequences, PTF/SNAPc has often been described as general (basal) transcription factor.54,56,79,110-112 However, in vitro transcription experiments with 7SK- or U6-promoter deletion mutants showed that accurate, RNAP III-dependent transcription initiation, elongation and termination can be directed by sequences located about 40 nucleotides upstream of the transcription start site only. Thus, basal in vitro transcription of these genes does not require the PSE or DSE, but is exclusively dependent on the TATA-box and possibly surrounding nucleotides.38,113 Taking these results into account, the TATA-box should be regarded as the only explicitly described promoter element for U6 and 7SK genes (and possibly for all RNAP III-transcribed genes with promoter elements 5′ of the TSS). Accordingly, the PSE and its interacting PTF/SNAPc in turn should be regarded as a pair of enhancer sequence and interacting transcriptional activator complex. In support of a possible transcriptional activating, rather than basal transcription factor-like function of PTF/SNAPc is the finding that at least one subunit of the SNAPc/PTF acts as (co-)activator of mRNA transcription. It was shown that the SNAPC1 (PTFγ), but not the SNACP4 (PTFα) subunit of this complex co-localizes with actively transcribed protein-coding genes in a manner that depends on active RNA polymerase II transcription elongation. Depletion of SNAPC1/PTFγ resulted in reduced levels of activated transcription, but did not affect basal transcription of these genes. Many of the genes depending on SNAPC1/PTFγ for activated transcription were regulated by the epidermal growth factor (EGF) and retinoic acid (RA) signaling pathways.114 These results indicate that at least parts of the SNAPc/PTF complex may act as a transcriptional (co-)activator rather than a basal transcription factor.
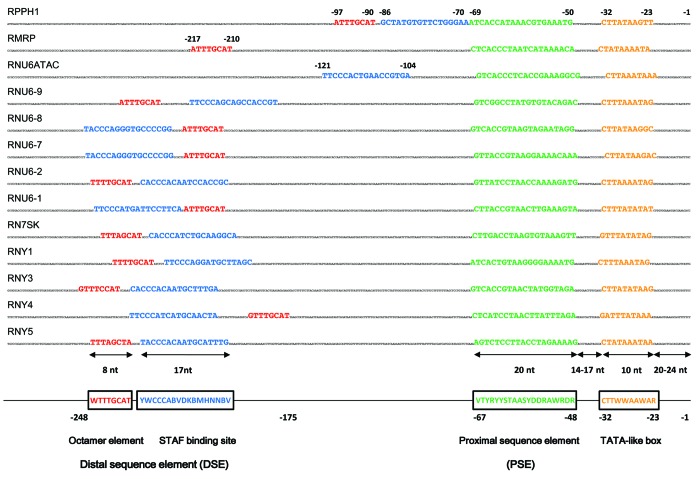
Figure 2. Schematic representation of regulatory elements of RNA polymerase III transcribed type 3 genes. The sequences and regulatory elements located 5′ of the transcription start site of the H1 (RPPH1), MRP (RMRP), U6ATAC (RNU6ATAC), U6–1 to U6–9 (RNU6–1, -2, -7, -8 and -9), 7SK (RN7SK), Y1 to Y5 (RNY-1, -3, -4 and -5) are depicted. Consensus sequences of the octamer element (red), STAF-binding site (blue), proximal sequence element (PSE; green) and TATA-like sequences (orange) are shown below. Nucleic acid nomenclature: B = T/C/G; D = A/T/G; H = A/C/T; V = C/A/G; K = G/T; M = A/C; S = G/C; W = A/T; Y = C/T; R = G/A; n = any of the four bases. Typical distances between the transcription start site (TSS) and regulatory boxes are indicated below the representation of the genes. The distances spanned by individual DNA elements, as well as those between the TSS and the TATA-like box and between the TATA-like box and the PSE are indicated by arrows and number of nucleotides (nt). The distal sequence element (DSE) is typically found 175 to 248 nt upstream of the TSS and composed of an octamer element and a STAF binding site. Octamer element and an inverted STAF-binding site are located closer to the transcription start site of the RPPH1 gene. The RMRP gene possesses an octamer element only at around -215 and the RNU6ATAC gene that comprises a potential STAF-binding site around -110. Sequences showing homology to inverted STAF-binding sites are located at -420 and -470, respectively, relative to the transcription start site of the RMRP gene (not shown). STAF (ZNF143) binding to the human U6 and H1 genes was demonstrated.94,95 The other STAF-binding sites have been deduced by sequence homology. PBP/PTF/SNAPc-binding to human U6 and 7SK genes has been demonstrated.26,54,55 Potential PSEs in the other gene-regulatory sequences have been deduced by sequence homology. OCT-1 binding and stimulation of in vitro transcription of human 7SK and U6 genes was demonstrated.54,135 Deletion of the octamer element caused moderate effects on in vitro transcription of the RPPH1 gene.136,137
RNAP III genes with promoters within the transcribed sequence
It is also an unresolved question which DNA elements represent the basal human RNAP III promoter elements of genes that do not require gene external promoter or enhancer elements. However, results of studies that have been conducted in the yeast S. cerevisiae shed some light on this question. In S. cerevisiae, the U6 gene comprises promoter elements that are located 5′ of the transcription start site (TATA-box), a weak A-box within the transcribed region as well as a B-box downstream of the transcription termination site (reviewed in115). Comparable to promoter recognition in tRNA genes, TFIIIC binds to the B-box downstream of the U6 gene and positions TFIIIB around the transcription start site, permitting the recruitment of RNAP III. Both S.cerevisiae U6 and tRNA genes utilize the A-box for determining the transcription start site in vivo. In the case of the U6 gene, a TATA-box contributes to transcription start site selection in vitro.116 The transcription of S. cerevisiae tRNA genes was also shown to be regulated by T/A enriched sequences upstream of the TSS.117 Thus, besides the fact that the B-box is either located within the transcribed region (tRNA genes) or downstream of the transcription termination site (U6 gene), the overall structure of gene regulatory elements is similar in both cases. Yeast TFIIIC is composed of two well characterized sub-modules, τA and τB, that respectively recognize the A- (τA) or B-box (τB).118 It was shown that the distance of A- and B-boxes can be increased from 74 to 365 nucleotides without affecting transcription efficiency. However, mutations in the A-box were not tolerated if the A- to B-box distance was increased, whereas the wild type promoter architecture was still active with the same A-box mutations.119 As a consequence, TFIIIC may be regarded as a complex that integrates functions of a general transcription factor (GTF; A-box binding; τA) and of a transcriptional activator (B-box binding; τB.)115 Importantly, it was also experimentally shown that the A-box of the SNR52 gene is sufficient for directing S. cerevisiae RNAP III transcription, supporting a functional specialization of submodules within TFIIIC into GTF (τA) and transcriptional activator (τB).120 In accordance with this model, the B-Box element within the transcribed regions of genes with type 2 promoter elements may be considered as an enhancer element. Since the subunit composition of yeast and human TFIIIC has been conserved during evolution,52 it can be assumed that this point of view may also hold true for human RNAP III transcription although this has not yet formally been shown.
RNAP III genes with promoter sequences within the gene and 5′ of the transcription start site
RNAP III transcribed genes containing not only promoter elements located 5′ of the TSS, but also regulatory elements within the transcribed region have been described. These genes have been identified in humans and other higher eukaryotes and their gene regulatory elements are also sometimes referred to as type 4 promoters (e.g., 7SL RNA [SRP], vault RNA [HVG], BC200 RNA [BCYRN1] and Epstein-Barr virus encoded EBER RNA genes; Figure 3).7,8,89,90,121 These genes possess not only canonical A- and B-boxes appropriately positioned within the transcribed region of the gene, but also promoter and enhancer elements, such as the TATA-box, PSE, DSE, ATF- or SP1-binding sites in several distinct individual combinations upstream of the TSS (Fig. 3). Promoter elements upstream and downstream of the transcription start site are of different importance for the transcription of these genes in vitro or in vivo. It was shown that efficient in vitro transcription of the mouse BC1 analog of human BC200 in HeLa nuclear extracts only requires gene internal promoter elements. In contrast, in vitro transcription with rat cortex nuclear extracts was additionally dependent on DNA elements upstream of the TSS.122 These results were confirmed by transfection of the prosimian G22 ortholog of human BC200 into HeLa cells123 or by analysis of 5′ regulatory element deletion mutants in transgenic mice.124 The first 22 nucleotides upstream of the TSS were indispensable for the expression of the G22 gene, demonstrating that they comprise promoter elements. The efficiency of BC1 transcription, however, was further increased if more than 22 nucleotides upstream of the transcription start site were included into the expression constructs, indicating that these sequences contained enhancer elements.123 In addition, it was demonstrated that sequences more than 250 nucleotides upstream of the TSS were important for brain-specific expression of the BC200 gene.124 In line with the importance of gene regulatory elements 5′ of the TSS for genes with type 4 promoters is the observation that the HVG4 (VTRNA3–1) pseudogene lacks such regulatory sequence elements and is not expressed in several cell lines although it does contain functional gene internal type 2 promoter elements.125
Mechanistic aspects of activated RNAP III transcription
Fundamentally, the mechanisms of transcription by RNAP III, and in particular the discrimination of basal and activated transcription, seem to be very similar to those that have been described for RNAP II transcription. However, an important difference between RNAP II and RNAP III transcription concerns the distance of enhancer elements to the transcription start site. This distance is very limited in RNAP III transcription and does not exceed several hundreds of base pairs (Figs. Two and 3).7 In contrast, in RNAP II transcription enhancer-promoter-distances of several tens of thousands of base pairs have been described and mechanisms including DNA looping have been proposed for bringing enhancer and promoter sequences in close vicinity. DNA looping may require the participation of protein-protein contacts as well as protein-DNA interactions of a multi-subunit transcriptional co-activator complex, the mediator.126 Thus, the difference in the localization of enhancer sequences relative to the promoter elements in transcription by RNA polymerases II or III may largely be explainable by the utilization of the mediator complex in RNAP II transcription and simple activator-basal transcription factor-interactions in the RNAP III transcription system. The lack of the participation of the mediator complex in RNAP III transcription may also explain the fact that no enhancer elements have been identified downstream of the transcribed region so far for human RNAP III genes that are regulated by promoters located upstream of the TSS.
Conclusions
In vitro transcription assays have been instrumental for the identification and characterization of the proteins that constitute the human RNA polymerase III transcription machinery. In addition, by employing promoter deletion constructs and partially purified reconstituted in vitro transcription systems, the minimal regulatory sequence requirements for human RNAP III genes with promoters 5′ of the transcription start site could be defined (sequences comprising about 40 nucleotides upstream of the TSS, including the TATA-box). In the case of gene internal promoters, the A-box is the only essential promoter element in S. cerevisiae and due to the high degree of conservation of the proteins required for tRNA transcription in yeast and humans, the situation is probably similar in humans. Therefore, TATA- and A-boxes may be regarded as the only clearly defined RNAP III promoter elements, whereas other regulatory DNA sequences associated with RNAP III transcription (including DSE, STAF-binding site, Octamer element, SP1-binding site, PSE and B-box) may rather be considered as RNAP III transcriptional enhancer elements. Accordingly, the function of PTF/SNAPc (or possibly some of the subunits of this protein complex) resembles more that of a transcriptional activator. Likewise, individual proteins (modules) within TFIIIC may primarily be regarded as general transcription factor (τA) or as activator/co-activator (τB), respectively. In that sense, TFIIIC may be comparable to the RNAP II transcription factor TFIID, which comprises not only the general transcription factor TBP, but also transcriptional co-activators, the TBP-associated factors (TAFs).127,128 In addition, in vitro transcription has helped to shed light on the regulation of RNAP III transcription by activators, co-activators and repressor proteins. In summary, in vitro transcription has not only been decisive for the identification of the proteins required to reconstitute basal and activated human RNA polymerase III transcription, but also crucial for defining minimal RNAP III promoters in eukaryotes. Collectively, past and very likely also future results obtained by in vitro transcription have provided and will continue to provide important information for understanding the mechanisms underlying human RNAP III transcription. Therefore, in vitro transcription assays will remain irreplaceable to complement our understanding of the regulation of RNAP III transcription that will be determined in more physiological systems in the future.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Dean N, Berk AJ. Separation of TFIIIC into two functional components by sequence specific DNA affinity chromatography. Nucleic Acids Res. 1987;15:9895–907. doi: 10.1093/nar/15.23.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carey MF, Peterson CL, Smale ST. DNase I footprinting. Cold Spring Harb Protoc. 2013;2013:469–78. doi: 10.1101/pdb.prot074328. [DOI] [PubMed] [Google Scholar]
- 3.Carey MF, Peterson CL, Smale ST. Electrophoretic mobility-shift assays. Cold Spring Harb Protoc. 2013;2013:636–9. doi: 10.1101/pdb.prot075861. [DOI] [PubMed] [Google Scholar]
- 4.Wu GJ. Adenovirus DNA-directed transcription of 5.5S RNA in vitro. Proc Natl Acad Sci U S A. 1978;75:2175–9. doi: 10.1073/pnas.75.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weil PA, Segall J, Harris B, Ng SY, Roeder RG. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979;254:6163–73. [PubMed] [Google Scholar]
- 6.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–22. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: themes and variations. Gene. 2012;493:185–94. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Segall J, Matsui T, Roeder RG. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980;255:11986–91. [PubMed] [Google Scholar]
- 10.Lassar AB, Martin PL, Roeder RG. Transcription of class III genes: formation of preinitiation complexes. Science. 1983;222:740–8. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- 11.Bieker JJ, Martin PL, Roeder RG. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985;40:119–27. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- 12.Moorefield B, Roeder RG. Purification and characterization of human transcription factor IIIA. J Biol Chem. 1994;269:20857–65. [PubMed] [Google Scholar]
- 13.Arakawa H, Nagase H, Hayashi N, Ogawa M, Nagata M, Fujiwara T, Takahashi E, Shin S, Nakamura Y. Molecular cloning, characterization, and chromosomal mapping of a novel human gene (GTF3A) that is highly homologous to Xenopus transcription factor IIIA. Cytogenet Cell Genet. 1995;70:235–8. doi: 10.1159/000134041. [DOI] [PubMed] [Google Scholar]
- 14.Drew PD, Nagle JW, Canning RD, Ozato K, Biddison WE, Becker KG. Cloning and expression analysis of a human cDNA homologous to Xenopus TFIIIA. Gene. 1995;159:215–8. doi: 10.1016/0378-1119(95)00145-V. [DOI] [PubMed] [Google Scholar]
- 15.Engelke DR, Ng SY, Shastry BS, Roeder RG. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980;19:717–28. doi: 10.1016/S0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- 16.Ginsberg AM, King BO, Roeder RG. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984;39:479–89. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- 17.Waldschmidt R, Jahn D, Seifart KH. Purification of transcription factor IIIB from HeLa cells. J Biol Chem. 1988;263:13350–6. [PubMed] [Google Scholar]
- 18.Hoffman A, Sinn E, Yamamoto T, Wang J, Roy A, Horikoshi M, Roeder RG. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID) Nature. 1990;346:387–90. doi: 10.1038/346387a0. [DOI] [PubMed] [Google Scholar]
- 19.Peterson MG, Tanese N, Pugh BF, Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990;248:1625–30. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- 20.Margottin F, Dujardin G, Gérard M, Egly JM, Huet J, Sentenac A. Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science. 1991;251:424–6. doi: 10.1126/science.1989075. [DOI] [PubMed] [Google Scholar]
- 21.Lobo SM, Lister J, Sullivan ML, Hernandez N. The cloned RNA polymerase II transcription factor IID selects RNA polymerase III to transcribe the human U6 gene in vitro. Genes Dev. 1991;5:1477–89. doi: 10.1101/gad.5.8.1477. [DOI] [PubMed] [Google Scholar]
- 22.Simmen KA, Bernués J, Parry HD, Stunnenberg HG, Berkenstam A, Cavallini B, Egly JM, Mattaj IW. TFIID is required for in vitro transcription of the human U6 gene by RNA polymerase III. EMBO J. 1991;10:1853–62. doi: 10.1002/j.1460-2075.1991.tb07711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldschmidt R, Wanandi I, Seifart KH. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 1991;10:2595–603. doi: 10.1002/j.1460-2075.1991.tb07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White RJ, Jackson SP, Rigby PW. A role for the TATA-box-binding protein component of the transcription factor IID complex as a general RNA polymerase III transcription factor. Proc Natl Acad Sci U S A. 1992;89:1949–53. doi: 10.1073/pnas.89.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobo SM, Tanaka M, Sullivan ML, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–40. doi: 10.1016/0092-8674(92)90397-U. [DOI] [PubMed] [Google Scholar]
- 26.Simmen KA, Waldschmidt R, Bernués J, Parry HD, Seifart KH, Mattaj IW. Proximal sequence element factor binding and species specificity in vertebrate U6 snRNA promoters. J Mol Biol. 1992;223:873–84. doi: 10.1016/0022-2836(92)90249-J. [DOI] [PubMed] [Google Scholar]
- 27.Taggart AK, Fisher TS, Pugh BF. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–28. doi: 10.1016/0092-8674(92)90396-T. [DOI] [PubMed] [Google Scholar]
- 28.White RJ, Jackson SP. Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell. 1992;71:1041–53. doi: 10.1016/0092-8674(92)90398-V. [DOI] [PubMed] [Google Scholar]
- 29.Chiang CM, Ge H, Wang Z, Hoffmann A, Roeder RG. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–62. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Roeder RG. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci U S A. 1995;92:7026–30. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–42. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–9. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 33.López-De-León A, Librizzi M, Puglia K, Willis IM. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–20. doi: 10.1016/0092-8674(92)90350-L. [DOI] [PubMed] [Google Scholar]
- 34.Kassavetis GA, Joazeiro CA, Pisano M, Geiduschek EP, Colbert T, Hahn S, Blanco JA. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–64. doi: 10.1016/0092-8674(92)90399-W. [DOI] [PubMed] [Google Scholar]
- 35.Ha I, Lane WS, Reinberg D. Cloning of a human gene encoding the general transcription initiation factor IIB. Nature. 1991;352:689–95. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- 36.Malik S, Hisatake K, Sumimoto H, Horikoshi M, Roeder RG. Sequence of general transcription factor TFIIB and relationships to other initiation factors. Proc Natl Acad Sci U S A. 1991;88:9553–7. doi: 10.1073/pnas.88.21.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teichmann M, Seifart KH. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J. 1995;14:5974–83. doi: 10.1002/j.1460-2075.1995.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teichmann M, Dieci G, Huet J, Rüth J, Sentenac A, Seifart KH. Functional interchangeability of TFIIIB components from yeast and human cells in vitro. EMBO J. 1997;16:4708–16. doi: 10.1093/emboj/16.15.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelter AR, Herchenbach J, Wirth B. The transcription factor-like nuclear regulator (TFNR) contains a novel 55-amino-acid motif repeated nine times and maps closely to SMN1. Genomics. 2000;70:315–26. doi: 10.1006/geno.2000.6396. [DOI] [PubMed] [Google Scholar]
- 40.Schramm L, Pendergrast PS, Sun Y, Hernandez N. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 2000;14:2650–63. doi: 10.1101/gad.836400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teichmann M, Wang Z, Roeder RG. A stable complex of a novel transcription factor IIB- related factor, human TFIIIB50, and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc Natl Acad Sci U S A. 2000;97:14200–5. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshinaga SK, Boulanger PA, Berk AJ. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci U S A. 1987;84:3585–9. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cromlish JA, Roeder RG. Human transcription factor IIIC (TFIIIC). Purification, polypeptide structure, and the involvement of thiol groups in specific DNA binding. J Biol Chem. 1989;264:18100–9. [PubMed] [Google Scholar]
- 44.Schneider HR, Waldschmidt R, Jahn D, Seifart KH. Purification of human transcription factor IIIC and its binding to the gene for ribosomal 5S RNA. Nucleic Acids Res. 1989;17:5003–16. doi: 10.1093/nar/17.13.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshinaga SK, L’Etoile ND, Berk AJ. Purification and characterization of transcription factor IIIC2. J Biol Chem. 1989;264:10726–31. [PubMed] [Google Scholar]
- 46.Kovelman R, Roeder RG. Purification and characterization of two forms of human transcription factor IIIC. J Biol Chem. 1992;267:24446–56. [PubMed] [Google Scholar]
- 47.L’Etoile ND, Fahnestock ML, Shen Y, Aebersold R, Berk AJ. Human transcription factor IIIC box B binding subunit. Proc Natl Acad Sci U S A. 1994;91:1652–6. doi: 10.1073/pnas.91.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagna G, Kovelman R, Sukegawa J, Roeder RG. Cloning and characterization of an evolutionarily divergent DNA-binding subunit of mammalian TFIIIC. Mol Cell Biol. 1994;14:3053–64. doi: 10.1128/mcb.14.5.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinn E, Wang Z, Kovelman R, Roeder RG. Cloning and characterization of a TFIIIC2 subunit (TFIIIC beta) whose presence correlates with activation of RNA polymerase III-mediated transcription by adenovirus E1A expression and serum factors. Genes Dev. 1995;9:675–85. doi: 10.1101/gad.9.6.675. [DOI] [PubMed] [Google Scholar]
- 50.Hsieh YJ, Wang Z, Kovelman R, Roeder RG. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol Cell Biol. 1999;19:4944–52. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh YJ, Kundu TK, Wang Z, Kovelman R, Roeder RG. The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol Cell Biol. 1999;19:7697–704. doi: 10.1128/mcb.19.11.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dumay-Odelot H, Marck C, Durrieu-Gaillard S, Lefebvre O, Jourdain S, Prochazkova M, Pflieger A, Teichmann M. Identification, molecular cloning, and characterization of the sixth subunit of human transcription factor TFIIIC. J Biol Chem. 2007;282:17179–89. doi: 10.1074/jbc.M611542200. [DOI] [PubMed] [Google Scholar]
- 53.Weser S, Gruber C, Hafner HM, Teichmann M, Roeder RG, Seifart KH, Meissner W. Transcription factor (TF)-like nuclear regulator, the 250-kDa form of Homo sapiens TFIIIB”, is an essential component of human TFIIIC1 activity. J Biol Chem. 2004;279:27022–9. doi: 10.1074/jbc.M312790200. [DOI] [PubMed] [Google Scholar]
- 54.Murphy S, Yoon JB, Gerster T, Roeder RG. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–61. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadowski CL, Henry RW, Lobo SM, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–48. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 56.Henry RW, Sadowski CL, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerase II and III. Nature. 1995;374:653–6. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 57.Sadowski CL, Henry RW, Kobayashi R, Hernandez N. The SNAP45 subunit of the small nuclear RNA (snRNA) activating protein complex is required for RNA polymerase II and III snRNA gene transcription and interacts with the TATA box binding protein. Proc Natl Acad Sci U S A. 1996;93:4289–93. doi: 10.1073/pnas.93.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoon JB, Roeder RG. Cloning of two proximal sequence element-binding transcription factor subunits (gamma and delta) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol Cell Biol. 1996;16:1–9. doi: 10.1128/mcb.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bai L, Wang Z, Yoon JB, Roeder RG. Cloning and characterization of the beta subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerases II and III. Mol Cell Biol. 1996;16:5419–26. doi: 10.1128/mcb.16.10.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henry RW, Ma B, Sadowski CL, Kobayashi R, Hernandez N. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 1996;15:7129–36. [PMC free article] [PubMed] [Google Scholar]
- 61.Wong MW, Henry RW, Ma B, Kobayashi R, Klages N, Matthias P, Strubin M, Hernandez N. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol. 1998;18:368–77. doi: 10.1128/mcb.18.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henry RW, Mittal V, Ma B, Kobayashi R, Hernandez N. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 1998;12:2664–72. doi: 10.1101/gad.12.17.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ittmann M, Ali J, Greco A, Basilico C. The gene complementing a temperature-sensitive cell cycle mutant of BHK cells is the human homologue of the yeast RPC53 gene, which encodes a subunit of RNA polymerase C (III) Cell Growth Differ. 1993;4:503–11. [PubMed] [Google Scholar]
- 64.Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–26. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 65.Hu P, Wu S, Sun Y, Yuan C-C, Kobayashi R, Myers MP, Hernandez N. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol Cell Biol. 2002;22:8044–55. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dumay-Odelot H, Durrieu-Gaillard S, Da Silva D, Roeder RG, Teichmann M. Cell growth- and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle. 2010;9:3687–99. doi: 10.4161/cc.9.18.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haurie V, Durrieu-Gaillard S, Dumay-Odelot H, Da Silva D, Rey C, Prochazkova M, Roeder RG, Besser D, Teichmann M. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc Natl Acad Sci U S A. 2010;107:4176–81. doi: 10.1073/pnas.0914980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong RC-B, Pollan S, Fong H, Ibrahim A, Smith EL, Ho M, Laslett AL, Donovan PJ. A novel role for an RNA polymerase III subunit POLR3G in regulating pluripotency in human embryonic stem cells. Stem Cells. 2011;29:1517–27. doi: 10.1002/stem.714. [DOI] [PubMed] [Google Scholar]
- 69.Gjidoda A, Henry RW. RNA polymerase III repression by the retinoblastoma tumor suppressor protein. Biochim Biophys Acta. 2013;1829:385–92. doi: 10.1016/j.bbagrm.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–9. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 71.Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, Hopper AK, Sentenac A, Boguta M. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5031–40. doi: 10.1128/MCB.21.15.5031-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reina JH, Azzouz TN, Hernandez N. Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS One. 2006;1:e134. doi: 10.1371/journal.pone.0000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell. 2007;26:367–79. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 74.Rollins J, Veras I, Cabarcas S, Willis I, Schramm L. Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int J Biol Sci. 2007;3:292–302. doi: 10.7150/ijbs.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goodfellow SJ, Graham EL, Kantidakis T, Marshall L, Coppins BA, Oficjalska-Pham D, Gérard M, Lefebvre O, White RJ. Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J Mol Biol. 2008;378:481–91. doi: 10.1016/j.jmb.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 76.Michels AA. MAF1: a new target of mTORC1. Biochem Soc Trans. 2011;39:487–91. doi: 10.1042/BST0390487. [DOI] [PubMed] [Google Scholar]
- 77.Kropotov A, Sedova V, Ivanov V, Sazeeva N, Tomilin A, Krutilina R, Oei SL, Griesenbeck J, Buchlow G, Tomilin N. A novel human DNA-binding protein with sequence similarity to a subfamily of redox proteins which is able to repress RNA-polymerase-III-driven transcription of the Alu-family retroposons in vitro. Eur J Biochem. 1999;260:336–46. doi: 10.1046/j.1432-1327.1999.00162.x. [DOI] [PubMed] [Google Scholar]
- 78.Humphrey GW, Englander EW, Howard BH. Specific binding sites for a pol III transcriptional repressor and pol II transcription factor YY1 within the internucleosomal spacer region in primate Alu repetitive elements. Gene Expr. 1996;6:151–68. [PMC free article] [PubMed] [Google Scholar]
- 79.Emran F, Florens L, Ma B, Swanson SK, Washburn MP, Hernandez N. A role for Yin Yang-1 (YY1) in the assembly of snRNA transcription complexes. Gene. 2006;377:96–108. doi: 10.1016/j.gene.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 80.Kurose K, Hata K, Hattori M, Sakaki Y. RNA polymerase III dependence of the human L1 promoter and possible participation of the RNA polymerase II factor YY1 in the RNA polymerase III transcription system. Nucleic Acids Res. 1995;23:3704–9. doi: 10.1093/nar/23.18.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.White RJ, Khoo BC, Inostroza JA, Reinberg D, Jackson SP. Differential regulation of RNA polymerases I, II, and III by the TBP-binding repressor Dr1. Science. 1994;266:448–50. doi: 10.1126/science.7939686. [DOI] [PubMed] [Google Scholar]
- 82.Kantidakis T, White RJ. Dr1 (NC2) is present at tRNA genes and represses their transcription in human cells. Nucleic Acids Res. 2010;38:1228–39. doi: 10.1093/nar/gkp1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moosmann P, Georgiev O, Thiesen HJ, Hagmann M, Schaffner W. Silencing of RNA polymerases II and III-dependent transcription by the KRAB protein domain of KOX1, a Krüppel-type zinc finger factor. Biol Chem. 1997;378:669–77. doi: 10.1515/bchm.1997.378.7.669. [DOI] [PubMed] [Google Scholar]
- 84.Selvakumar T, Gjidoda A, Hovde SL, Henry RW. Regulation of human RNA polymerase III transcription by DNMT1 and DNMT3a DNA methyltransferases. J Biol Chem. 2012;287:7039–50. doi: 10.1074/jbc.M111.285601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bark C, Weller P, Zabielski J, Janson L, Pettersson U. A distant enhancer element is required for polymerase III transcription of a U6 RNA gene. Nature. 1987;328:356–9. doi: 10.1038/328356a0. [DOI] [PubMed] [Google Scholar]
- 86.Carbon P, Murgo S, Ebel JP, Krol A, Tebb G, Mattaj LW. A common octamer motif binding protein is involved in the transcription of U6 snRNA by RNA polymerase III and U2 snRNA by RNA polymerase II. Cell. 1987;51:71–9. doi: 10.1016/0092-8674(87)90011-0. [DOI] [PubMed] [Google Scholar]
- 87.Kunkel GR, Pederson T. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev. 1988;2:196–204. doi: 10.1101/gad.2.2.196. [DOI] [PubMed] [Google Scholar]
- 88.Murphy S, Pierani A, Scheidereit C, Melli M, Roeder RG. Purified octamer binding transcription factors stimulate RNA polymerase III--mediated transcription of the 7SK RNA gene. Cell. 1989;59:1071–80. doi: 10.1016/0092-8674(89)90763-0. [DOI] [PubMed] [Google Scholar]
- 89.Howe JG, Shu MD. Epstein-Barr virus small RNA (EBER) genes: unique transcription units that combine RNA polymerase II and III promoter elements. Cell. 1989;57:825–34. doi: 10.1016/0092-8674(89)90797-6. [DOI] [PubMed] [Google Scholar]
- 90.Bredow S, Sürig D, Müller J, Kleinert H, Benecke BJ. Activating-transcription-factor (ATF) regulates human 7S L RNA transcription by RNA polymerase III in vivo and in vitro. Nucleic Acids Res. 1990;18:6779–84. doi: 10.1093/nar/18.23.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–4. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 92.Myslinski E, Schuster C, Huet J, Sentenac A, Krol A, Carbon P. Point mutations 5′ to the tRNA selenocysteine TATA box alter RNA polymerase III transcription by affecting the binding of TBP. Nucleic Acids Res. 1993;21:5852–8. doi: 10.1093/nar/21.25.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schuster C, Myslinski E, Krol A, Carbon P. Staf, a novel zinc finger protein that activates the RNA polymerase III promoter of the selenocysteine tRNA gene. EMBO J. 1995;14:3777–87. doi: 10.1002/j.1460-2075.1995.tb00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Myslinski E, Krol A, Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J Biol Chem. 1998;273:21998–2006. doi: 10.1074/jbc.273.34.21998. [DOI] [PubMed] [Google Scholar]
- 95.Rincon JC, Engler SK, Hargrove BW, Kunkel GR. Molecular cloning of a cDNA encoding human SPH-binding factor, a conserved protein that binds to the enhancer-like region of the U6 small nuclear RNA gene promoter. Nucleic Acids Res. 1998;26:4846–52. doi: 10.1093/nar/26.21.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stünkel W, Kober I, Kauer M, Taimor G, Seifart KH. Human TFIIIA alone is sufficient to prevent nucleosomal repression of a homologous 5S gene. Nucleic Acids Res. 1995;23:109–16. doi: 10.1093/nar/23.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kundu TK, Wang Z, Roeder RG. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–15. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mertens C, Roeder RG. Different functional modes of p300 in activation of RNA polymerase III transcription from chromatin templates. Mol Cell Biol. 2008;28:5764–76. doi: 10.1128/MCB.01262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–57. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 100.Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci U S A. 2007;104:14917–22. doi: 10.1073/pnas.0702909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stünkel W, Kober I, Seifart KH. A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol Cell Biol. 1997;17:4397–405. doi: 10.1128/mcb.17.8.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao X, Pendergrast PS, Hernandez N. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol Cell. 2001;7:539–49. doi: 10.1016/S1097-2765(01)00201-5. [DOI] [PubMed] [Google Scholar]
- 103.Boyd DC, Greger IH, Murphy S. In vivo footprinting studies suggest a role for chromatin in transcription of the human 7SK gene. Gene. 2000;247:33–44. doi: 10.1016/S0378-1119(00)00134-7. [DOI] [PubMed] [Google Scholar]
- 104.Yuan C-C, Zhao X, Florens L, Swanson SK, Washburn MP, Hernandez N. CHD8 associates with human Staf and contributes to efficient U6 RNA polymerase III transcription. Mol Cell Biol. 2007;27:8729–38. doi: 10.1128/MCB.00846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ge H, Roeder RG. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–23. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 106.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–34. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 107.Wang Z, Roeder RG. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–57. doi: 10.1016/S1097-2765(00)80074-X. [DOI] [PubMed] [Google Scholar]
- 108.Rosonina E, Willis IM, Manley JL. Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III. Mol Cell Biol. 2009;29:2308–21. doi: 10.1128/MCB.01841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tavenet A, Suleau A, Dubreuil G, Ferrari R, Ducrot C, Michaut M, Aude J-C, Dieci G, Lefebvre O, Conesa C, et al. Genome-wide location analysis reveals a role for Sub1 in RNA polymerase III transcription. Proc Natl Acad Sci U S A. 2009;106:14265–70. doi: 10.1073/pnas.0900162106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yoon JB, Murphy S, Bai L, Wang Z, Roeder RG. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol. 1995;15:2019–27. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jawdekar GW, Hanzlowsky A, Hovde SL, Jelencic B, Feig M, Geiger JH, Henry RW. The unorthodox SNAP50 zinc finger domain contributes to cooperative promoter recognition by human SNAPC. J Biol Chem. 2006;281:31050–60. doi: 10.1074/jbc.M603810200. [DOI] [PubMed] [Google Scholar]
- 112.Gu L, Husain-Ponnampalam R, Hoffmann-Benning S, Henry RW. The protein kinase CK2 phosphorylates SNAP190 to negatively regulate SNAPC DNA binding and human U6 transcription by RNA polymerase III. J Biol Chem. 2007;282:27887–96. doi: 10.1074/jbc.M702269200. [DOI] [PubMed] [Google Scholar]
- 113.Murphy S, Di Liegro C, Melli M. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell. 1987;51:81–7. doi: 10.1016/0092-8674(87)90012-2. [DOI] [PubMed] [Google Scholar]
- 114.Baillat D, Gardini A, Cesaroni M, Shiekhattar R. Requirement for SNAPC1 in transcriptional responsiveness to diverse extracellular signals. Mol Cell Biol. 2012;32:4642–50. doi: 10.1128/MCB.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gabrielsen OS, Sentenac A. RNA polymerase III (C) and its transcription factors. Trends Biochem Sci. 1991;16:412–6. doi: 10.1016/0968-0004(91)90166-S. [DOI] [PubMed] [Google Scholar]
- 116.Burnol AF, Margottin F, Schultz P, Marsolier MC, Oudet P, Sentenac A. Basal promoter and enhancer element of yeast U6 snRNA gene. J Mol Biol. 1993;233:644–58. doi: 10.1006/jmbi.1993.1542. [DOI] [PubMed] [Google Scholar]
- 117.Giuliodori S, Percudani R, Braglia P, Ferrari R, Guffanti E, Ottonello S, Dieci G. A composite upstream sequence motif potentiates tRNA gene transcription in yeast. J Mol Biol. 2003;333:1–20. doi: 10.1016/j.jmb.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 118.Ducrot C, Lefebvre O, Landrieux E, Guirouilh-Barbat J, Sentenac A, Acker J. Reconstitution of the yeast RNA polymerase III transcription system with all recombinant factors. J Biol Chem. 2006;281:11685–92. doi: 10.1074/jbc.M600101200. [DOI] [PubMed] [Google Scholar]
- 119.Fabrizio P, Coppo A, Fruscoloni P, Benedetti P, Di Segni G, Tocchini-Valentini GP. Comparative mutational analysis of wild-type and stretched tRNA3(Leu) gene promoters. Proc Natl Acad Sci U S A. 1987;84:8763–7. doi: 10.1073/pnas.84.24.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guffanti E, Ferrari R, Preti M, Forloni M, Harismendy O, Lefebvre O, Dieci G. A minimal promoter for TFIIIC-dependent in vitro transcription of snoRNA and tRNA genes by RNA polymerase III. J Biol Chem. 2006;281:23945–57. doi: 10.1074/jbc.M513814200. [DOI] [PubMed] [Google Scholar]
- 121.Englert M, Felis M, Junker V, Beier H. Novel upstream and intragenic control elements for the RNA polymerase III-dependent transcription of human 7SL RNA genes. Biochimie. 2004;86:867–74. doi: 10.1016/j.biochi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 122.Martignetti JA, Brosius J. BC1 RNA: transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol Cell Biol. 1995;15:1642–50. doi: 10.1128/mcb.15.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ludwig A, Rozhdestvensky TS, Kuryshev VY, Schmitz J, Brosius J. An unusual primate locus that attracted two independent Alu insertions and facilitates their transcription. J Mol Biol. 2005;350:200–14. doi: 10.1016/j.jmb.2005.03.058. [DOI] [PubMed] [Google Scholar]
- 124.Khanam T, Rozhdestvensky TS, Bundman M, Galiveti CR, Handel S, Sukonina V, Jordan U, Brosius J, Skryabin BV. Two primate-specific small non-protein-coding RNAs in transgenic mice: neuronal expression, subcellular localization and binding partners. Nucleic Acids Res. 2007;35:529–39. doi: 10.1093/nar/gkl1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Zon A, Mossink MH, Schoester M, Scheffer GL, Scheper RJ, Sonneveld P, Wiemer EA. Multiple human vault RNAs. Expression and association with the vault complex. J Biol Chem. 2001;276:37715–21. doi: 10.1074/jbc.M106055200. [DOI] [PubMed] [Google Scholar]
- 126.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–72. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dynlacht BD, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–76. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 128.Zhou Q, Lieberman PM, Boyer TG, Berk AJ. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–74. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- 129.Majowski K, Mentzel H, Pieler T. A split binding site for TFIIIC on the Xenopus 5S gene. EMBO J. 1987;6:3057–63. doi: 10.1002/j.1460-2075.1987.tb02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Veldhoen N, You Q, Setzer DR, Romaniuk PJ. Contribution of individual base pairs to the interaction of TFIIIA with the Xenopus 5S RNA gene. Biochemistry. 1994;33:7568–75. doi: 10.1021/bi00190a009. [DOI] [PubMed] [Google Scholar]
- 131.Nielsen JN, Hallenberg C, Frederiksen S, Sørensen PD, Lomholt B. Transcription of human 5S rRNA genes is influenced by an upstream DNA sequence. Nucleic Acids Res. 1993;21:3631–6. doi: 10.1093/nar/21.16.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–232. doi: 10.1017/S1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–62. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–40. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Danzeiser DA, Urso O, Kunkel GR. Functional characterization of elements in a human U6 small nuclear RNA gene distal control region. Mol Cell Biol. 1993;13:4670–8. doi: 10.1128/mcb.13.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hannon GJ, Chubb A, Maroney PA, Hannon G, Altman S, Nilsen TW. Multiple cis-acting elements are required for RNA polymerase III transcription of the gene encoding H1 RNA, the RNA component of human RNase P. J Biol Chem. 1991;266:22796–9. [PubMed] [Google Scholar]
- 137.Myslinski E, Amé JC, Krol A, Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–9. doi: 10.1093/nar/29.12.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Howe JG, Shu MD. Upstream basal promoter element important for exclusive RNA polymerase III transcription of the EBER 2 gene. Mol Cell Biol. 1993;13:2655–65. doi: 10.1128/mcb.13.5.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kickhoefer VA, Emre N, Stephen AG, Poderycki MJ, Rome LH. Identification of conserved vault RNA expression elements and a non-expressed mouse vault RNA gene. Gene. 2003;309:65–70. doi: 10.1016/S0378-1119(03)00507-9. [DOI] [PubMed] [Google Scholar]
- 140.Stadler PF, Chen JJ-L, Hackermüller J, Hoffmann S, Horn F, Khaitovich P, Kretzschmar AK, Mosig A, Prohaska SJ, Qi X, et al. Evolution of vault RNAs. Mol Biol Evol. 2009;26:1975–91. doi: 10.1093/molbev/msp112. [DOI] [PubMed] [Google Scholar]