Abstract
Chemically synthesized DNA can carry small RNA sequence information but converting that information into small RNA is generally thought to require large double-stranded promoters in the context of plasmids, viruses and genes. We previously found evidence that circularized oligodeoxynucleotides (coligos) containing certain sequences and secondary structures can template the synthesis of small RNA by RNA polymerase III in vitro and in human cells. By using immunoprecipitated RNA polymerase III we now report corroborating evidence that this enzyme is the sole polymerase responsible for coligo transcription. The immobilized polymerase enabled experiments showing that coligo transcripts can be formed through transcription termination without subsequent 3′ end trimming. To better define the determinants of productive transcription, a structure-activity relationship study was performed using over 20 new coligos. The results show that unpaired nucleotides in the coligo stem facilitate circumtranscription, but also that internal loops and bulges should be kept small to avoid secondary transcription initiation sites. A polymerase termination sequence embedded in the double-stranded region of a hairpin-encoding coligo stem can antagonize transcription. Using lessons learned from new and old coligos, we demonstrate how to convert poorly transcribed coligos into productive templates. Our findings support the possibility that coligos may prove useful as chemically synthesized vectors for the ectopic expression of small RNA in human cells.
Keywords: In vitro transcription, RNA polymerase III, coligo, small RNA, RNA expression vector, miRNA, siRNA, RNA hairpin
Introduction
Small RNA sequences can exhibit a wide range of interesting cellular functions, including those found to occur naturally in cells and those engineered for therapeutic or experimental intervention.1-4 The problem of delivering intact RNA to cells, however, continues to delay realization of the potential of small RNA therapeutics.5,6 The instability of unmodified small RNA and the additional cost and difficulty of chemical RNA synthesis, compared with DNA synthesis,7 add to the difficulties of using small RNA in therapeutic applications. In comparison, DNA oligonucleotides are simpler to make, more stable in biological fluids than RNA, and can carry the same sequence information. Converting DNA sequence information into RNA however requires comparatively large transcriptional promoters integrated into viral or plasmid vectors, an impediment that prevents the use of the more easily synthesized and more stable synthetic DNA.
We are developing promoterless transcription vectors made from the template strand of the cDNA encoding a desired small, usually hairpin, RNA. We have found that, in circularized form, and with certain sequence and secondary structure features, RNA polymerase III (RNAP III) can use such templates to make discrete transcripts in human cells and cell extracts.8 We refer to these chemically synthesized vectors as coligos, for circular oligonucleotides. Circularization imparts resistance to cellular8 and serum9 exonucleases, and appears to be structurally required for productive transcription, perhaps through stabilizing a template secondary structure that mimics a natural intermediate on the pathway of promoter-based transcription.8 To establish the extent of their usefulness, it will be necessary to understand the sequence and structural features that determine transcription by RNAP III, including initiation, termination and possible processing by cellular ribonucleases. Since in vitro coligo transcription appears to faithfully represent the intracellular transcription of coligos in transfected cells,8 we herein make use of two in vitro transcription systems—one using whole cell extract (WCE) and another using RNAP III purified by immunoprecipitation (IP) via a stably transfected FLAG-tagged RNAP III subunit—to explore the initiation, elongation, termination and processing of coligo transcripts made by RNAP III.
Results
Coligo transcription by immunoprecipitated RNA polymerase III
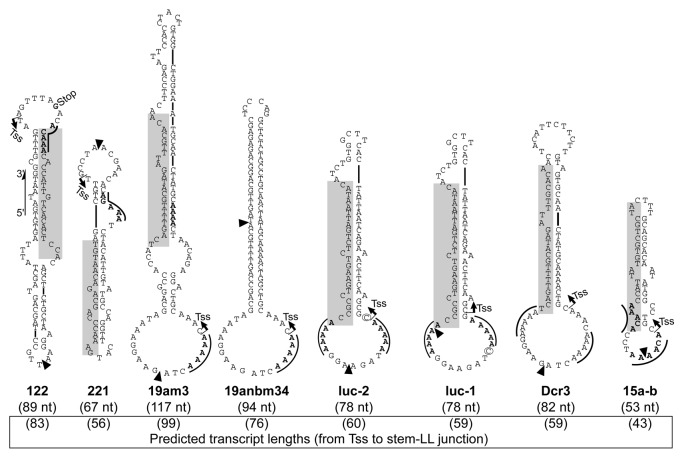
Using the transcription inhibitor α-amanitin, against which the three mammalian RNA polymerases are differently susceptible, we previously identified RNAP III as the main and likely sole RNA polymerase responsible for coligo transcription in mammalian cells and cell extracts.8 Since RNAP III generally appears to circumtranscribe once around a coligo before terminating, rather than multiple times as do bacterial and bacteriophage RNA polymerases,10-12 we have found coligo-derived transcripts laborious to sequence due to interference by abundant endogenous small RNA.8 To facilitate coligo transcript sequencing and to corroborate by a different method that RNAP III is the enzyme responsible for transcribing coligos, we used a HEK293-based cell line stably expressing a FLAG-tagged RNAP III F-subunit (HEK293/POLR3F).13 To evaluate the coligo-transcribing ability of RNAP III immunoprecipitated (IP’d) via this subunit, we used a variety of coligos encoding pre-miRNA-like hairpin RNAs, and which have the general coligo features previously found8 to favor promoterless transcription (Fig. 1, structures predicted using the mfold program14). Specifically, the transcription start site (TSS) is predicted at a pyrimidine, usually flanked by purines, near the 3′ end of the stem-larger loop junction. Transcription termination is predicted to occur at or close to the 5′ end of the stem-larger loop junction. Where present, RNAP III termination signals15,16 are indicated in the figures by a line over the pentamer sequence. Total coligo size and predicted transcript lengths are also given.
Figure 1. Sequence and predicted secondary structure of the coligos used in Figure 2. Coligo 122 transcription start site (TSS, arrow) and transcript 3′ end site (Stop) were identified previously by cDNA sequencing.8 TSS of coligo 19am3 and 19anbm34 were predicted based on their close similarity to coligo 19aTAR transcript sequencing data.8 The TSS for coligos 221, luc-2, luc-1, Dcr3, and 15a-b are predicted to be at a pyrimidine close to the 3′ end of the DNA stem, near its junction with the larger loop. Transcript 3′ end sites, resulting from termination or termination with nuclease trimming, are predicted to be at the 5′ end of the DNA stem, based on previous observations.8 Coligo luc-2 and luc-1 differ only by one nucleotide (circled) in the larger terminal loop region. Shaded regions indicate cDNA of mature miRNA in those coligos designed from human genomic miRNA loci. Black triangles denote the 3′ and 5′ ends of the linear precursor deoxyoligonucleotides, where the sequences were ligated to form a circle. Coligo sizes are shown in nucleotides (nt); solid lines above sequences indicate known RNA RNAP III termination sequences.
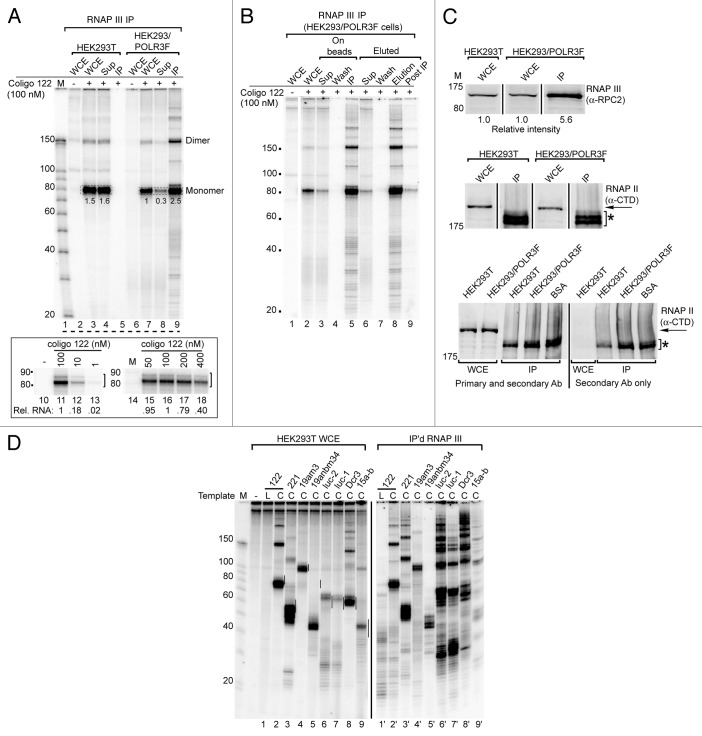
Coligo 122 (Fig. 1), based on pre-miRNA 122, was previously found to be transcribed into an 83 nucleotide (nt) transcript encompassing the pre-miRNA.8 Immunoprecipitate from HEK293/POLR3F whole cell extract (WCE), but not that from untransfected HEK293T WCE, displayed robust in vitro coligo transcription activity (Fig. 2A, lanes 5 and 9). In vitro transcription by the FLAG-RNAP III complex appeared identical whether it took place on the IP beads or after elution using the FLAG peptide (Fig. 2B).
Figure 2. In vitro transcription (IVT) of coligo templates by immunoprecipitated RNA RNAP III compared with IVT in whole cell extract (WCE). (A) IVT of coligo 122 using the RNAP III complex immunoprecipitated (IP) from HEK293/POLR3F WCE or from unmodified HEK293T cells. Lanes 10–18: titration of coligo 122 shows that it reaches saturation in the IP experiments at 50–100 nM, as found previously for WCE. Sup, IP supernatant. Numbers indicate relative intensity of single circumtranscription (monomer) gel bands. (B) IVT of coligo 122 by IP’d RNAP III complex on beads or after elution from beads. (C) western blot analysis of RNAP III subunit B (POLR3B, aka RPC2) (upper gel) and the RNAP II largest subunit RPB1 C-terminal domain (CTD) (lower two gels) in WCE and IP’d RNAP III. Asterisk in lower gels denotes bands originating from the commercial IP beads and reacting directly with the secondary antibody. The IP and WCE were loaded on the gel in the same proportion as used in IVT. BSA was used in a control IP in place of WCE. (D) IVT profiles for a set of eight different coligos using either HEK293T WCE or IP’d RNAP III complex from HEK293/POLR3F WCE. Coligo sequences are shown in Figure 1. Vertical black lines mark a 10 nt window centered on the predicted size for monomer transcripts as specified in Figure 1 legend and in text.
The pattern of in vitro transcripts from coligo 122 by IP’d RNAP III was very similar to the pattern produced in HEK293T WCE. The observation that the main ~83 nt transcript was produced by both in vitro systems indicates that this transcript is formed directly by termination, rather than by post-transcriptional end trimming of a longer, initially formed transcript. There were however small differences in the overall transcript patterns. The IP’d enzyme produced more apparently abortive transcripts, i.e., those failing to complete one circumtranscription (e.g., compare Figure 2A lane 3 and Figure 2B lane 5). Two other differences in the IP’d enzyme were: slightly less efficient termination, leading to the production of more dimer and higher order multimer transcripts, and the appearance of weak transcripts in between the main transcripts (e.g., Figure 2A, lane 9 vs. lane 4). (Note that multimer transcripts appear over-represented as they have more incorporated label.)
Once we had established the protocol for in vitro transcription by IP’d RNAP III, to assess its activity we estimated the relative amount of the polymerase present in reactions using the IP’d enzyme and reactions using WCE from HEK293T and HEK293/POLR3F cells (Fig. 2C). Using an antibody specific for the RNAP III RPC2 subunit, we found that the IP’d enzyme contained ~5.6-fold more RPC2 per in vitro reaction than did the WCE reactions from either type of cells. No RNAP II could be detected in the RNAP III immunoprecipitate using an antibody against the RNAP II largest subunit C-terminal domain (Fig. 2C, middle and bottom) In comparison, only 2.5-fold more coligo 122 monomer transcript was produced by the IP’d enzyme than by the enzyme in WCE from HEK293/POLR3F cells (Fig. 2A), and 1.7-fold (2.5/1.5) more 122 monomer transcript was produced by the IP’d enzyme compared with WCE from HEK293T cells. Since the enzyme (and not the template, see Figure 2A, lanes 10–18) is limiting over this concentration range, the IP’d enzyme appears to be slightly less active than the WCE enzyme on a per RPC2 subunit basis. This observation argues against the possibility that single-stranded (ss) DNA-binding proteins strongly compete for, and reduce transcription from, 122’s larger loop in WCE, since their removal should have led to greater coligo transcription activity. In contrast, three coligos (luc-1, luc-2, Dcr3) did appear to show significantly increased transcription activity by the IP’d enzyme (Fig. 2D). These coligos also exhibited greater read-through and consequently greater tandem multimer transcript production, despite having canonical RNAP III termination signals. One possible interpretation is that for these three coligos, removing cellular proteins does increase RNAP III access to the larger loop and decreased termination by alleviating steric obstruction of the elongating polymerase. Thus, removing most other cellular proteins by RNAP III IP may for some coligos increase the activity of the polymerase, but this effect appears to depend on the coligo sequence. These observations raise the possibility that some stem-loop junction sequences may favor transcription initiation by RNAP III more than others do, so that optimization of the larger loop for initiation and termination might be possible.
The transcript pattern similarities and differences between the endogenous and IP’d enzymes also applied to coligos 221, 19am3, luc-2, luc-1 and Dcr3 (Fig. 2D). The main transcripts produced by endogenous RNAP III in WCE were also found in the transcripts produced by the IP’d enzyme. As noted above, the reduced termination efficiency was exaggerated for luc-1, luc-2 and Dcr3, the same coligos that showed increased transcription. Overall, the results shown in Figure 2D demonstrate that, for a variety of coligo templates, transcription by IP’d RNAP III and WCE from untransfected cells produced similar though not always identical transcript patterns. In combination with the lack of detectable RNAP II in the IP’d RNAP III (Fig. 2C), and with the previous α-amanitin findings,8 these results provide corroborating evidence that RNAP III is the main and likely sole coligo-transcribing RNA polymerase, and that the IP’d enzyme complex behaves similarly to the endogenous complex.
Coligo transcript termination and release from RNAP III and template
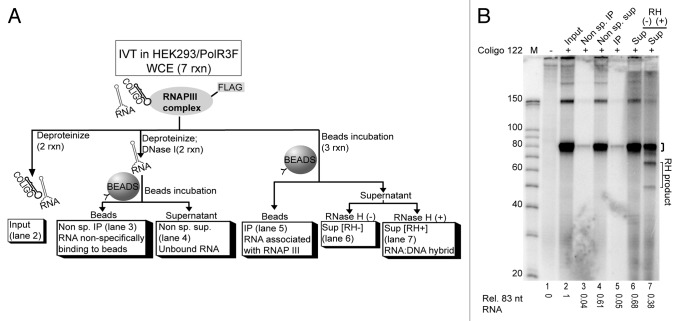
Immobilizing RNAP III through the FLAG tag enabled us to examine whether the transcription products were released from the polymerase or whether transcription had merely paused at the time the transcripts were isolated. A scaled up in vitro transcription reaction was performed and divided as depicted in Figure 3A. Coligo transcripts were not found to be associated with the IP’d RNAP III (Fig. 3B, lane 5), but instead had been released into the supernatant, suggesting that termination had taken place.
Figure 3. Coligo termination and transcript fate after in vitro transcription. (A) Schematic depiction of an experiment designed to detect coligo transcript fate at the end of IVT. A scaled up IVT in HEK293/POLR3F WCE was divided and processed as shown. (B) DPAGE analysis of the uniformly labeled coligo 122 transcript after processing. Small bracket on gel indicates coligo 122 monomer transcript band used for quantification; RH, RNase H; RH product, RNase H degraded transcript products; M, decade marker; Sup., supernatant.
It was recently found that a polyU sequence immediately downstream of a transcript hairpin can trigger RNAP III termination.15 Coligos are single-stranded and made exclusively from the non-coding strand of a cDNA, so might behave differently than ds templates. Nevertheless, we had independently noticed that RNAP III tends to terminate coligo transcription following the completion of a hairpin, rather than within a stem.8 By chance and by design, most of the coligos we have tested so far have had a RNAP III termination signal in the larger loop or near the larger loop-stem junction. In the two cases we have sequenced, 122 and 19aTAR,8 termination appeared to occur either just before or just after the termination sequence, but not in the same position with respect to the termination sequence, leaving its contribution unclear. In the coligos depicted in Figure 1, the termination sequences (indicated by a solid line over the pentamer sequence) occur at different locations within or near the larger loop. The sizes of the transcripts observed correlate well with the number of nucleotides between the predicted TSS and the stem-larger loop junction (Fig. 2D, vertical lines on gel), though in some cases termination at the 2nd nt of the termination pentamer17 would create a monomer transcript of similar size. (Coligo 19anbm34 was an exception for reasons discussed below.) Termination at a termination sequence followed by 3′-5′ exonuclease trimming too rapid to observe may be possible for some coligos (e.g., luc-1, luc-2, Dcr3, Figure 2D), though as described above it is not universal (e.g., 122, 221, 19am3, Figure 2D). We cannot yet be sure how termination depends on the coligo secondary structure and sequence, but suspect that termination sequences and completion of the hairpin stem both contribute.
The terminated transcripts from coligo 122 were also probed with RNase H to assess persistent hybridization between the transcript and coligo template. Transcript release is desirable because stable coligo:transcript hybrids (i) may prevent multiple rounds of transcription from the same coligo, (ii) may lead to endogenous RNase H degradation of the coligo transcripts and (iii) may prevent coligo transcripts from folding into functional RNAs. As previously found for transcripts made in WCE, about half of the RNA dissociated from the coligo (Fig. 3B, compare lanes 6 and 7), an outcome likely favored by the strong intramolecular base pairing in both coligo and transcript.8
Bulges and internal loops facilitate coligo transcription elongation
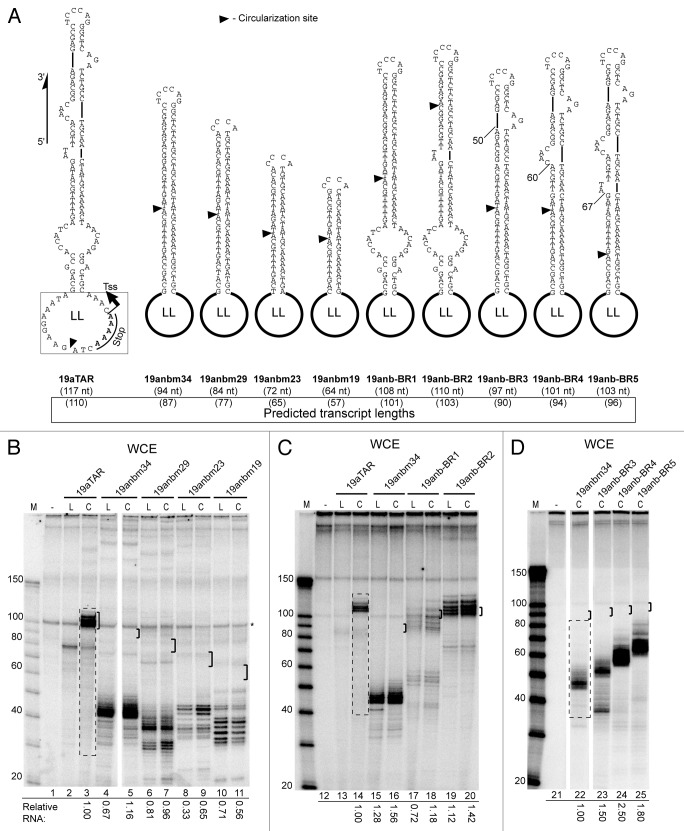
Since a hairpin appears necessary for RNAP III termination,15 we wondered if fully base-paired intramolecular hairpin stems such as are found in short hairpin RNA (shRNA)17-19 might promote termination as well as transcript release from coligo templates, and if shRNA could be made from coligos. We therefore made an shRNA-type coligo by deleting all of the unpaired nucleotides in the stem of coligo 19aTAR (Fig. 4A), a well-transcribed coligo based on miR19a,8 producing coligo 19anbm34. To test the variable of perfectly base-paired stem length on transcription efficiency, we made a homologous series with shorter stems (19anbm29, -23 and -19). Each of these coligos contains the larger loop of 19aTAR, where transcription initiates and terminates in that coligo. The 19aTAR loop appeared to initiate significant amounts of transcription in this context, but none of the coligos were transcribed into the expected full-length RNA hairpins (Fig. 4B, brackets indicate full-length monomer size). Instead, transcription appeared to proceed only half-way around the coligo (19anmb34 and -29) or slightly more (-23 and -19), with substantial amounts of smaller abortive transcripts evident. (Note that the linear forms have the intact initiation loop as well, and were transcribed similarly.) This experiment revealed that internal loops and bulges are necessary for coligo transcription into full-length monomer transcripts.
Figure 4. Coligos require unpaired nucleotides near the larger loop for successful circumtranscription. (A) Sequence and predicted secondary structure of coligos used for IVT in Panels B, C, and D. Transcription start sites (TSS) and 3′ end stop sites for 19aTAR were previously determined.8 All other TSS, stop sites and expected transcript sizes are predicted based on similarity to 19aTAR, and as described in Figure 1 legend and in text. Coligo 19anbm34 is a modified 19aTAR lacking all internal bulges and loops. The original internal loops and bulges of 19aTAR were incrementally re-introduced to form coligo 19anb-BR1 and -BR2, -BR3, -BR4, and -BR5. LL, larger terminal loop from coligo 19aTAR; black triangle, circularization sites; nt, nucleotides. (B) HEK293T WCE IVT for coligos lacking internal bulges and loops. Brackets mark a 10 nt window centered on the predicted size for monomer transcripts using the 19aTAR initiation site. (C and D) HEK293T WCE IVT of coligos with re-introduced internal loops and bulges from 19aTAR. M, marker; L, linear precursor; C, coligo. WCE, whole cell extract; IVT, in vitro transcription. Dashed line boxes indicate gel regions (for every lane in gel but shown only for the first lane) used for transcript quantification.
We next added back the internal loops and bulges, either from the direction of the larger loop (LL), closer to the expected initiation site, or from the direction of the smaller loop, which encodes the apical loop of the hairpin transcript (Fig. 4A). Adding back the internal loops from the direction of the larger loop first partially (19anb-BR1), then fully (-BR2) restored full-length transcription (Fig. 4C), but adding back the bulges to the other end of the stem (19anb-BR3, -BR4, -BR5) did not restore full-length transcription. In the latter three cases, transcription appeared to proceed well until the start of the perfectly base-paired shRNA stem was encountered from the direction of the smaller loop, where transcription appeared to stop abruptly (Fig. Four A, D; lanes 23–25; 50, 60 and 67 nt counting from predicted TSS). Thus, throughout the 19anb series shown in Figure 4, transcription appeared to initiate well in the larger loop and transcribe at least to the smaller loop or a little beyond. These results indicate that shRNA-encoding coligos will likely only be useful if some mismatched nucleotides can be introduced into the stem close to the larger loop, perhaps in the form of dA-dC mismatches to produce rG-rU base pairs in the shRNA.
Linear vs. circular templates, and the point of circularization
The results shown in Figure 4B and C contained an unexpected observation: the linear templates were transcribed nearly as well as their circular counterparts. This result was particularly unexpected for coligo 19anb-BR2 because both linear and circular forms gave rise to full-length monomer transcripts (Fig. 4C, lanes 19, 20). In contrast, previous work indicated that the circular topology of the coligo was necessary for the production of discrete full-length monomer transcripts by RNAP III.8 In previously studied coligos, like 19aTAR (Fig. 4A) and 122 (Fig. 1), we had circularized the linear templates in terminal loop regions using TS2126 RNA ligase, which does not require structural juxtaposition of the 5′ and 3′ ends within a helix,20,21 but each of the new coligos in Figure 4A was circularized in a ds region using T4 DNA ligase. Moving the ligation site to a helical region gives the linear form of the sequence a predicted coligo-like secondary structure, including an intact larger loop, where transcription appears to initiate. Transcription beginning in the larger loop of the linear form should however “run off” at the nick (Fig. 4A, black triangles), producing transcripts that cannot be as long as the intact coligo’s full-length monomer transcripts. We considered two possible explanations: either the polymerase could jump, i.e., transcribe through, the nick, as T7 RNA polymerase can do,22,23 or a DNA ligase in the WCE was first circularizing the linear form to produce the coligo in situ, followed by coligo transcription. Because circularity has been a defining coligo feature, we tested for in situ circularization.
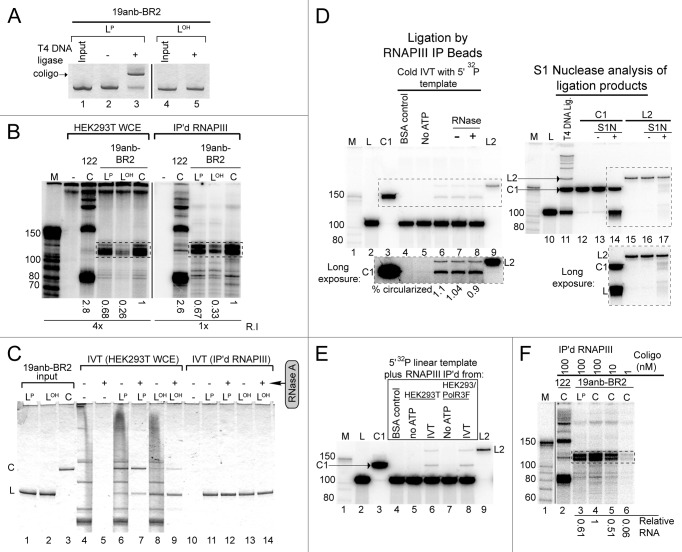
We made linear versions of 19anb-BR2 with and without the 5′ phosphorylation required for DNA ligase (Fig. 5A, LP and LOH), then performed in vitro transcription in WCE and IP’d RNAP III (Fig. 5B). In WCE, all three versions of the 19anb-BR2 sequence gave rise to full-length monomer transcripts. LOH was the least efficient template, leading us to suspect that in WCE linear oligonucleotides with this type of secondary structure can be sequentially 5′ phosphorylated and ligated by endogenous enzymes before undergoing transcription by WCE RNAP III. The lag time taken to prepare the coligo in situ would reduce the time available for transcription; hence, less RNA is made when the in vitro reaction begins with the linear templates.
Figure 5. Coligo linear precursors nicked in a helical region are transcribed like intact coligos. (A) T4 DNA ligase assay to verify dephosphorylation of the 19anb-BR2 linear precursor. LOH, dephosphorylated linear precursor; LP, 5′ phosphorylated linear precursor. DNA visualized by Stains-All after DPAGE. (B) IVT of 100 nM 19anb-BR2 templates (LP, LOH, C) using HEK293T WCE or IP’d RNAP III. Coligo 122 was used as a positive IVT control. The relative amounts of monomer transcripts (boxed) were calculated separately for IP’d RNAP III and HEK293T WCE and normalized within each gel to coligo templated transcripts. R.I., relative exposure intensity of image. (C) Input templates were recovered from HEK293T WCE IVT reactions and visualized by Stains-All after DPAGE. RNase A was used to clarify results by degrading endogenous RNA. (D) About 1% of LP19anb-BR2 is circularized by IP’d RNAP III beads. 5′ 32P labeled 19anb-BR2 linear precursor DNA was examined at the end of an IP’d RNAP III IVT reaction (lanes 1–9). C1 is a gel mobility standard for coligo 19anb-BR2; L2 is a linear ligation dimer. BSA negative control substituted BSA protein for WCE during IP. No ATP lane is a negative control for DNA Ligase activity on the IP’d RNAP III beads. S1 Nuclease treatment (S1N) of C1 and L2 isolated from a T4 DNA ligase reaction on 5′-32P labeled linear precursor was used to check for circularization (lanes 10–17). Insets are longer exposures of boxed gel regions. (E) Control IP from unmodified HEK293T cell WCE also contained a contaminating DNA Ligase activity, indicating that the contaminating ligase activity is not RNAP III -dependent. (F) IP’d RNAP III IVT as a function of coligo 19anb-BR2 concentration. The 1 nM lane (lane 6, 0.06 relative RNA produced) represents the amount of circularized linear precursor formed by contaminating DNA ligase activity during IP’d RNAP III IVT. It cannot account for the amount of transcript formed by 100 nM linear template (lane 3, 0.61 relative RNA).
Surprisingly, IP’d RNAP III, which should have no appreciable 5′ kinase and ligase activities remaining, gave a similar result (Fig. 5B, right). To examine the fate of the oligonucleotides, we scaled up the in vitro transcription reactions and stained the recovered linear and coligo templates after resolution on a denaturing polyacrylamide gel (DPAGE) (Fig. 5C). RNase A treatment was used to clarify the results by removing cellular RNA. The linear versions had indeed been mostly (LP) or partially (LOH) circularized in WCE, consistent with our explanation for their transcriptional behavior in WCE. Since little or no circularization was found in the IP’d RNAP III reactions, despite the similar transcription results, a more sensitive technique was used to look for circularization. We examined the DNA at the end of an IVT reaction using IP’d RNAP III and the linear, 5′ phosphorylated 19anb-BR2 precursor sequence containing a trace of 5′-32P label (Fig. 5D). This experiment revealed small amounts of two new ATP-dependent products, including one (C1) confirmed by gel mobility and S1 Nuclease nicking conditions24 to be coligo 19anb-BR2 (Fig. 5D). Coligo 19anb-BR2 formed in situ by IP’d RNAP III amounted to only ~1% of the input linear oligo. The other product, L2 (Fig. 5D) had the same gel mobility as one demonstrated by S1 Nuclease nicking conditions to be the linear dimer (trace amounts of minor linear and circular ligation multimers can be formed by T4 DNA ligase, see Figure 5D, lane 11). A control immunoprecipitation from untransfected HEK293T WCE contained the same ligase activity (Fig. 5E), showing that the ligase activity was a contaminant adhering non-specifically to the FLAG IP beads, rather than an activity associated with the IP’d RNAP III. We next asked if 1% (i.e., 1 nM coligo) of the 100 nM linear 5′ phosphorylated 19anb-BR2 input − the amount of coligo 19anb-BR2 estimated to form in situ − could account for the amount of transcript produced by linear 19anb-BR2 (Fig. 5F). This amount of coligo (1 nM) produced 10-fold less RNA than did the linear coligo precursor at 100 nM (Fig. 5F, lane 3 vs. 6). Thus, the amount circularized in situ does not account for the amount of full-length monomer transcripts observed from linear 19anb-BR2 in the IP’d RNAP III IVT reaction. Although transcribing through the nick seems to us less likely, we cannot rule it out, and both processes likely contribute to the unexpected transcription of the linear 19anb-BR2 sequence.
Improving transcription from poorly transcribed coligos
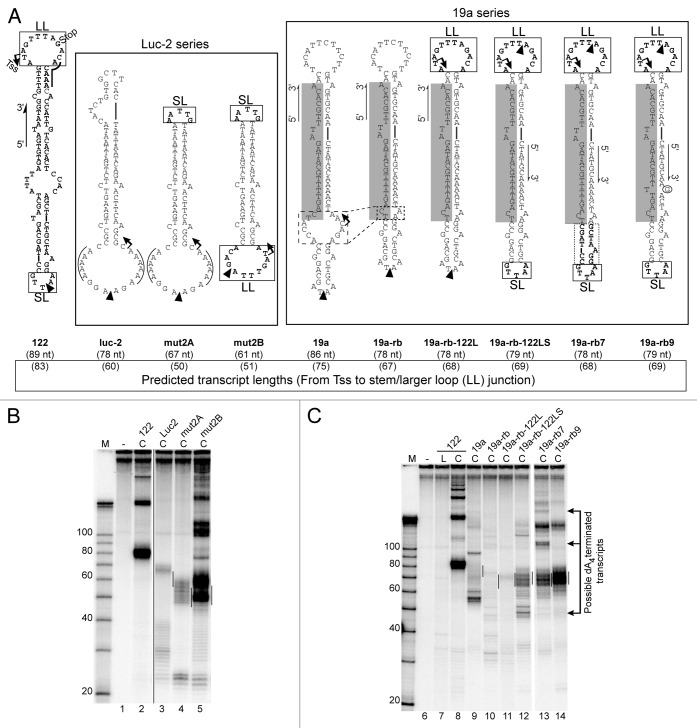
Coligos would prove useful if they could serve as a platform technology for expressing RNA hairpins in cells. We therefore asked whether some of the features of coligo 122 (Fig. 6A), our best-transcribed example, could be used to promote the transcription of other hairpins. Transcription of coligo 122 begins and ends in its larger loop.8 We took coligo luc-2, which transcribed poorly in WCE, and replaced its smaller and larger loops with those from 122 (Fig. 6A). When both loops were added, transcription was markedly improved (Fig. 6B). However, the pattern was not as homogeneous as that for 122 itself, possibly because the dAAACA RNAP III termination signal at the stem-loop junction of 122 was not preserved in creating mut2A and mut2B.
Figure 6. Conversion of poorly transcribed coligos into productive templates. (A) Predicted secondary structures of coligos used in Panels B and C. Luc-2 series: The terminal loops from coligo 122 were used to replace those of coligo luc-2. 19a series: Stepwise changes made to poorly transcribed coligo 19a, including removal of its large internal loop, replacement of its loops with those from 122, and mutation of the dA4 RNAP III termination sequence in the 19a stem (circled G). TSS, transcription start site; Stop, predicted transcript 3′ end site; black triangles, circularization sites; nt, nucleotides; SL, small terminal loop; LL, large terminal loop; lines over sequence indicate known RNAP III termination sequences. (B) IVT of 100 nM Luc-2 series coligos in HEK293T WCE. (C) IVT of 100 nM 19a series coligos in HEK293T WCE. Lines on gel mark a 10 nt window centered on the predicted size for monomer transcripts based on the 122 large loop TSS. Arrows to the right indicate transcripts suspected to terminate at the dA4 sequence in the coligo stem, and which disappear when dA4 is changed to d(AAGA). Coligo 122 used as a positive coligo control. M, marker; L, linear precursor; C, coligo.
We next applied, with additional modifications, a similar strategy to another poorly transcribed coligo, 19a.8,21 This coligo has a large A- and C-rich internal loop which we suspected might be providing unwanted internal initiation sites. Preliminary evidence from an RLM-RACE experiment8 indicated transcription initiation at this site in the closely related coligo 19aTAR (see Tss arrow on the 19a sequence, Figure 6A). We reduced the size of this internal loop to form coligo 19a-rb (Fig. 6A), and this change led to the loss of most transcripts (Fig. 6C, lane 10). We then replaced 19a’s larger loop with the 122 larger loop (coligo 19a-rb-122L). This change further reduced the background of shorter transcripts, but also produced a low level of transcripts at the expected size of ~69 nt, though unexpectedly, in and of itself, this loop did not confer the strong transcription it initiates in 122 (Fig. 6C, lane 11 vs. 8). We then swapped the smaller 19a loop for that from 122, which changed the smaller loop sequence from AATA to AATTG, to form coligo 19a-rb-122LS (Fig. 6A, boxed smaller loop). This change led to a significant increase in monomer transcript production which, given the modesty of the sequence change, we tentatively attribute to increasing the loop size from 4 to 5 nt. Coligo 19a-rb-122LS also produced smaller, possibly abortive transcripts around 50 nt, which if transcription was initiating in the 122 larger loop, would correspond to termination at the dA4 site in the stem. We increased the amount of sequence taken from the smaller 122 loop region, and placed it such that the cDNA of the mature miR19a (shaded in Figure 6A) was not altered (coligo 19a-rb7, dotted box). This change did little to improve the amount of monomer transcripts, strengthening our suspicion that the important feature of the 122 smaller loop is that it is 5 nt, rather than 4 nt which, when clamped by one or more G-C base pairs, might be too small to allow efficient read-through. It also appeared to change use of the dA4 termination site from monomer use (~50 nt) to dimer (~128 nt) and trimer (~206 nt) use (Fig. 6C, lane 13, see arrows to the right of the gel). Since the increased amount of the 122 smaller loop sequence proved not to be critical, we went back to 19a-rb-122LS and changed its possible dA4 termination site in the stem to dAAGA, to produce coligo 19a-rb9. This final change left the cDNA encoding the mature 19a intact, and produced RNA having a length consistent with mainly monomer hairpin transcripts (~69 nt), appearing to begin and end in the 122 larger loop. Destroying the dA4 site by this small change also led to the loss of all transcripts in 19a-rb-122LS and 19a-rb7 putatively resulting from use of the dA4 termination site. This transformation of an untranscribed coligo encoding a pre-miRNA-like hairpin to a strongly transcribed coligo shows that there are multiple considerations in coligo structure that must be taken into account to design a productive coligo. One unexpected consideration was that a RNAP III termination signal in the stem can in fact be used by the polymerase –it appeared not to be used in coligo 19aTAR (Fig. 4) – causing unwanted termination there, and also appearing generally to reduce the level of coligo transcription. We note that to be observed at the end of an in vitro transcription reaction in extracts, transcripts likely have to meet two requirements: (1) transcription initiation and termination by RNAP III, and (2) a hairpin structure conferring resistance against nucleases. Termination at a dA4 in the stem may not allow completion of a hairpin, leading to unstable transcripts that are quickly degraded.
Coligo transcript processing by Dicer
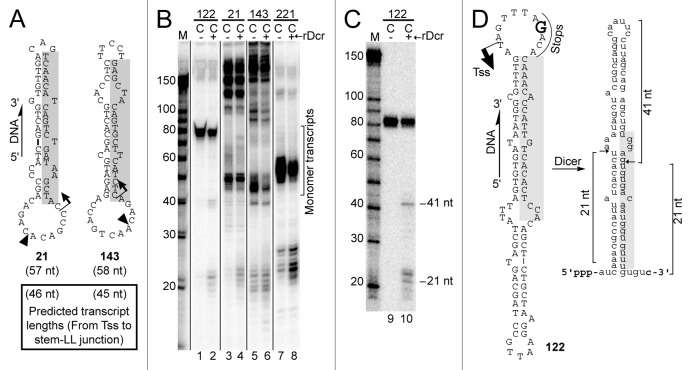
If coligos are to be suitable for use as vectors for the ectopic expression of miRNA or siRNA, they will need to template the production of pre-miRNA hairpin mimics that can be processed by the enzyme Dicer.25,26 We previously found evidence supporting the expectation that coligo transcripts carry 5′ triphosphates.8 In some organisms, Dicer requires a 5′ triphosphate for complete processing,27 but human Dicer substrates, the products of Drosha processing, carry a 5′ monophosphate.28,29 It is not known if cleavage by human Dicer will be affected by a 5′ triphosphate,30 but shRNAs made from RNAP III promoters undergo processing to functional siRNA.17 We have observed in some cases what might be processing products in the 19–24 nt range (e.g., Figure 2D, 221; Figure 6B, mut2A, mut2B) but in many cases we have not observed any possible Dicer products, or have been unable to discern processing due to abortive transcription products. In an initial effort to assess the possibility of Dicer processing, we transcribed four miRNA-encoding coligos using HEK293T WCE in the presence of recombinant Dicer (rDicer) IP’d from transiently transfected HEK293T cells. The tested coligos included two based on miR-21 and miR-143 (Fig. 7A). Supplementing endogenous Dicer in this way led to modest increases in putative Dicer products (19–24 nt) for three of the four coligos (Fig. 7B, lanes 2, 4 and 8). To further clarify this result, we gel purified the transcript from 122 and treated it with the IP’d rDicer (Fig. 7C). Products having sizes consistent with Dicer processing were observed (Fig. 7D). Thus, hairpin-encoding coligos can give rise to Dicer substrates, raising the possibility that coligos can be used to produce RNAs that enter the siRNA or miRNA processing pathways.
Figure 7. Coligo transcript processing by Dicer. (A) Sequences and predicted secondary structures of two additional coligos used for IVT in the presence of recombinant human Dicer. Shaded regions indicate cDNA of mature miRNA 21 and 143. TSS, predicted transcription start site; LL, larger loop; black triangle, circularization site; nt, coligo sizes in nucleotides. (B) HEK293T WCE IVT in the presence or absence of supplemental recombinant Dicer (rDcr). Bracket indicates gel region containing expected monomer transcripts. M, marker; L, linear precursor; C, coligo. (C) DPAGE analysis of an in vitro processing reaction between coligo 122’s purified monomer transcript and FLAG-Dicer immunoprecipitated from transiently transfected HEK293T cells. (D) Predicted secondary structures of coligo 122 and its monomer transcript, with deduced Dicer cut sites indicated.
Discussion
Small RNA expression vectors can provide an alternative to chemically synthesized small RNA, for example siRNA and miRNA mimics, in experimental and therapeutic applications.17,31-33 Here we continue our development of small RNA expression vectors able to use a cell’s own RNAP III to make small RNA hairpins from information contained in chemically synthesized DNA. We previously described the basic anatomy of an RNA hairpin-encoding coligo: an imperfectly base-paired stem capped by two ss loops, the larger of which is purine-rich and provides a transcription initiation site at a pyrimidine near the stem-loop junction.8 Here we extend and refine the determinants of coligo transcription. One important conclusion is that, while important, the larger loop is not the sole determinant of transcription. For the larger loop to function optimally, it is important (1) to avoid large internal loops that can provide secondary initiation sites, (2) to avoid the dA ≥ 4 termination sequence16 in the stem, (3) to ensure that the smaller loop is at least 5 nt, and (4) that unpaired nt be included in the stem, especially near the larger loop. When swapping loops, we also note that it is important to use a program like mfold14 to ensure that the coligo structure is maintained, since changing loop sequences can sometimes change the basic coligo structure substantially. Overall, the coligos tested here afford new insights into the design of productive hairpin-encoding RNAP III templates, though more work remains to be done to understand how the stem-larger loop junction affects the final 5′ and 3′ end formation precision.
Coligos are by definition covalently closed circular oligonucleotides. Previous work demonstrated that circularization conferred stability against exonucleases and appeared to promote transcription initiation at a small number of sites near the stem-larger loop junction. Coligo linear precursors discontinuous in the larger loop were poorly transcribed and suffered degradation in WCE during in vitro transcription.8 Surprisingly, when the precursor’s point of discontinuity was moved from the unstructured larger loop to a helical region in the stem, the linear and coligo templates gave similar full-length monomer transcription products (Fig. 4). This behavior might be explained in the WCE case by the finding that linear coligo precursor sequences were circularized by a cellular ligase in the extract (with, presumably, 5′ phosphorylation when starting with the unphosphorylated precursor), after which they underwent coligo transcription. The same pre-transcription events occurred to a lesser extent on IP’d RNAP III, due to small amounts of kinase and ligase activities adhering to the IP beads, but these did not produce enough coligo to explain the amount of transcripts formed by the linear template. This discrepancy would be resolved if RNAP III is able to transcribe through a nick in the template strand DNA, or at least run-off at the nick and then resume with priming by the nascent transcript. Our observation that IP’d RNAP III produced more full-length transcript from a 5′ phosphorylated linear precursor than from an unphosphorylated precursor supports the sequential phosphorylation, circularization and coligo transcription on the IP beads, but this explanation alone cannot account for all of full-length monomer transcript made from the nicked form of coligo 19anb-BR2. Our results do not allow us to rule out the possibility that RNAP III transcribes across the nick while that part of the DNA is functioning as the template strand. These two processes are not mutually exclusive, and each may contribute to the apparent circumtranscription of the linear 19anb-BR2 template by IP’d RNAP III. In any case, our results show that the linear precursor of the coligo sequence can be stabilized in WCE by placing the discontinuity in a helix (Fig. 5C), and raise the possibility that the circularization step, to produce the coligo, like the transcription step, to produce the RNA, may be left to cellular enzymes if coligo precursors nicked in a helix can be stabilized in serum and cells.
In this work we made use of IP’d RNAP III in vitro to confirm that RNAP III is the coligo-transcribing polymerase, and to determine whether coligo transcripts pause or terminate in vitro. Removing the endogenous cellular small RNA background that has hampered our efforts to sequence coligo transcripts may in addition provide the basis for a coligo transcript-specific sequencing protocol. IP’d RNAP III behaved much like the endogenous enzyme, confirming the identity of the coligo-transcribing polymerase, but produced slightly different transcript patterns for some coligos tested (Fig. 2). For example, the IP’d enzyme produced a higher background of abortive transcripts and a reduced tendency to terminate at the end of one circumtranscription. These differences could result from the loss during precipitation of one or more loosely held polymerase subunits contributing to elongation and termination, respectively. Alternatively, abortive transcripts might appear to be more abundant if immunoprecipitation removed exonucleases that degrade them in WCE. Regarding the difference in termination, ss DNA-binding proteins in WCE might bind to the coligo larger loop and increase termination by steric obstruction of RNAP III. One might also expect ss DNA binding proteins to compete with initiating RNAP III for access to the larger loop. These behaviors were in fact observed in Figure 2D for some coligos (luc-1, -2 and Dcr3) but not for others, raising the possibility that ss DNA binding proteins can, for better or worse, modulate coligo transcribability in some cases.
Though the IP’d RNAP III appeared to release terminated transcripts instead of pausing, we note that our in vitro transcription reactions are performed for 90 min, which may be too long to distinguish between pausing and true termination. Nevertheless, on ds DNA templates RNAP III normally has a strong tendency to terminate directly after a hairpin followed by a polyU sequence.15 Though we have not sequenced the new coligo transcripts presented here, the observed size of the monomer transcripts were generally consistent with coligo transcription initiating and terminating in the larger loop, with termination taking place just after completion of the encoded hairpin stem, as found previously by sequencing 122’s main monomer transcript.8 Most of our coligos have strong or weak RNAP III termination pentamers in or near the larger loop. It is reasonable to expect they can play a role in termination because coligos lacking such sequences, e.g., mut2B (Fig. 6), 21 and 143 (Fig. 7), produced more tandem multimer transcripts in WCE than those that have termination sequences. Yet, even the coligos lacking a termination pentamer showed significant termination after one circumtranscription. It is therefore likely that for hairpin-encoding coligos, a termination signal is not absolutely required, but can increase termination efficiency when it is close to or overlapping with the end of the hairpin stem, as in coligos 122 and apparently in 221. Trimming that is too rapid to observe by our methods from an initial ss 3′ extension back to the hairpin stem is also a possibility, and may be responsible for some differences in the transcript patterns in the IP’d polymerase, but some coligos, like 122 and 221, showed almost no difference between the WCE and IP’d polymerase sources, indicating that trimming cannot be universal.
In conclusion, the in vitro transcription of a new set of coligos based mostly on pre-miRNA hairpins and shRNA has allowed us to further characterize the determinants of promoterless transcription of this new and unique type of chemically synthesized small RNA expression template. In addition to developing a much-needed sequencing protocol exploiting the IP’d RNAP III, future work will aim to find larger-loop sequences preferred by RNAP III, and to optimize the precision of initiation and termination, which may in turn lead to optimal processing by structure-dependent nucleases like Dicer.
Materials and Methods
Coligo preparation
5′ phosphorylated synthetic linear DNA Ultramer® oligonucleotides were purchased from Integrated DNA Technologies. Coligo secondary structures were predicted using the mfold program.14 Circularization of coligo precursors discontinuous in regions predicted to be ss (122, 19aTAR, 221, 19am3, luc-1, luc-2, Dcr3, 15a-b, 19a, mut2A, mut2B, 19a-rb, 19a-rb-122L, 19a-rb-122LS, 19a-rb5, 19a-rb7, 19a-rb9) was performed using TS2126 RNA ligase enzyme as previously described.21 5′ phosphorylated nicked dumbbell or dumbbell-like linear DNA Ultramers (19anbm19, 19anbm23, 19anbm29, 19anbm34, 19anb-BR1, 19anb-BR2, 19anb-BR3, 19anb-BR4, 19anb-BR5) were circularized using T4 DNA ligase enzyme (New England BioLabs, NEB, M0202S) and purified as described below. Unphosphorylated coligo precursors used in Figures 5A-C were made by dephosphorylating the purchased Ultramer oligo using Calf intestinal alkaline phosphatase (Promega, M182A) according to the manufacturer’s instructions. Similarly, 5′ [γ-32P]-ATP labeling of dephosphorylated linear Ultramer for Figures 5D and 5E was done using T4 Polynucleotide kinase (NEB, M0201S) according to the manufacturer’s instructions.
Whole cell extract (WCE) preparation
HEK293T cell culture was performed as described.8 HEK293/POLR3F cells were grown under puromycin selection as described.13 To prepare the extract, 500 μl of chilled WCE lysis buffer [10mM HEPES pH 7.9, 1.5mM MgCl2, 10mM KCl, 0.05% NP40, 0.2 mM EDTA, 0.3M NaCl, 15% Glycerol, and Protease Inhibitor Cocktail (Roche, 04693159001)] was added per 10 cm dish and the extract was prepared exactly as described.8 Three plates of either cell type yielded ~1400 μl WCE with a total protein concentration of ~3–4 μg/μl compared with bovine serum albumin (BSA) as the standard.
Immunoprecipitation of RNAP III complex
HEK293/POLR3F cells were kindly provided by Dr. James Chen (U.T. Southwestern). FLAG® Immunoprecipitation Kit (FLAGIPT1, Sigma-Aldrich) was used for immunoprecipitation of the RNAP III complex. 40 μl of drained ANTI-FLAG M2-Agarose Affinity Gel were used per 1400 μl of WCE (HEK293T or HEK293/POLR3F). The beads were pre-washed 3X with 500 μl 1X Sigma wash buffer. The WCE was added to the beads and gently rocked at 4 °C for 2 h. The supernatant (“Sup” in Figure 2A, 2B) and immunoprecipitated (IP’d) RNAP III complex were separated by centrifugation. The IP’d RNAP III was washed 3X with 500 μl of WCE lysis buffer. The final wash was stored (“Wash” in Figure 2B) and the RNAP III bound beads were resuspended in 160 μl (200 μl final) of WCE lysis buffer. For elution of RNAP III complex from the beads, after the final wash the IP’d beads were resuspended in 80 μl of elution buffer (WCE lysis buffer with 190 ng/μl FLAG peptide) and incubated for 1 h with gentle rocking at 4 °C. The supernatant containing the RNAP III complex was collected by centrifugation and transferred to a new pre-chilled microfuge tube. The beads were again resuspended in 80 μl of WCE lysis buffer, centrifuged and the supernatants were pooled. The volume of eluted RNAP III complex was finally adjusted to 200 μl by adding WCE lysis buffer and stored at -80 °C. After elution, the beads (Post IP in Figure 2B) were resuspended in WCE lysis buffer to a final volume of 200 μl and stored at -80 °C. For BSA controls experiments (Fig. 2C, 5D, 5E) an identical immunoprecipitation procedure was followed but BSA was used in place of WCE.
In vitro transcription (IVT)
IVT using the linear or circular templates, WCE or immunoprecipitated RNAP III complex, and uniform label incorporation with [α-32P]-UTP was performed as follows. A typical 20 μl reaction contained 4 μl (~16 μg) of WCE (0.28% of an entire 1400 μl WCE preparation), 8 U RNasin® RNase inhibitor (N2515, Promega), 0.6 mM each ATP, GTP, CTP, 0.2 mM UTP, ~2 μCi [α-32P]-UTP, 40 mM TRIS-HCl pH 7.9 (at 25 °C), 6 mM MgCl2, 10 mM DTT, 2 mM spermidine, and 100 nM DNA template unless otherwise indicated. For IVT performed using immunoprecipitated RNAP III complex (IP, elution, post IP), 4 μl was used per IVT reaction which corresponds to 2% of an entire 1400 μl WCE preparation. For IVT reactions done cold without uniform [α-32P]-UTP labeling (Figs. 5C, 5D, and 5E), the [α-32P]-UTP was replaced with an equivalent amount of cold UTP. Transcription reactions were incubated for 90 min at 37 °C followed by extraction with TRIzol® reagent (Invitrogen, 15596–026) and precipitation following manufacturer’s instructions. Precipitated RNA and/or 5′ labeled coligo linear precursors were separated on a 10% denaturing polyacrylamide electrophoresis gel (DPAGE) including 40% formamide (Fig. 5D and 5E) or without formamide (all other gels), and visualized by exposing the dried gel to a Molecular Dynamics Phosphorimager Screen. Image quantification was performed using MD ImageQuant software.
Western blotting
Western blotting of RPC2 shown in Figure 2C was performed as previously described.8 The WCE and IP’d RNAP III samples used for western blotting were from a different but identically prepared batch than the one used for the IVT in Figure 2A and 2B, and the IVT activity was assayed and found to be identical before using for western blotting (not shown). The amount of RNAP III (WCE or IP beads) normally used for IVT was doubled for western blotting. Rabbit anti-RPC2 (Bethyl Labs Inc. A301–855A) and goat anti-rabbit (Calbiochem, DC03L) were used as primary and secondary antibodies respectively for RPC2. Similarly, mouse monoclonal anti-RPB1 CTD (Covance 8WG16) and goat anti-mouse (Calbiochem DC08L) were used as primary and secondary antibodies respectively for detection of the RNA Polymerase II (RNAP II) RPB1 CTD. The goat anti-mouse antibody recognizes mouse anti-FLAG antibodies shed from IP beads, producing 8WG16-independent signals on the RPB1 blot, denoted by asterisk in Figure 2C.
T4 DNA ligase circularization
Preparative T4 DNA ligase reaction for circularization of dumbbell or dumbbell-like linear precursors was performed as follows (500 μl final reaction volume): 500 pmol of linear 5′ phosphorylated precursor sequence was incubated in 1X T4 DNA ligase buffer at 94 °C for 3 min and slow-cooled to 50 °C, then put at room temperature and then at 18 °C. 1000 units of T4 DNA ligase and 1 mM final ATP was added and left at 18 °C for 2 h. DNA was extracted using phenol:choloroform:isoamyl alcohol (PCI) and ethanol precipitated. Uncircularized precursor was removed by Exonuclease III (M181A, Promega) treatment using the manufacturer’s protocol, or by gel purification of the circularized product as previously described.21 The small-scale analytical T4 DNA ligase reaction shown in Figure 5A was done similarly but in 20 μl containing 10 pmol of linear template and 40 units of the enzyme. Extracted DNA was visualized by staining with Stains-all (7423–31–6, Acros) as previously described.8 Similar conditions were used for circularization of 5′ radiolabeled linear 19anb-BR2 template as shown in Figure 5D and 5E without staining.
Template recovery from IVT reactions
As shown in Figure 5C, each reaction was scaled up 4-fold compared with typical IVT reaction described above to ensure visualization of the templates by Stains-all dye. After IVT reaction, each sample was PCI extracted and ethanol precipitated. The samples were further treated +/− RNase A for additional 30 min at 37 °C. Nucleic acids were isolated again by PCI extraction and ethanol precipitation, run on 10% DPAGE and stained with Stains-all as previously described.8
S1 nuclease Assay
The circular topology of the coligos used for IVT reactions was verified as previously described21 but with following differences: 50 μl reaction volume containing 15 pmol of circular oligodeoxynucleotide was treated with 5 units of S1 Nuclease (70019, USB). The topology of gel-purified products from T4 DNA ligase reaction of 5′ radiolabeled linear 19anb-BR2 (Fig. 5D) was also verified similarly using S1 Nuclease. The bands were visualized and quantified by exposure to Phosphorimager screen and MD ImageQuant software.
Purification of recombinant Dicer complex
For purification of recombinant human Dicer (rDicer), a FLAG-tagged Dicer plasmid (Addgene plasmid 19881, pFRT/TO/FLAG/HA-DEST DICER34) was transfected into HEK293T cells: 2.4x106 cells per 10 cm dish were seeded the day before transfection and transfected with 8 µg of the purified rDicer plasmid using 80 µl PolyFect transfection reagent (Qiagen, 301105) according to the manufacturer's protocol. 48 h after transfection, the dishes were washed twice with ice-cold 1X PBS, and rDicer was purified using Sigma's FLAG-IP kit (FLAGIPT1, Sigma-Aldrich). Briefly, the cells were lysed in 0.8 ml ice cold lysis buffer supplemented with Protease Inhibitor Cocktail (PIC) (04693132001, Roche). The cells were scraped with a chilled cell-scraper, transferred into pre-cooled 1.5 ml tubes and incubated on ice for 15 min with three vortex pulses every 5 min. The lysate was spun for 10 min at 12000 x g at 4 °C and the supernatant was transferred to a new, chilled tube. To immunoprecipitate FLAG-tagged rDicer, 50 µl of anti-FLAG-M2-agarose affinity resin suspension (around 25 µl of packed bead volume) per 10 cm dish were washed five times with ice cold wash buffer in order to remove all traces of storage solution with centrifugation at 6000 x g for 30 s at 4 °C in between. After the last wash, beads were resuspended in 200 µl lysis buffer per 10 cm dish supplemented with PIC and added to the cell extract. The extract was rotated for at least 4 h or overnight at 4 °C. The beads-rDicer complex was spun for 30 s at 6000 x g, 4 °C, and then washed 5 times with ice cold wash buffer (1.2 ml per wash per 10 cm plate). The rDicer-beads complex was resuspended in 60 µl lysis buffer supplemented with PIC and 10% Glycerol and stored at -20 °C up to four months. As negative control (-rDcr lanes, Figure 7B), cells were transfected with empty pcDNA3.1 vector and treated in exactly the same way as the rDicer-transfected dishes.
In vitro transcription in the presence of recombinant Dicer
In vitro transcription in the presence of rDicer-beads was performed exactly as described8 but with the addition of 3 µl purified rDicer-beads to the 20 µl IVT reaction volume. During the 90 min incubation, the tubes were flicked every 15–20 min to resuspend beads. RNA extraction and DPAGE was performed as described.8
Dicer processing assay for coligo-generated transcripts (Fig. 7C)
The body-labeled coligo 122 monomer transcript from a 4-fold scaled up IVT reaction (see above) in HEK293T WCE was purified by 10% DPAGE and extracted from the gel. Half of the RNA was then treated with bead-bound FLAG-Dicer immunoprecipitated from whole cell extract from HEK293T cells transiently transfected with the plasmid expressing FLAG-Dicer. The processing reaction was performed in Dicer IP assay buffer (10 mM Tris pH 7.5, 5 mM MgCl2, 1mM DTT, 0.4U/μl RNasin, 1 mM ATP)35 for 60 min at 37 °C. RNA was extracted, ethanol precipitated and fractionated on 10% DPAGE.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grant 5SC1GM083754 from the National Institutes of Health (NIH). Additional infrastructural support was provided by the NIH National Center for Research Resources (2G12RR03060–26) and the National Institute on Minority Health and Health Disparities (8G12MD007603–27). We thank James Chen (Univ. Texas, SW) for providing the HEK293/POLR3F cell line.
Glossary
Abbreviations:
- RNAP III
RNA polymerase III
- POLR3
RNA polymerase III cDNA or gene
- WCE
whole cell extract
- HEK293
human embryonic kidney 293 cells
- HEK293T
HEK293 cells transformed with SV40 large T antigen
- POLR3F
subunit F of RNA polymerase III
- IP
immunoprecipitate
- IP’d
immunoprecipitated
- TSS
transcription start site
- ss
single-stranded
- ds
double-stranded
- nt
nucleotide(s)
- IVT
in vitro transcription
- RPC2
subunit B of RNA polymerase III
- RPB1
largest subunit of RNA polymerase II
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–9. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10:282–9. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61:746–59. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Davidson BL, McCray PB., Jr. Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–40. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guzman-Villanueva D, El-Sherbiny IM, Herrera-Ruiz D, Vlassov AV, Smyth HD. Formulation approaches to short interfering RNA and MicroRNA: challenges and implications. J Pharm Sci. 2012;101:4046–66. doi: 10.1002/jps.23300. [DOI] [PubMed] [Google Scholar]
- 7.Somoza A. Protecting groups for RNA synthesis: an increasing need for selective preparative methods. Chem Soc Rev. 2008;37:2668–75. doi: 10.1039/b809851d. [DOI] [PubMed] [Google Scholar]
- 8.Seidl CI, Lama L, Ryan K. Circularized synthetic oligodeoxynucleotides serve as promoterless RNA polymerase III templates for small RNA generation in human cells. Nucleic Acids Res. 2013;41:2552–64. doi: 10.1093/nar/gks1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rumney S, Kool ET. DNA recognition by hybrid oligoether-oligodeoxynucleotide macrocycles. Angew Chem Int Ed Engl. 1992;31:1617–9. doi: 10.1002/anie.199216171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubendiek SL, Ryan K, Kool ET. Rolling-Circle RNA Synthesis: Circular Oligonucleotides as Efficient Substrates for T7 RNA Polymerase. J Am Chem Soc. 1995;117:7818–9. doi: 10.1021/ja00134a032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diegelman AM, Kool ET. Generation of circular RNAs and trans-cleaving catalytic RNAs by rolling transcription of circular DNA oligonucleotides encoding hairpin ribozymes. Nucleic Acids Res. 1998;26:3235–41. doi: 10.1093/nar/26.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohmichi T, Maki A, Kool ET. Efficient bacterial transcription of DNA nanocircle vectors with optimized single-stranded promoters. Proc Natl Acad Sci U S A. 2002;99:54–9. doi: 10.1073/pnas.012589099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–91. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–80. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orioli A, Pascali C, Quartararo J, Diebel KW, Praz V, Romascano D, Percudani R, van Dyk LF, Hernandez N, Teichmann M, et al. Widespread occurrence of non-canonical transcription termination by human RNA polymerase III. Nucleic Acids Res. 2011;39:5499–512. doi: 10.1093/nar/gkr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 18.Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505–8. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 19.Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23:227–31. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 20.Blondal T, Thorisdottir A, Unnsteinsdottir U, Hjorleifsdottir S, Aevarsson A, Ernstsson S, Fridjonsson OH, Skirnisdottir S, Wheat JO, Hermannsdottir AG, et al. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res. 2005;33:135–42. doi: 10.1093/nar/gki149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seidl CI, Ryan K. Circular single-stranded synthetic DNA delivery vectors for microRNA. PLoS One. 2011;6:e16925. doi: 10.1371/journal.pone.0016925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rong M, Durbin RK, McAllister WT. Template strand switching by T7 RNA polymerase. J Biol Chem. 1998;273:10253–60. doi: 10.1074/jbc.273.17.10253. [DOI] [PubMed] [Google Scholar]
- 23.Zhou W, Reines D, Doetsch PW. T7 RNA polymerase bypass of large gaps on the template strand reveals a critical role of the nontemplate strand in elongation. Cell. 1995;82:577–85. doi: 10.1016/0092-8674(95)90030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diegelman AM, Kool ET. Chemical and Enzymatic Methods for Preparing circular Single-Stranded DNAs. Protocols in Nucleic Acid Chemistry. 2000:5.2.1–5.2.27. doi: 10.1002/0471142700.nc0502s00. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 26.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–74. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol. 2007;14:604–10. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- 28.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 30.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–5. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–7. doi: 10.1016/S1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 32.Devroe E, Silver PA. Retrovirus-delivered siRNA. BMC Biotechnol. 2002;2:15. doi: 10.1186/1472-6750-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–8. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 34.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–96. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perron MP, Landry P, Plante I, Provost P. Detection of human Dicer and Argonaute 2 catalytic activity. Methods Mol Biol. 2011;725:121–41. doi: 10.1007/978-1-61779-046-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]