Abstract
Transcription through chromatin by different RNA polymerases produces different biological outcomes and is accompanied by either nucleosome survival at the original location (Pol II-type mechanism) or backward nucleosome translocation along DNA (Pol III-type mechanism). It has been proposed that differences in the structure of the key intermediates formed during transcription dictate the fate of the nucleosomes. To evaluate this possibility, structure of the key intermediate formed during transcription by Pol III-type mechanism was studied by DNase I footprinting and molecular modeling. The Pol III-type mechanism is characterized by less efficient formation of the key intermediate required for nucleosome survival (Ø-loop, Pol II-type mechanism), most likely due to steric interference between the RNA polymerase and DNA in the Ø-loop. The data suggest that the lower efficiency of Ø-loop formation induces formation of a lower nucleosomal barrier and nucleosome translocation during transcription by Pol III-type mechanism.
Keywords: epigenetics, transcription, chromatin, structure, histones, mechanism
INTRODUCTION
Chromatin is remodeled prior to and during transcription initiation (by ATP-dependent remodelers) and also during transcript elongation by RNA polymerases (RNAPs). Two strikingly distinct pathways of transcription through chromatin (Pol II- and Pol III-type mechanisms) have been described in [1–4]. The signatures of the Pol III-type mechanism (used by bacteriophage SP6 RNAP and yeast Pol III) are a lower nucleosomal barrier to transcription and transfer of the entire histone octamer during transcription [2, 5]. In contrast, the Pol II-type mechanism is characterized by selective loss or exchange of H2A/H2B dimer(s) during transcription [3,6–9], a stronger barrier to transcription, and survival of the remaining histone hexamer at the original position on DNA [3, 4, 10–12].
It has been proposed that the Pol II- and Pol III-type mechanisms are designed to maintain selective exchange of histones H2A/H2B or all core histones, respectively [4, 8, 13, 14]. Furthermore, it has been proposed that the high nucleosomal barrier formed during transcription through chromatin by Pol II is involved in regulation of gene expression [15, 16]. However, the molecular mechanisms determining the differences between the Pol II- and Pol III-type mechanisms of transcription through chromatin remain to be determined.
Our previous data have identified a key intermediate (a small intranucleosomal DNA loop, Ø-loop) that is formed at the position +49 (the numbers indicate the distances (in base pairs) from the promoter-proximal boundary of the nucleosome) and is possibly required for nucleosome survival during transcription by Pol II through chromatin, and suggested that strong nucleosomal pausing guarantees efficient nucleosome survival [4, 17]. Specifically, it has been suggested that during transcription by Pol III-type mechanism the Ø-loop is less stable and is formed on a smaller fraction of the templates than in the case of Pol II-type mechanism [4]. Less efficient formation of the Ø-loop is expected to result in a lower nucleosomal barrier to transcription and more efficient nucleosome translocation along DNA.
To evaluate these predictions, the structure of the key intermediate formed at the position +49 during transcription by Pol III-type mechanism was studied by DNase I footprinting and molecular modeling. These studies revealed several distinct features of the elongation complex formed at this position (EC+49): less efficient formation of the Ø-loop and possible steric interference between RNA polymerase and DNA during formation of the Ø-loop. These features of the EC+49 possibly result in the observed more “open” structure of the complex. The lower efficiency of the Ø-loop formation possibly explains other features of the Pol III-type mechanism, such as nucleosome translocation and a lower nucleosomal barrier formed during transcription through chromatin.
MATERIALS AND METHODS
Protein purification
Hexahistidine-tagged T7 RNAP was purified as described in [18]. In short, expression of His-tagged T7 RNAP was induced in the presence of 400 µM IPTG in cultured PVM1/BL21 cells. Cells were then lysed. Proteins were precipitated by saturated ammonium sulfate. Pure His-tagged T7 RNAP was purified on the affinity (Ni-agarose) and ion-exchange columns (P11 cellulose). The -H1 chicken erythrocyte chromatin as the donor of histones for nucleosome assembly was purified as described in [19].
DNA templates and nucleosome reconstitution
The templates, T7P2-603+49 and T7-603+49, contain T7 promoter, a 50-bp linker DNA and the 603 nucleosome positioning sequence [4]. T7-603+49 is a modified version of the T7P2-603+49 [20] that allows stalling of T7 RNAP at the position -5 in the presence of incomplete mix of NTPs (ATP, CTP and GTP). The template was prepared by annealing pairs of long overlapping oligonucleotides and filling-in with the Klenow fragment of DNA polymerase I as described in [4]. The complete DNA templates were then amplified by PCR reaction and purified by gel electrophoresis using gel extraction kit (Omega Bio-Tek). The sequence of the template will be provided upon request. Nucleosomes were reconstituted on the DNA templates by histone octamer transfer [19].
Transcription, nucleosome fate and DNase I footprinting
T7 RNAP was first incubated with templates (DNA or nucleosome containing T7 promoter) in 1× T7 transcription buffer (50 mM Tris-acetate, pH 7.0, 10 mM magnesium acetate, 5 mM dithiothreitol (DTT), 100 mM sodium acetate) at 20 °C. T7 RNAP then formed 45-mer (EC-5), either sequentially by walking to 19-mer and 27-mer on the T7P2-603+49 template, or in one step by addition of 250 µM ATP, GTP and 50 µM CTP on the T7-603+49 template at 20 °C. Complexes were immobilized on Ni-NTA-agarose, extensively washed, and T7EC-5 were eluted from Ni-NTA beads in the presence of 100 mM imidazole. Transcription was continued in the presence of 400 mM NTPs at 0 °C for 3 minutes in the transcription buffer TB40 (20 mM Tris HCl, pH 8.0, 5 mM MgCl2, 2 mM β-mercaptoethanol, 40 mM KCl). In experiments with labeled RNA, the 45-mer was pulse-labeled in the presence of [α-32P]-CTP (3000 Ci/mmol, PerkinElmer Life Sciences) and analyzed by denaturing PAGE. In time-course experiments, transcription was performed for different time periods (2, 5, 10, 30, 60 or 180 seconds) at 0 °C [2]. The nucleosome fate was analyzed by native PAGE after transcription with DNA-labeled templates [2]. DNase I footprinting was conducted as described in [4]. EC+49 complexes were formed in the presence of 1 µM ATP on the 603-49 templates at 0 °C in TB40. Labeled DNA was purified from native PAGE and analyzed by denaturing PAGE. The data were quantified using ImageQuant software.
Restriction enzyme mapping
The in vitro transcription was performed as described above using 32P-labled template. EC-5, EC+49 and transcribed templates were incubated in the presence of an excess of MseI or ClaI restriction enzymes for 30 minutes at 20 °C in TB100 [4]. MseI and ClaI have single intranucleosomal sites at the positions −66 and +101, respectively. Labeled DNA templates were then purified and analyzed by denaturing PAGE. The amounts of digested DNA were quantified and plotted as fraction of all templates. Averages from three experiments and standard deviations were determined.
Modeling EC+49 and analysis of the structural features of the modeled complex
A model of Pol II EC+49 complex in a nucleosome ([4] and unpublished) was used as a starting structure. The Pol II structure was deleted from the complex, and the T7 polymerase elongation complex structure (PDB ID 1S77 [21]) was used to replace the Pol II structure as described in [4]. The orientation of T7 EC structure within the +49 histone-DNA Ø-loop complex was determined by aligning the 10-bp long RNA-DNA duplex in the active site of T7 EC with the corresponding duplex in the Pol II EC+49 complex. The model was displayed by PyMOL script (http://www.pymol.org).
RESULTS
T7 RNAP utilizes the Pol III-type mechanism of transcription through chromatin
Traversal of the nucleosome by RNA polymerase II (Pol II) results in loss of one H2A/2B dimer, but the nucleosome remains in place on the template [3, 8]. In contrast, during transcription by Pol III-type mechanism, a much lower nucleosomal barrier to transcription was detected; nucleosomes remain intact, but are translocated from their original locations [1, 2, 5, 15]. It has been proposed that differences in the structure of the key intermediate formed during transcription through the +45 region of nucleosomal DNA dictate the different nucleosome fates during transcription by the Pol II- and Pol III-type mechanisms [4].
The intermediate that is critical for nucleosome survival during transcription by Pol II-type mechanism contains a very small intranucleosomal DNA loop (Ø-loop) formed at the position +49 on nucleosomal DNA (elongation complex +49, EC+49 [4]). Formation of the Ø-loop by Pol II induces recoiling of partially and transiently uncoiled nucleosomal DNA on the surface of histone octamer upstream of the transcribing enzyme; this recoiling is coupled with uncoiling of DNA from the octamer in front of the enzyme, allowing further transcription and nucleosome survival at the same time [4]. It has been proposed that the structures of the EC+49 formed during transcription by Pol III- and Pol II-type mechanisms are likely to be different and this difference could dictate the nucleosome fate on transcription [4].
Previously we have shown that the Pol III-type mechanism is utilized by bacteriophage SP6 RNA polymerase (RNAP) and by yeast Pol III [1, 2, 15, 22]. However, high-resolution structures of SP6 RNAP and Pol III are unknown. Therefore, to conduct structural analysis of the Pol III-type mechanism we used structurally defined T7 RNAP [23] that is highly homologous to SP6 RNAP [24]. First, we characterized the mechanism of transcription through chromatin by T7 RNAP using the approaches and positioned 603 mononucleosomal templates utilized previously for analysis of transcription by Pol II and SP6 RNAP [2, 15]. Transcription by hexahistidine-tagged T7 RNAP was initiated on T7603+49 template containing a strong T7 promoter [20], a –U track sequence downstream of the promoter and 603+49 nucleosome positioning sequence [4] (Fig. 1A). Then the elongation complexes (ECs) were immobilized on Ni-NTA beads or transcribed by T7 RNAP in presence of all NTPs. The results of transcription were analyzed by denaturing or native PAGE. Active EC-5 complexes were detected on DNA and nucleosomal templates and then chased upon addition of all NTPs to release histone-free DNA or primarily a nucleosome, respectively (not shown). Thus complete nucleosome remains associated with DNA after transcription by T7 RNAP, as was shown for the Pol III-type [1, 2], but not for the Pol II-type mechanism where an H2A/H2B dimer is displaced during transcription [3, 8]. The data suggest that, as expected, T7 RNAP uses the Pol III-type mechanism for transcription through the nucleosome.
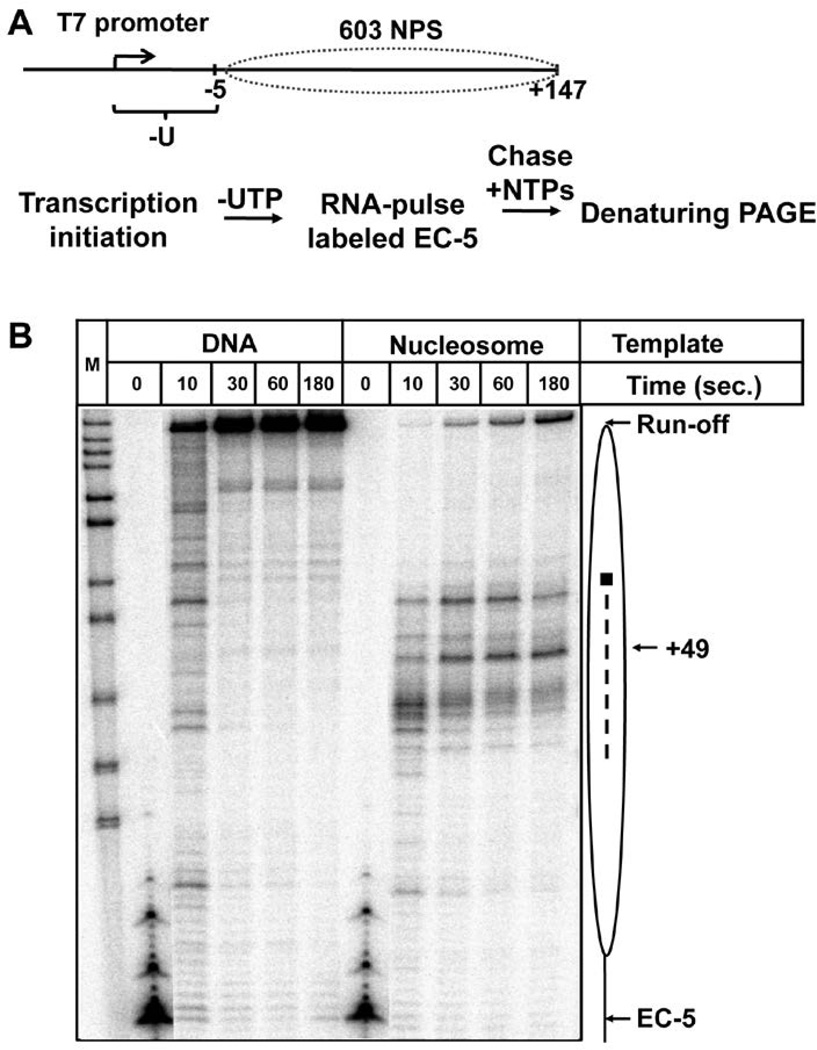
Figure 1. 603 nucleosome forms a barrier to transcribing T7 RNAP.
A. The schematic diagram of the template and the experimental approach for analysis of transcription of nucleosomal templates by T7 RNAP. The locations of the 603 nucleosome-positioning sequence (NPS, dotted oval) and the T7 promoter are shown. After transcription initiation, T7 RNAP was stalled at the position -5 relative to the boundary of 603 nucleosome forming EC-5. The transcripts were pulse-labeled. Then transcription was continued using unlabeled NTPs at 40 mM KCl and terminated after different time periods (from 10 to 180 seconds). Labeled RNA was analyzed by denaturing PAGE. B. Analysis of labeled transcripts by denaturing PAGE. The locations of the nucleosome (oval), the nucleosome dyad (square), the EC-5, position +49 and the run-off transcripts are shown. Dashed line: nucleosome-specific pausing. M: MspI digest of pBR322 plasmid. Note that a considerable fraction of elongation complexes completes transcription after 180 seconds, suggesting that the nucleosomal barrier is relatively low.
To further evaluate the mechanism of transcription through the nucleosome by T7 RNAP, two characteristic properties - nucleosomal pausing during transcription and nucleosome positioning after transcription were examined. Transcription on DNA or nucleosome template was conducted at 40 mM KCl for different time periods ranging from 10 seconds to 3 minutes (Fig. 1B). The nucleosomal barrier to T7 transcription was much lower than the barrier encountered by Pol II: up to 70% of T7 RNAP molecules synthesized run-off transcript under these low-salt conditions; the observed pausing pattern is similar to the one characteristic for the Pol III-type mechanism (Fig. 1B and [15]). To determine the positions of the nucleosome after transcription, the nucleosomes before (T7 EC-5) or after transcription were digested by selected restriction enzymes (Fig. 2A) [1]. After digestion, DNA was purified and analyzed by denaturing PAGE (Fig. 2B). Before transcription nearly all nucleosomes are fully sensitive to digestion by MseI and resistant to digestion by ClaI suggesting that, as expected, nucleosomes occupy the promoter-distal end of the template (Fig. 2B, C). In contrast, after transcription a large fraction of nucleosomes (~50–60%) becomes sensitive to digestion by ClaI and resistant to digestion by MseI (Fig. 2B, C) suggesting that on a considerable fraction of the templates nucleosomes were translocated to the promoter-proximal end of the template after transcription. Similar results were obtained after digestions by other restriction enzymes (not shown).
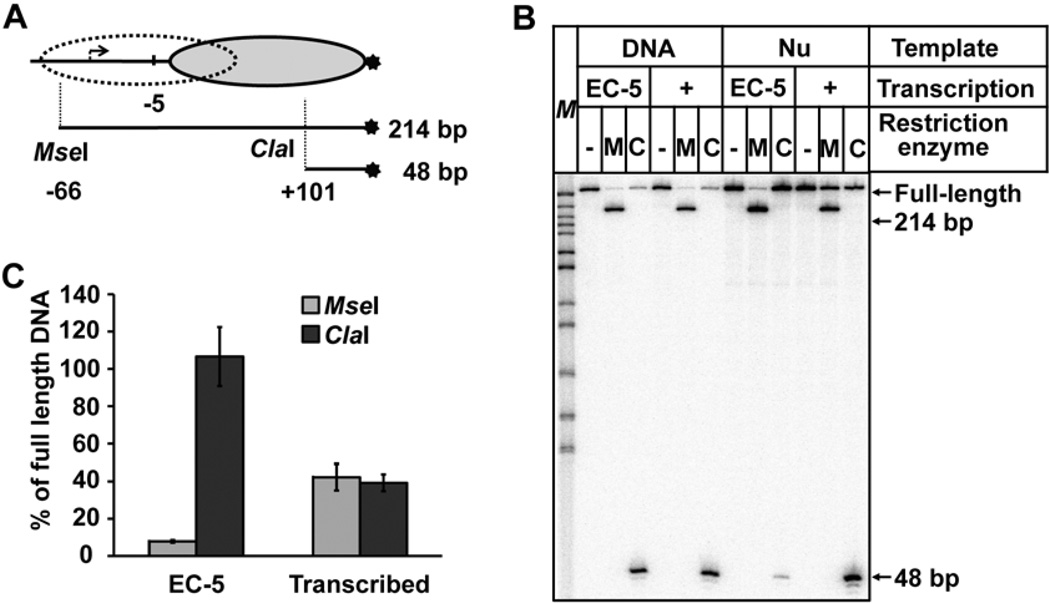
Figure 2. Nucleosomes are translocated after transcription by T7 RNAP.
A. The strategy for mapping of nucleosome positions before or after transcription using restriction enzymes. DNA is end-labeled (star). The locations of the nucleosome before (oval) and after transcription (dotted oval), and the positions of the sites for the restriction enzymes MseI or ClaI are shown. B. Analysis of DNA fragments after digestion with the restriction enzymes. DNA or nucleosomal templates were transcribed by T7 RNAPs after formation of the EC-5, and digested with restriction enzymes MseI (M) or ClaI (C). DNA was purified and analyzed by denaturing PAGE. C. Quantitation of the data shown in Fig. 2B. Averages from three experiments and standard deviations are shown. Note that MseI site becomes partially protected after transcription, indicating that nucleosome translocation occurred on a considerable fraction of the templates.
In summary, the overall height of the nucleosomal barrier and the patterns of nucleosomal pausing detected during transcription of the 603 nucleosomes by T7 RNAP were similar to the ones characteristic for SP6 RNAP and yeast Pol III that utilize the Pol III-type mechanism [2,15]. Furthermore, complete nucleosomes were translocated from in front to behind the transcribing T7 RNAP, further confirming that this enzyme utilizes the Pol III-type mechanism of transcription through chromatin.
Spatial constraints could affect formation of the Ø-loop by T7 RNAP
Stable EC+49 is efficiently formed during Pol II transcription in part because there are no steric clashes between Pol II and nucleosome. Therefore we hypothesized that the structures of the EC+49 complexes formed during transcription by Pol III- and Pol II-type mechanisms could be different (possibly due to presence of steric clashes within the complexes formed during Pol III-type transcription), and this difference could cause different stabilities of the Ø-loop, different pausing and the different fates of the nucleosomes on transcription by Pol III- and Pol II-type mechanisms.
In our previous work, a complex (Pol II EC+39) similar to the critical Ø-loop-containing intermediate has been modeled using a docking approach [4]. In the current work, the intermediate containing the intranucleosomal Ø-loop (Pol II EC+49) was first modeled using a similar approach [4]. Then the Pol II structure was deleted from the complex, and the T7 polymerase elongation complex structure (PDB ID 1S77 [21]) was used to replace the Pol II structure in the Ø-loop complex. The orientation of T7 EC structure with +49 histone-DNA Ø-loop complex was determined by aligning the RNA-DNA duplex (10 bases long upstream of the catalytic site) in the active site of T7 EC with the corresponding duplex in the Pol II EC+49. The resulting structure (T7 EC+49) has been evaluated for the presence of steric clashes (Fig. 3). This analysis revealed steric clashes between the protein body of T7 RNAP and the DNA at several points: downstream of the active site, and at two regions within the transcription bubble (Fig. 3). These clashes cannot be avoided by docking T7 RNAP at different positions along nucleosomal DNA. The failure to model the T7 EC+49 Ø-loop-containing complex that does not contain steric clashes suggests that the EC+49 complexes could have different structures during transcription by Pol III- and Pol II-type mechanisms.
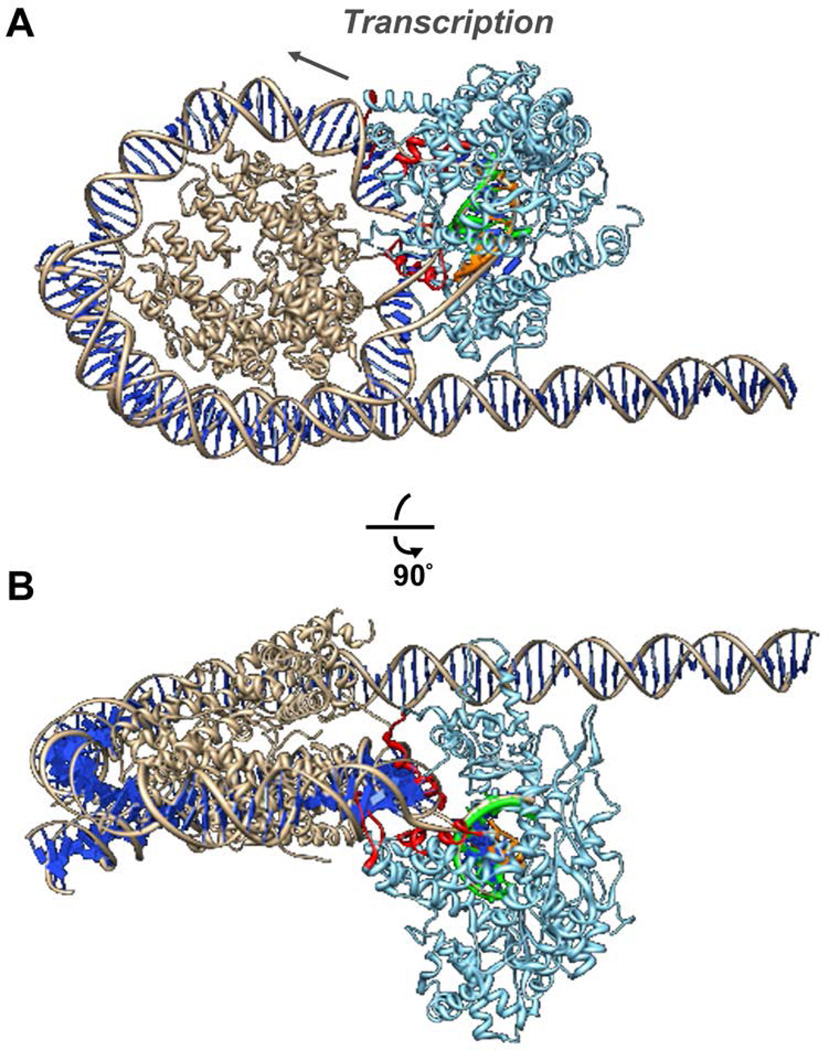
Figure 3. Structure of hypothetical intranucleosomal T7 EC+49 containing the Ø-loop: computational modeling.
A. Front and B. side views of the complex reveal steric clashes (in red) between T7 RNAP and nucleosomal DNA. The octamer and nucleosomal DNA helix (light brown), DNA bases (blue), RNA (green) and T7 RNAP (cyan) are shown. The DNA within the T7 EC is depicted in orange. The direction of transcription is indicated by arrow.
T7 RNAP does not form the Ø-loop at the position +49
The structure of T7 elongation complex stalled at the position +49 in a nucleosome (EC+49) has been analyzed by DNase I footprinting using the same 603 nucleosome positioning sequence that has been utilized previously to study the structure of EC+49 formed by yeast Pol II [4]. T7 RNAP was stalled at the position +49 on DNA or nucleosomal templates in the presence of a limited combination of NTPs (Fig. 4A). The vast majority of T7 RNAP molecules were stalled as productive complexes at the expected position on the templates, as evidenced by analysis of RNA transcripts (Fig. 4B). The T7 EC+49 complexes were homogenous and capable to continue transcription upon addition of all NTPs (Fig. 4B).
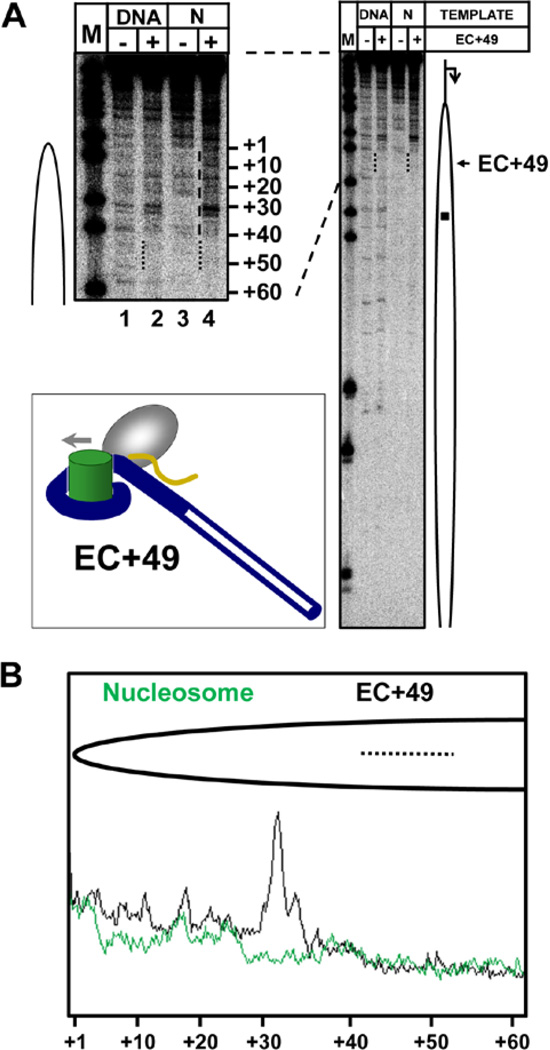
Figure 4. The intranucleosomal EC+49 formed by T7 RNAP is homogeneous and stable.
A. The experimental approach. The template allows progression and stalling of T7 ECs at −5 and +49 positions (relative to promoter-proximal boundary of the nucleosome) upon addition of different partial combinations of NTPs. B. Homogenous and active EC+49 complex is formed by T7 RNAP. T7 EC+49s were formed on the DNA or nucleosome and then extended in the presence of all NTPs. Analysis of pulse-labeled RNA by denaturing PAGE. Note that the EC+49 is functional and contains RNA, which is extendable upon addition of all NTPs.
Then the homogenous and stable T7 EC+49 was incubated in the presence of DNase I for a short time (30 seconds) to produce on average a single DNA break, and analyzed by denaturing PAGE (Fig. 5A). The representative lanes on the gel were scanned using a phosphorimager and scans were quantified (Fig. 5B). DNA in front of T7 RNAP is fully protected by the histone octamer to the extent comparable to the DNA in the original 603 nucleosome, suggesting that it is bound with the histone octamer remaining at the original position on DNA. At the same time, nucleosomal DNA upstream of the T7 EC+49 is fully accessible to DNase I, suggesting that it is largely uncoiled from the histone octamer (see the drawing in Fig. 5A insert). Thus, consistent with the results of computational modeling, T7 RNA polymerase cannot form a stable Ø-loop-containing complex on the majority of the templates. Instead, nucleosomal DNA upstream of T7 RNAP in EC+49 tends to uncoil from the octamer, producing an “open” complex that is unable to maintain nucleosome survival at the original position on DNA upon further transcription through the nucleosome.
Figure 5. DNA is uncoiled from the octamer behind T7 EC+49.
A. Analysis of the structure of T7 EC+49 formed on DNA or nucleosomal templates (N) by DNase I footprinting. Analysis of end-labeled DNA by denaturing PAGE. A part of the gel is shown at a higher magnification on the left. The footprints of the EC+49 (dotted line), the uncoiled DNA region (dashed line), the position of the active center (EC+49, arrow) and the nucleosome dyad (square) are shown. The suggested structure of the EC+49 complex formed on the 603 nucleosomal template is shown in the inset. Note that DNA behind T7 RNAP in the EC+49 is more accessible to DNase I than in the intact nucleosome, indicating its partial uncoiling from the octamer. B. Quantitative analysis of DNase I footprinting (Fig. 5A). Lanes 3 (603 nucleosome) and 4 (T7 EC+49 in the 603 nucleosome) in Fig. 5A were scanned and scans were aligned pairwise.
In summary, the structures of the EC+49 complexes formed during transcription by Pol III- and Pol II-type mechanisms through identical 603 nucleosomes are very different. In the first case, the Ø-loop is formed at this position on nucleosomal DNA, allowing recoiling of nucleosomal DNA upstream of transcribing Pol II and nucleosome survival at the original position on the template [4]. In contrast, during transcription by the Pol III-type mechanism steric clashes between T7 RNAP and nucleosomal DNA (Fig. 3) possibly prevent recoiling of nucleosomal DNA upstream of the transcribing enzyme and formation of the Ø-loop (Fig. 5). The lack of the Ø-loop formation, in turn, possibly restricts the ability of the nucleosome to survive at the original position on DNA and therefore favors the nucleosome translocation pathway (see Discussion).
DISCUSSION
In summary, the data obtained using uniquely positioned nucleosomes and structurally defined T7 RNAP suggest that this enzyme utilizes the Pol III-type pathway of transcription through chromatin. In particular, the overall height of the nucleosomal barrier, the patterns of nucleosomal pausing (Fig. 1) and the fate of nucleosome on transcription by T7 RNAP (Fig. 2) were similar to the ones characteristic for SP6 RNAP and yeast Pol III that utilize the Pol III-type mechanism of transcription through chromatin [2,15]. Computational modeling of the key intermediate determining the fate of nucleosomes on transcription (Ø-loop-containing EC+49 complex) revealed several steric clashes between the protein body of T7 RNAP and the DNA (Fig. 3), suggesting that proper Ø-loop-containing complexes cannot be formed during transcription by the Pol II-type mechanism. Finally, analysis of the structure of T7 elongation complex stalled in the position +49 in a nucleosome (EC+49) using DNase I footprinting revealed that Ø-loop cannot be formed during transcription by the Pol II-type mechanism (Figs. 4, 5). Overall, the new data suggest that the structures of the key intermediates determining the fate of a nucleosome on transcription are considerably different during transcription through chromatin by the Pol II- and Pol III-type mechanisms.
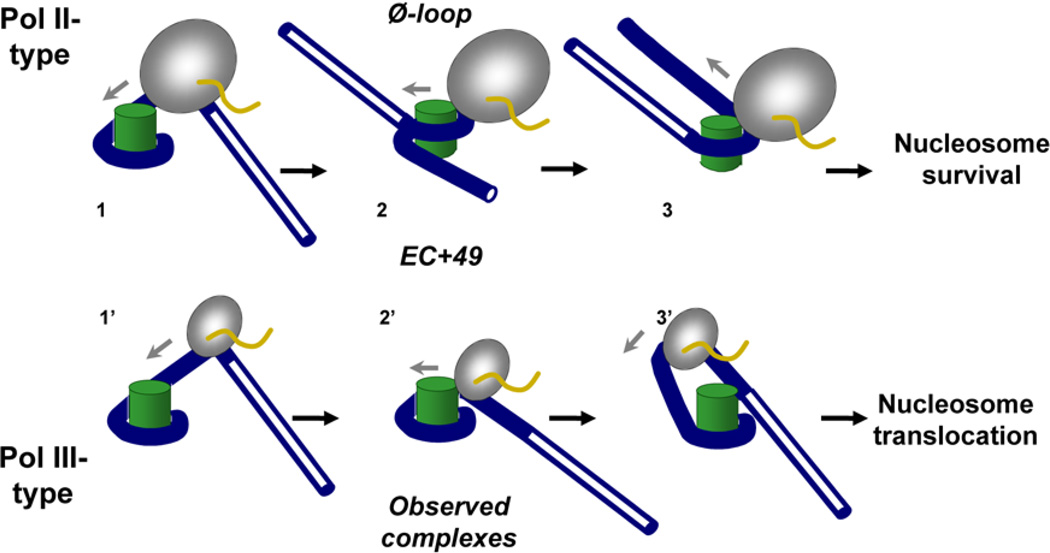
Based on our data and previous studies, we propose the following model explaining the observed relationship between the intensity of +45 pausing, Ø-loop formation and nucleosome fate during transcription by the Pol II- and Pol III-type mechanisms (Fig. 6). During transcription by the Pol II-type mechanism [4], the enzyme enters the nucleosome and partially uncoils promoter-proximal DNA from the octamer (complex 1). As Pol II approaches the +45 region, it encounters a region of strong nucleosome-specific pausing that persists before the Ø-loop is formed at the position +49 (EC+49, complex 2). Formation of the Ø-loop induces uncoiling of DNA in front of the enzyme (complex 3). As Pol II proceeds further along the template, DNA behind the enzyme is progressively re-wrapped around the histone octamer and the nucleosome is recovered at the original position of the template [4]. In contrast, during transcription by the Pol III-type mechanism DNA behind of RNAP in the EC+49 is uncoiled from the octamer and therefore does not form the Ø-loop (EC+49, complex 2’), although it remains coiled around the octamer in front of the enzyme (Fig. 5). Further transcription by the Pol III-type mechanism results in partial uncoiling of DNA in front of the enzyme and formation of a larger intranucleosomal DNA loop (complex 3’) that induces backward nucleosome translocation observed experimentally during further transcription [1, 2, 22].
Figure 6. Proposed pathways of transcription through a nucleosome by Pol II- or Pol III-type mechanisms: the structure of the EC+49 dictates the fate of the nucleosome on transcription.
As different RNAPs enter the nucleosome and pause within the +45 region (complex 1 or 1’), they form structurally different complexes at the position +49. Pol II forms a stable intranucleosomal Ø-loop-containing intermediate 2. Further transcription by Pol II results in uncoiling of the downstream nucleosomal DNA (complex 3) and nucleosome survival. In contrast, during transcription by Pol III-type mechanism DNA is uncoiled behind the enzyme in the EC+49 and the Ø-loop forms with much lower efficiency (complex 2’). In this case, further transcription would result in partial uncoiling of DNA from the octamer in front of the enzyme (complex 3’), formation of a larger intranucleosomal DNA loop and nucleosome translocation.
How does the transition between the intermediates 2’ and 3’ (Fig. 6) occur? It has been shown that during ongoing transcription by the Pol III-type mechanism DNA is partially uncoiled from the octamer immediately downstream of the transcribing enzyme [22]. Thus during transcription by Pol III-type mechanism a larger octamer surface is exposed to solution (compare complexes 1 and 1’ in Fig. 6). This observation possibly explains the higher probability of capturing upstream DNA by the open octamer surface and transfer of the histone octamer to DNA upstream of the EC during transcription by the Pol III-type mechanism, as we have proposed previously [4]. In contrast, the lack of DNA uncoiling in front of Pol II would make formation of the Ø-loop and accompanying displacement of the promoter-distal end of nucleosomal DNA more efficient and would prevent nucleosome translocation.
Typically, in our in vitro studies of different mechanisms of transcription through chromatin ([2, 3, 8] and described here), short (250–300 bp) mononucleosomal templates were used. At the same time, transcription of longer (650 to 3900 bp) templates containing a single mononucleosome by Pol II-type mechanism results in nucleosome translocation from in front to behind the transcribing enzyme [10, 12]. Thus during transcription by Pol II-type mechanism on the shorter templates nucleosomes remain at the original position on DNA; on the longer templates nucleosomes are translocated. The difference in the fates of nucleosomes on transcription of shorter and longer templates is most likely explained by differential interaction of the upstream DNA with the transiently exposed surface of the histone octamer after partial uncoiling of nucleosomal DNA during transcription [4,10]. Which of the in vitro experimental mononucleosomal models better recapitulates the in vivo scenario for the Pol II-type mechanism? On moderately transcribed genes (representing the majority of eukaryotic genes), nucleosomes are present immediately in front and behind of transcribing Pol II [25]. Therefore existence of extended histone-free DNA regions on moderately transcribed genes in vivo is unlikely, and the shorter mononucleosomal templates accurately recapitulate the mechanism of moderate-level transcription through chromatin in vitro [10].
We have previously proposed that formation of the high nucleosomal barrier is coupled with formation of the intranucleosomal Ø-loop [4, 17]. Current work provides further evidence for this concept. Indeed, the Pol III-type mechanism is characterized by formation of a lower nucleosomal barrier than the Pol II-type mechanism [2, 3, 5, 15]. Thus there is an apparent correlation between the ability to form the intranucleosomal Ø-loop and formation of a stronger nucleosomal barrier to transcription, both characteristic for the Pol II-type, but not for the Pol III-type mechanism. Therefore the structures of the key intermediates formed during transcription through the +45 nucleosomal region possibly dictate both the fate of histones and the height of the nucleosomal barrier to transcription.
CONCLUSION
In summary, our work shows that the structures of the intermediates formed during transcription by different RNA polymerases and the dynamics of coiling/uncoiling of the nucleosomal DNA from the octamer determine multiple critical outcomes of this process. For example, the Pol II-type mechanism allows survival of the original histones H3/H4 and their modifications during transcription and possible formation of regulatory nucleosomal barrier, while the Pol III-type mechanism more likely induces more extensive displacement/exchange of all core histones [4, 26]. Further studies will reveal further details and functional significance of these very fundamental biological processes.
ACKNOWLEDGEMENTS
We thank W. McAllister for plasmid expressing T7 RNAP. This work was supported by the National Institutes of Health (GM58650) to V.M.S. and Russian Foundation for Basic Research (12-04-31942) to A.K.S..
REFERENCES
- 1.Studitsky VM, Clark DJ, Felsenfeld G. Cell. 1994;76:371. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 2.Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Science. 1997;278:1960. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 3.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Mol. Cell. 2002;9:541. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 4.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Nat. Struct. Mol. Biol. 2009;16:1272. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Studitsky VM, Clark DJ, Felsenfeld G. Cell. 1995;83:19. doi: 10.1016/0092-8674(95)90230-9. [DOI] [PubMed] [Google Scholar]
- 6.Kimura H, Cook PR. J. Cell. Biol. 2001;153:1341. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiriet C, Hayes JJ. Genes Dev. 2005;19:677. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulaeva OI, Hsieh FK, Studitsky VM. Proc. Natl. Acad. Sci. USA. 2010;107:11325. doi: 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C. Nat. Struct. Mol. Biol. 2011;18:1394. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulaeva OI, Studitsky VM. Transcr. 2010;1:85. doi: 10.4161/trns.1.2.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C. Cell. 2012;151:738. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Science. 2009;325:626. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaykalova DA, Nagarajavel V, Bondarenko VA, Bartholomew B, Clark DJ, Studitsky VM. Nucleic Acids Res. 2011;39:3520. doi: 10.1093/nar/gkq1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh FK, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM. Proc. Natl. Acad. Sci. USA. 2013;110:7654. doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Mol. Cell. 2006;24:469. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Mol. Cell. 2005;18:97. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh FK, Fisher M, Ujvari A, Studitsky VM, Luse DS. EMBO Rep. 2010;11:705. doi: 10.1038/embor.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He B, Rong M, Lyakhov D, Gartenstein H, Diaz G, Castagna R, McAllister WT, Durbin RK. Protein Expr. Purif. 1997;9:142. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 19.Walter W, Kireeva ML, Tchernajenko V, Kashlev M, Studitsky VM. Methods Enzymol. 2003;371:564. doi: 10.1016/S0076-6879(03)71042-8. [DOI] [PubMed] [Google Scholar]
- 20.Protacio RU, Widom J. J. Mol. Biol. 1996;256:458. doi: 10.1006/jmbi.1996.0101. [DOI] [PubMed] [Google Scholar]
- 21.Yin YW, Steitz TA. Cell. 2004;116:393. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 22.Bednar J, Studitsky VM, Grigoryev SA, Felsenfeld G, Woodcock CL. Mol. Cell. 1999;4:377. doi: 10.1016/s1097-2765(00)80339-1. [DOI] [PubMed] [Google Scholar]
- 23.Durniak KJ, Bailey S, Steitz TA. Science. 2008;322:553. doi: 10.1126/science.1163433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAllister WT, Raskin CA. Mol. Microbiol. 1993;10:1. doi: 10.1111/j.1365-2958.1993.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 25.De Bernardin W, Koller T, Sogo JM. J. Mol. Biol. 1986;191:469. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- 26.Kulaeva OI, Hsieh FK, Chang HW, Luse DS, Studitsky VM. Biochim. Biophys. Acta. 2013;1829:76. doi: 10.1016/j.bbagrm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]