Abstract
Mitochondrial (mt) sequences are frequently used for phylogenetic reconstruction and for identification of species of molluscs. This study expands the phylogenetic range of Hygrophila (Panpulmonata) for which such sequence data are available by characterizing the full mt genome of the invasive freshwater snail Physella acuta (Physidae). The mt genome sequences of two P. acuta isolates from Stubblefield Lake, New Mexico, USA, differed in length (14,490 vs 14,314 bp) and showed 11.49% sequence divergence, whereas ITS1 and ITS2 sequences from the nuclear genome differed by 1.75%. The mt gene order of P. acuta (cox1, P, nad6, nad5, nad1, D, F, cox2, Y, W, nad4L, C, Q, atp6, R, E, rrnS, M, T, cox3, I, nad2, K, V, rrnL, L1, A, cytb, G, H, L2, atp8, N, nad2, S1, S2, nad4) differs considerably from the relatively conserved gene order within Panpulmonata. Phylogenetic trees show that the 13 protein-encoding mt gene sequences (equivalent codons) of P. acuta group according to gastropod phylogeny, yet branch lengths and dN/dS ratios for P. acuta indicate elevated amino acid substitutions relative to other gastropods. This study indicates that mt sequences of P. acuta are phylogenetically informative despite a considerable intraspecific divergence and the atypical gene order in its mt genome.
INTRODUCTION
Mitochondrial (mt) gene sequences are commonly used to reconstruct phylogenetic relationships (Boore, 1999; Valles & Boore, 2006), but obtaining entire mitochondrial genomes provides greater amounts of sequences for analysis, identification of mt gene order and discovery of novel mt gene rearrangements. Comparative mitogenomic analyses can inform on animal phylogeny (Rokas & Holland, 2000; Knudsen et al., 2006; Jex et al., 2010; Kayal et al., 2013).
The classes of Mollusca display diverse sets of mt gene orders (Kurabayashi & Ueshima, 2000; Boore, Medina & Rosenberg, 2004; Grande, Templado & Zardoya, 2008). Within the Gastropoda, a generally standard order of mt genes has been recorded in Panpulmonata (Knudsen et al., 2006; White et al., 2011), a clade established by Jörger et al. (2010). Still, the mt genomes of Panpulmonata are no exception to frequent, but minor, gene rearrangements that mainly involve modest numbers of tRNA genes but occasionally also single protein-encoding genes, as seen in Cepea nemoralis (Terrett, Miles & Thomas, 1996), Pyramidella dolabrata (Grande et al., 2008), Siphonaria gigas (White et al., 2011) and S. pectinata (Grande et al., 2008).
Our current insights are restricted by the incomplete phylogenetic coverage that is provided by the 24 panpulmonate species from which mt genomes have been sequenced completely. Panpulmonata contains the medically important clade Hygrophila; many of these freshwater snails are intermediate host for flatworm parasites and transmit infectious diseases of human and veterinary importance such as fascioliasis (Mas-Coma, Valero & Bargues, 2009), clonorchiasis and paragonimiasis (Rozendaal, 1997), cercarial dermatitis and schistosomiasis (Morgan et al., 2002). Based on phylogenetic analysis of 16S, 18S and CO1 mt gene sequences, Hygrophila was divided into five families: Acroloxidae, Chilinoidae, Planorbidae, Lymnaeidae and Physidae (Dayrat et al., 2011). Perhaps because the mt genomes of freshwater panpulmonates are considered difficult to sequence (White et al., 2011), so far complete mt genomes are available only for two families of Hygrophila: Planorbidae [Biomphalaria glabrata (DeJong, Emery & Adema, 2004) and B. tenagophila (Jannotti-Passos et al., 2010)] and Lymnaeidae [Radix balthica (Feldmeyer, Hoffmeier & Pfenninger, 2010) and Galba pervia (Liu et al., 2012)]. No mt genome sequences have previously been available for the family Physidae.
Physids are the most abundant and diverse freshwater gastropods native to North America and due to their invasive nature occur throughout the world (Burch, 1989). The phylogeny of Physidae is complex but 16S and COI mt sequences combined with morphological features have been used to reorganize taxonomy of North American physids (Wethington & Lydeard, 2007). Physella acuta (Draparnaud, 1805), frequently designated Physa acuta, is a widely used model snail that is widely distributed, readily obtainable and can be maintained with ease in the laboratory. This species serves as an aquatic biomarker due to its ability to live in polluted water (Sánchez-Argüello, Fernández & Tarazona, 2009; Lee et al., 2011), it has high salinity thresholds (Kefford & Nugegoda, 2005) and it is used in population and mating studies (Bousset et al., 2004; Dillon, Wethington & Lydeard, 2011). As an invasive species, P. acuta has been studied for competitiveness with indigenous gastropod fauna (Madsen & Frandsen, 1989; Albrecht et al., 2009). Here, we characterize the mt genome of P. acuta.
In this study, 16S and COI mt sequences (Wethington & Lydeard, 2007) are used for species identification of laboratory maintained physid snails. In addition, sequences from the nuclear genome, internal transcribed spacer (ITS)1 and ITS2, are also employed. These ITS sequences are often used for species identifications at lower taxonomic levels (Armbruster & Korte, 2006), including species identification within Hygrophila (DeJong et al., 2001; Correa et al., 2010). The mt genomes from two isolates of P. acuta (A and B) are characterized and compared. The mt genes and gene order from these physid snails are compared with those of other panpulmonates. Finally we perform a rate analysis and determine dN/dS ratios of mt protein-encoding genes of P. acuta to investigate the rate of genome evolution relative to other panpulmonates.
MATERIAL AND METHODS
Snail isolates, DNA extraction and species identification
In 2010, freshwater panpulmonate snails, morphologically identified as physids (sinistral shells, digitations on mantle collar; Paraense & Pointier, 2003) were collected from Stubblefield Lake in northern New Mexico (USA) and maintained in aquariums at room temperature. Separate lines of laboratory-cultured physid snails were initiated with hatchlings from recently deposited single egg masses that were isolated in different tanks. This approach was used to separate morphologically similar yet genetically distinct lineages (Wethington & Lydeard, 2007) and to avoid pre-existing (trematode) parasite infections in the parental snails that were collected from the field. Two separate lines of physids were established, designated as isolates A and B.
Total DNA was extracted from whole body tissues from individual snails (4–6 mm shell length) using a cetyltrimethyl-ammonium bromide (CTAB)-based method (Winnepenninckx, Backeljau & De Wachter, 1993). For taxonomic identification, PCR (AmpliTaq Gold, Applied Biosystems) was performed to amplify sequences fragments from the phylogenetically informative mt genes 16S (Palumbi et al., 1991) and COI (Folmer et al., 1994) as described by Wethington & Lydeard (2007). Primers are listed in Table 1. The complete nuclear ITS1 and ITS2 regions were amplified using the following primers: ITS1 5′TAACAAGGTTTCCGTATGTGAA3′ (Armbruster & Bernhard, 2000) and ITS2R 5′GGTTTCACGTACTCTTGAAC3′ (provided by J. Nekola, modified from that published by Wade & Mordan, 2000). Termini of ITS regions were assigned by identifying flanking ribosomal DNA gene boundaries according to DeJong et al. (2001). Thermal cycling consisted of 10 min at 94°C (initial denaturation), 25 cycles of 30 s at 94°C, 30 s at primer annealing temperatures (50°C for 16S and COI, 48°C for ITS regions) , 1 min at 72°C, and 7 min 72°C final extension. Amplicons were purified (QIAquick PCR purification Kit, Qiagen) and sequenced directly on both strands (Big Dye 3.1, Applied Biosystems). Extension products were read on an ABI 3130 automated DNA sequencer. Sequences were edited by eye and assembled into contigs using Sequencher v. 5.0 (Gene Codes Corporation). The sequences were compared with the GenBank database using BLAST (Altschul et al., 1997) for gene identification. Phylogenetic analyses of COI and 16S sequences from the P. acuta isolates were performed using neighbor joining (NJ), maximum parsimony (MP) and maximum likelihood (ML) (Gamma distribution + invariant sites) to place the experimentally-obtained nucleotide sequences in the context of separate pre-existing COI- and 16S-based phylogenies of Physidae, which also included members of Lymnaeidae and Planorbidae as outgroups (NCBI popset: 164430598 and NCBI popset: 164430551, respectively; Wethington & Lydeard, 2007) with 1,000 replicates using MEGA v. 5.05 (Tamura et al., 2011).
Table 1.
Primers used to characterize the mitochondrial genomes of Physella acuta.
| Primer (5′–3′) | 3′ Position of primer (A/B) | |||
|---|---|---|---|---|
| 1 | Pa16SF | TAAAGTGGTATTAGATCTGACGA | 10780/10598 | |
| *H3080 | ACGTGATCTGAGTTCAGACCGG | 10915/10733 | ||
| PaCYBF | GGAGATCACATACTTGCCAAGACC | 11200/11017 | ||
| (7) | PaCYBR | TCAAAAGATCTGGCGATATTAGCC | 11296/11114 | |
| 2 | ATP8JF | AATTCCATAAGTGGGGCTGAG′ | 12610/12431 | |
| ND3JR | TCTTGAAAGTGTCGTGATCCT | 13040/12608 | ||
| ATP8JFC | CCTCTTGATATACCTCTGGATCG | 13080/12902 | ||
| ND4JR (B) | ATGTCCAACTGACGAATACGC | 13986/13810 | ||
| *LCO1490 | GGTCAACAAATCATAAAGATATTGG | 38 | ||
| A_CO1JRC | AAACCTGTACCGACCAATCC | 90 | ||
| B_CO1JRC | CAAAAGCATGTGCTGTAACG | 159 | ||
| 3 | PaCO1F | GTTTGATCGGTGTTAATTACTGCA | 564 | |
| *LCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | 694 | ||
| CO1JFC | CGAGCTTATTTTACAGCAGCAAC′ | 911 | ||
| ND5JRC | GACGTGATTCCTAATCCATCTCAC | 2371 | ||
| 4 | ND5JF | TAAGGCAATGCTTTTCATGG | 2939 | |
| ND5JR | GGAATACCCATTAATGAAAGTCCAC | 3042 | ||
| ND5JFC | ATCGGTTCCGTAAACACGTC | 3249 | ||
| CO2JRC | CCTCCTGAATAGGTGATGCTG | 4701/4699 | ||
| 5 | PaCO2F | AACAAGTGCTGACGTATTGCATGC | 5127/5125 | |
| CO2JR2 | CAATGACAGGCACTAATATCTGC | 5562/5367 | ||
| A_ND4LJFC | TTTGGTGGCAGATATGTAGTGC | 5576/– | ||
| B_ND4LJFC | GCCCTGGGACTGACCTTG | –/5698 | ||
| A_ATP6JF2 | AAGCTCAAATCTTTTTGTGCAAC | 6064/5869 | ||
| 12SJRC | GTGGGGCACAAATGTAGGAC | 7428/7237 | ||
| 6 | CO3JF | GTTATGGGCCCAATAGCTTC | 7679/7469 | |
| CO3JR | ACCACGTTGGATTCTTAGCC′ | 7855/7666 | ||
| CO3JFC | CCTCAATGGCATGATGAGC | 8127/7685 | ||
| ND2JRC | GACTTTCGGGTAAAACAACAGG | 9381/9195 | ||
| 7 | ND2JF | CCTGTTGTTTTACCCGAAAGTC | 9402/9216 | |
| 16SJR2 | ATACTTTTCCCCGCTATCCAG | 10051/9863 | ||
| N2G16SJFC | CCTTTCAAATTTTGTGATAGCTG | 10053/9865 | ||
| *L2510 | CGCCTGTTTATCAAAAACAT | 10418/10232 |
Lines to the left of the primers delineate the seven overlapping long distance PCR amplicons that were cloned and sequenced to confirm data obtained by direct sequencing. Amplicons 1 and 7 overlap due to the circular nature of the mt genome, (7) indicates the end of the 7th fragment. Asterisks indicate conserved 16S and COI primers for species identification (Wethington & Lydeard, 2007).
The uncorrected p-distances (proportion of nucleotide sites at which sequences differ; Nei & Kumar, 2000) were calculated for each of 16S, COI, ITS1 and ITS2 sequences and the full length mt genome from the two isolates of P. acuta, and for several publicly-available sequences to determine and compare ranges of intra- and interspecific sequence differences. Intraspecific differences were determined for 16S sequences of P. acuta (NCBI popset: 164430551; Wethington & Lydeard, 2007) and Biomphalaria glabrata (NCBI popset: 15717799; DeJong et al., 2001), COI sequences from P. acuta (Albrecht et al., 2009), and ITS1 and ITS2 sequences from 12 species of Biomphalaria (NCBI popset: 15717841; DeJong et al., 2001). Interspecific differences among entire mt genomes or selected genes from four genera were determined for Aplysia: A. californica (GenBank: NC005827; Knudsen et al., 2006); A. dactlyomela (GenBank: NC015088; Medina et al., 2011) and A. vaccaria (GenBank: DQ991928; Medina et al., 2011); Biomphalaria: B. glabrata (GenBank: NC005439; DeJong et al., 2004) and B. tenagophila (GenBank: NC010220; Jannotti-Passos et al., 2010); Onchidella: O. borealis (GenBank: DQ991936; Medina et al., 2011) and O. celtica (GenBank: NC012376; Grande et al., 2008); and Siphonaria: S. gigas (GenBank: NC016188; White et al., 2011) and S. pectinata (GenBank: NC012383; Grande et al., 2008).
Full mitochondrial genome sequencing
Complete mt genomes were characterized from single individual snails, one each from P. acuta isolates A and B. PCR primers (Table 1) were designed and optimized using Primer3 (Rozen & Skaletsky, 2000) to target conserved regions of mt genes that were identified in alignments of previously reported complete mt genome sequences from panpulmonate species and EST data available from GenBank (Lee et al., 2011; White et al., 2011). High fidelity, long distance (LD)-PCR (Advantage Genomic LA Polymerase Mix, Clontech) was used to generate overlapping amplicons that encompassed the complete mt genome. Amplicons were sequenced directly by primer walking (see above) at double coverage or higher. Chromatograms were edited by eye and assembled into contigs using Sequencher v. 5.0. Once mt genome sequences of isolates A and B were characterized completely, primers listed in Table 1 were used to generate seven overlapping PCR fragments (range 1931–2624 bp) from the same original genomic DNA templates, which completely covered the mt genomes. High fidelity LD-PCR amplicons were cloned (TOPO TA-cloning, Invitrogen) and sequenced completely to confirm the mt sequence data.
Annotation and comparison of P. acuta mitochondrial genomes
BLAST was used to identify protein-encoding and rRNA mt genes of P. acuta isolates A and B. Gene termini were designated based on open reading frame (ORF) analyses to minimize overlap with adjacent genes, considering alternative start and stop codons, and finally checking predictions against RNA-SEQ data from P. acuta (J.R. Nolan & C.M. Adema, unpubl.). The mapping of tRNA genes was based on identification of anticodons surrounded by sequences that formed secondary structures, similar to DeJong et al. (2004). The predicted secondary structures of tRNAs were visualized with RNAviz2 (De Rijk, Wuyts & De Wachter, 2003). Codon usage was determined using MEGA v. 5.2 (Tamura et al., 2011). To predict the location of the potential origin of replication (POR), the following were considered: (1) noncoding regions >40 bp in length containing high, localized AT richness and predictive 5′ TATA sequence repeats as seen in Drosophila (Kilpert & Podsiadlowski, 2006); (2) regions with high GC skew [(G-C)/(G + C)] (Xia, 2012) using window size 2500 nt and step size 72 nt, (CGview; Stothard & Wishart, 2005); (3) POR locations as hypothesized for other panpulmonates (Grande et al., 2008; White et al., 2011). Mt genomes were depicted graphically using Artemis (Rutherford et al., 2000). The mt genomic sequences from P. acuta isolates A and B were compared for length, indels, nucleotide content and predicted amino acid composition using Sequencher v. 5.0.
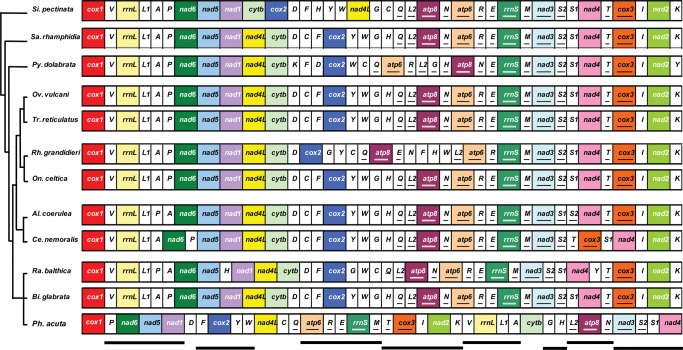
Mitochondrial gene order: P. acuta vs other panpulmonates
Starting with cox1, the order of mt genes recorded from P. acuta isolates A and B was depicted in linear fashion and aligned with mt genomes of basal and derived Panpulmonata, as inferred from 18S, 28S, 16S and CO1 sequence data (Jörger et al., 2010): S. pectinata (basal) (GenBank: NC012383; Grande et al., 2008); Salinator rhamphidia (Amphiboloidea; GenBank: NC016185; White et al., 2011); Ovatella vulcani (GenBank: NC016175) and Trimusculus reticulatus (GenBank: NC016193) (both Ellobiidae; White et al., 2011); Rhopalocaulis grandidieri (Veronicellidae; GenBank: NC016183; White et al., 2011) and O. celtica (Onchidiidae; GenBank: NC012376; Grande et al., 2008) (both Systellomatophora); Albinaria caerulea (GenBank: NC001761; Hatzoglou, Rodakis & Lecanidou, 1995) and C. nemoralis (GenBank: NC001816; Yamazaki et al., 1997) (both Stylommatophora); Pyramidella dolabrata (Pyramidellidae; GenBank: NC012435; Grande et al., 2008); and from members of two sister families of the Physidae within the Hygrophila, B. glabrata (Planorbidae; GenBank: NC005439; DeJong et al., 2004) and Radix balthica (Lymnaeidae; GenBank: HQ330989; Feldmeyer et al., 2010).
Substitution rates of mitochondrial genomes of P. acuta vs other gastropods
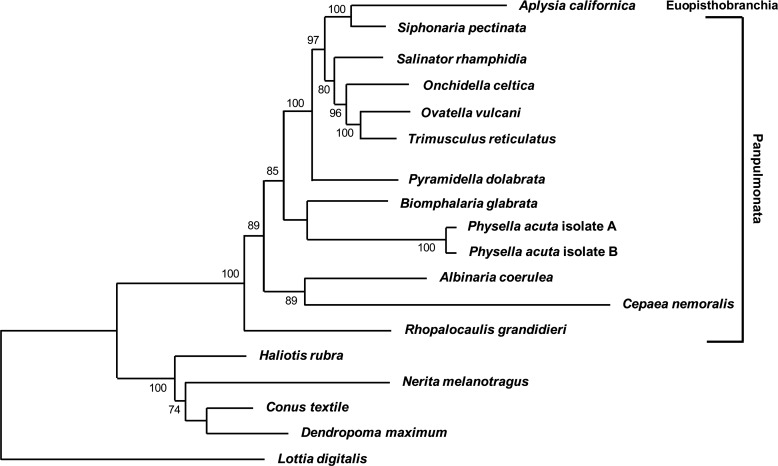
NJ, MP and ML analyses were performed to investigate the phylogenetic relationship of P. acuta with other gastropods and to determine branch lengths as a measure of divergence. Complete nucleotide sequences for protein-encoding genes were obtained from Genbank for the panpulmonates listed above. The phylogenetic range was expanded by also including sequences from Aplysia californica (GenBank: NC005827; Knudsen et al., 2006), a euopisthobranch, and basal outgroups Dendropoma maximum (GenBank: NC014583; Rawlings et al., 2010), Conus textile (GenBank: NC008797; Bandyopadhyay et al., 2008), Haliotis rubra (GenBank: NC005940; Maynard et al., 2005), Nerita melanotragus (GenBank: GU810158; Castro & Colgan, 2010) and Lottia digitalis (GenBank: NC007782; Simison, Lindberg & Boore, 2006). The mt genome sequence of the lymnaeid R. balthica was not used because of the low quality of the 454-reads with respect to length of mononucleotide tracts (Feldmeyer et al., 2010). The protein-encoding gene sequences of the 16 gastropods and P. acuta isolates A and B were individually translated, aligned and cropped by hand to remove highly divergent, nonalignable gap-columns using COBALT (Papadopoulous & Agarwala, 2007) and Bioedit (Hall, 1999). Gene sequences were then concatenated for each gastropod and phylogenetic analyses were performed with MEGA v. 5.05, using integrated utilities for selection of model as best fit for the data. Phylogenetic NJ (Jones-Taylor-Thornton model with gamma distributed rate variation among sites), MP (10 initial trees by random addition) and ML (WAF + F + G, 5 gamma categories) analyses were performed with 1,000 bootstrap replicates.
The relative rate test (Tajima, 1993) was performed in MEGA v. 5.05 to test the mt genomes of P. acuta for accelerated nucleotide and amino acid substitution rates relative to B. glabrata (with Pyramidella dolabrata as an outgroup) using aligned sequences with gaps removed.
The GA-Branch program was used through the Datamonkey portal (Kosakovsky, Pond & Frost, 2005) to identify terminal branches with significantly different dN/dS ratios in the gastropod ML tree. The dN/dS ratios were generated from 12 of the 13 protein-encoding gene sequences from the two isolates of P. acuta along with selected gastropods to investigate substitution rates of P. acuta compared to other gastropods. Due to short length of alignable codons, atp8 was excluded in dN/dS analyses. The gastropod species A. californica, C. nemoralis and N. melanotragus appeared to have undergone rate acceleration and were excluded from this analysis. The nucleotide sequences of individual protein-encoding genes were translated, aligned and gap columns were removed to analyse dN/dS ratios for each gene and also for the concatenated gene sequences to identify amino acid substitutions across the mt genomes as a whole.
RESULTS
Species identification
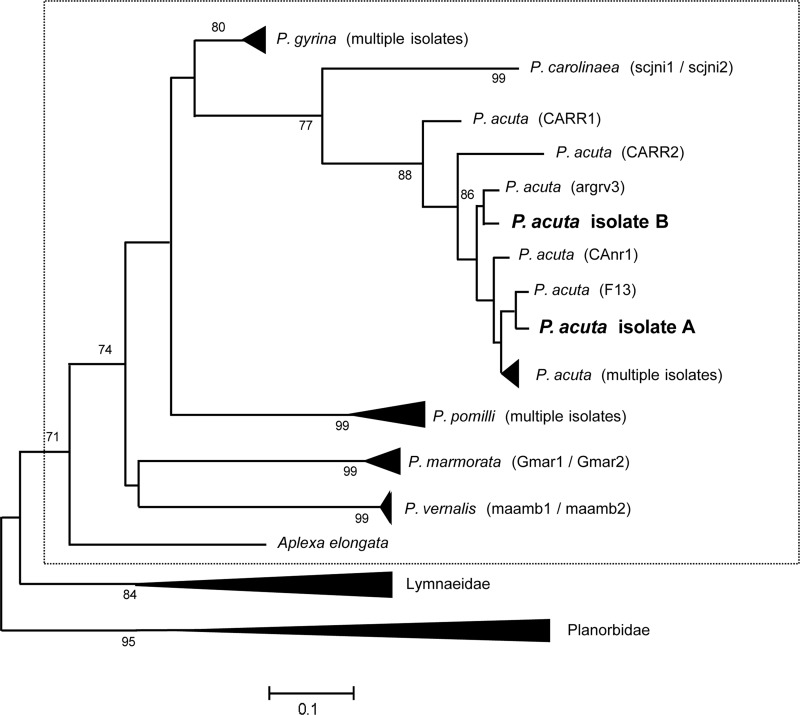
The initial morphology-based identification as physid snails was confirmed when BLAST searches revealed greatest similarities of sequence from both isolates A and B to GenBank entries of ITS1, ITS2, 16S and COI from Physella acuta. The ITS1 sequences (A: 495 bp, GenBank: KF316327; B: 497 bp, GenBank: KF316329) differed at 10 nucleotide (nt) positions and ITS2 sequences (A: 301 bp, GenBank: KF316328; B: 302 bp, GenBank: KF316326) differed by 4 nt (for alignments see Supplementary material Tables S1 and S2). The combined ITS regions differed in nt sequence by 1.75% between P. acuta A and B. This value falls within the intraspecific divergence for the combined ITS fragments of 12 different Biomphalaria species, ranging from 0% (B. alexandrina) to 2.70% (B. glabrata) (DeJong et al., 2001). The mt genome-derived sequences (GenBank accession numbers in following paragraph) from isolates A and B showed a higher divergence. The amplicons from the 16S genes were 496 bp (A) and 500 bp (B) with a 5.38% nt difference evident from the sequence alignment (length 502 bp with gaps). The COI sequence fragments, 655 bp for both isolates, displayed a 4.27% nt difference. The sequences were confirmed from sibling snails of both isolates. Combining both 16S and COI, the total sequence difference was 4.75% over 1151 bp. Based on a threshold of <6% difference in these combined sequences, as defined by Wethington & Lydeard (2007), both isolates are representatives of the species P. acuta. This divergence between mt sequences of P. acuta A and B is less than the maximum intraspecific divergence calculated at 7.0% for 16S sequences from B. glabrata (DeJong et al., 2001) and at 11.9% from COI sequences reported for P. acuta elsewhere (Albrecht et al., 2009). Accordingly, analysis of the COI sequences relative to a previously reported phylogeny of physid snails (Wethington & Lydeard, 2007) placed isolates A and B within the clade of P. acuta, with the two isolates representing separate genetic lineages of the species (Fig. 1). Similar results were obtained with 16S sequences (data not shown).
Figure 1.
Phylogenetic placement of Physella acuta isolates A and B within Physidae. Experimentally derived sequences were incorporated into NCBI popset 164430598 (Wethington & Lydeard, 2007) COI sequences from snails of the family Physidae (boxed) to generate a ML tree; NJ and MP yielded the same results. Original identifiers of strains or isolates of P. acuta are indicated in brackets. The outgroup includes sequences from Lymnaeidae and Planorbidae. Isolates A and B, which coexist in Stubblefield Lake, NM (bolded), cluster with different clades of P. acuta. The tree has been simplified for clarity, bootstrap values are indicated from 1,000 replicates.
General features of mitochondrial genome of P. acuta
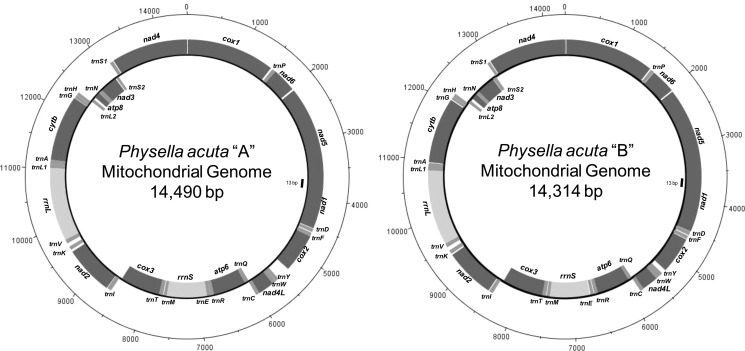
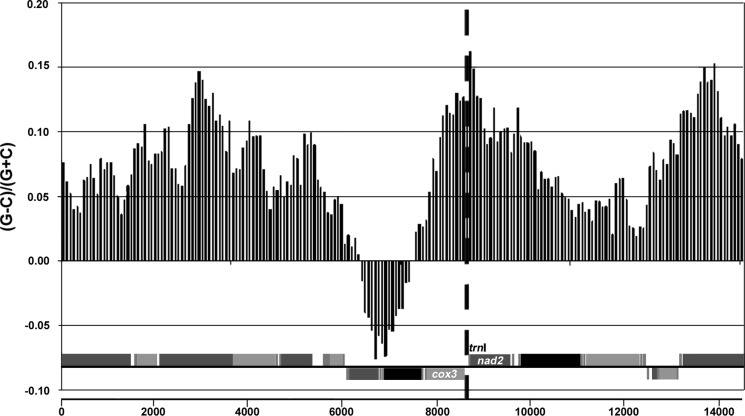
The complete mt genomes of isolates A and B were characterized (A: GenBank: JQ390525; B: GenBank: JQ390526) and while they differed considerably in sequence composition (see below), the following features are held in common. Physella acuta has the standard metazoan complement of mt genes consisting of 13 protein-encoding genes, 2 ribosomal RNA (rRNA) genes and 22 transfer RNA (tRNA) genes. The genomes have an AT-content of 69.22% for isolate A and 69.69% for isolate B. The mt gene order is as follows: cox1, P, nad6, nad5, nad1, D, F, cox2, Y, W, nad4L, C, Q, atp6, R, E, rrnS, M, T, cox3, I, nad2, K, V, rrnL, L1, A, cytb, G, H, L2, atp8, N, nad3, S2, S1 and nad4 (Fig. 2). The underlined genes are located on the negative strand of the circular genome. Intergenic regions are evident but the genes are generally spaced closely together. The protein-encoding gene nad4 has an incomplete stop codon (T_ _); inspection of cDNA transcripts confirmed that this stop codon is completed by mRNA polyadenylation (not shown). Several genes overlap partially; nad5 and nad1 overlap by 13 bp, nad4L and trnC by 2 bp, trnY and trnW by 7 bp, trnL1 and trnA by 4 bp, and finally trnC and trnQ (two tRNAs that are located on opposite strands) overlap by 6 bp. The location of the potential origin of replication (POR) is predicted in the intergenic region between cox3 and trnI, upstream of nad2. This is one of the largest intergenic regions, 45/48 bp with 84.1%/87.5% AT-richness (P. acuta isolate A/B, respectively) and contains predictive 5′ TATA sequence repeats. Additionally, this intergenic region is near a change from low to high G/C skew (Fig. 3), as measured using the method of Xia (2012), showing average values over sequence intervals of 2,500 nt along the mt genome (step size 72), and it has been predicted to contain the POR for other panpulmonates (Grande et al., 2008; White et al., 2011).
Figure 2.
The mitochondrial genomes of Physella acuta isolates A and B. The outer circle represents the positive strand, the inner circle the negative strand. Protein-encoding genes are darkened to distinguish from rRNA genes. Bars (with length in bp) indicate location of sequence overlap between protein-encoding genes. Note the size difference of the mt genomes of the two P. acuta isolates, especially the indel beginning in cox2 following the intergenic region upstream of trnY.
Figure 3.
Potential origin of replication (POR), location by GC skew analysis. GC skew [(G − C)/(G + C)] ratios plotted in a bar graph relative to a linear representation of the mt genome of P. acuta (isolate A shown). Positive values indicate greater G content and negative values indicate increased C content. The vertical dotted line indicates the predicted location of the POR; note the GC skew maximum at 0.162 that further supports this prediction. This high peak is the origin of the sequence interval (window size 2,500 nt) with the highest GC skew, transitioning from low GC skew upstream (Xia, 2012). Shading of protein-encoding and RNA genes as in Figure 2.
Differences between mitochondrial sequences of P. acuta isolates A and B
The mt genomes from isolates A and B of the same species P. acuta are dissimilar in both size (14,490 vs 14,314 bp) and in sequence content. With the exception of the tRNAs I, M, and P, every other mt gene homologue differed in sequence composition and/or size (Supplementary material Table S3). The intergenic regions range from 1–226 bp in length, with the latter only recorded from isolate A. The nucleotide composition of the mt genome sequence from the two isolates differ by 9.92% (1,416 nt in 14,275 bp), gaps excluded, this value increases to 11.49% (1,670 nt in 14,529 bp) with the inclusion of indel positions.
A total of 37 indels contribute to the size difference of the two mt genomes. A 193 bp indel occurs in the intergenic region between cox2 and trnY; the 3′ coding region of the cox2 gene of isolate A contains a 39 bp extension followed by a 154 bp addition to the noncoding region between cox2 and trnY. No indels created frame shifts within protein-encoding gene sequences. Further indels contributed one additional amino acid codon (3 bp) in each of isolate A's atp6 and nad1, an additional one bp in rrnS of isolate A, and an additional nine bp in rrnL of isolate B. The remaining indels occur in intergenic regions.
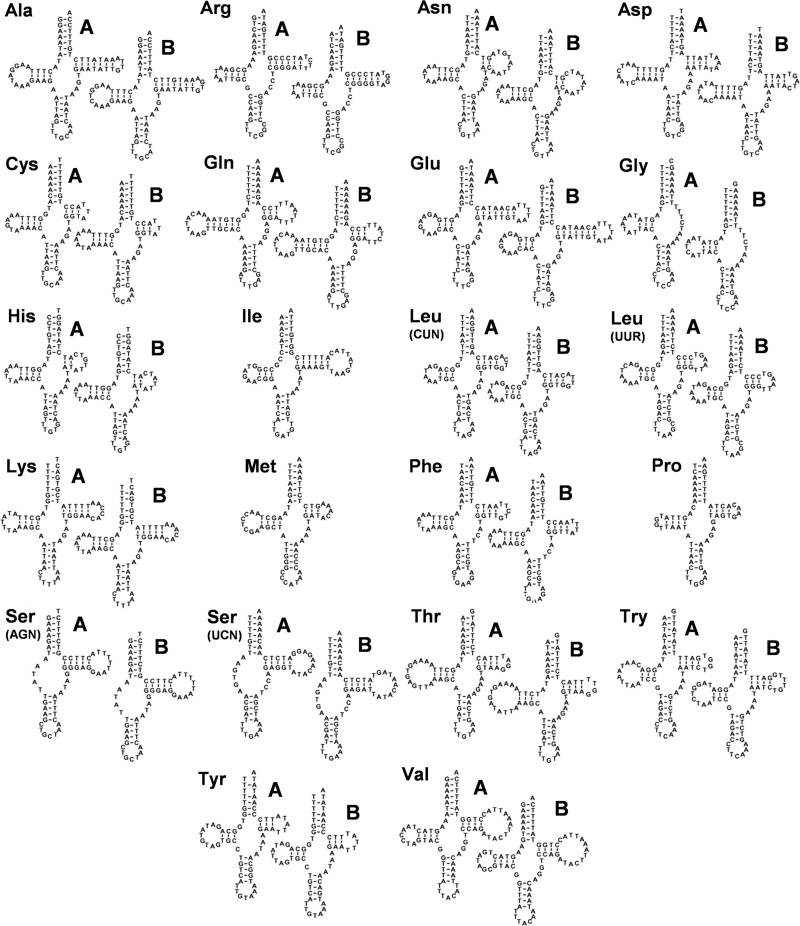
Differences in sequence composition occur in 19 of the 30 intergenic regions, both rRNAs, and in 19 of the 22 tRNAs. The nt substitutions between the tRNAs from the isolates A and B typically affect the loops and rarely the stems of the predicted clover-leaf structures (Fig. 4). The protein-encoding genes between the two isolates have a broad range of synonymous and nonsynonymous nt substitutions (Supplementary Material Table S3). Nucleotide sequence differences ranged from 5.26% (atp8) to 29.75% (nad4L). This affected overall codon usage, with the greatest difference recorded for Leucine (L1): CUA was the dominant codon in isolate A vs CUU in isolate B (Supplementary Material Table S4), but this was not significant (CUA χ2 = 0.087, P = 0. 77; CUU χ2 = 0.98, P = 0.32). Additionally, (alternative) start codons and stop codons vary between atp8, cox2 and cytb gene homologues. The amino acid substitutions ranged from 0.59% (cox1) to 25.81% (nad4L). With the exception of cox2 (increased length due to indel), the similarity of protein sequences of P. acuta A and B was ≥90% due to a majority of synonymous replacements (Supplementary Material Table S3).
Figure 4.
Physella acuta isolates A and B: tRNA sequence and structure. Predicted secondary structures of the 22 tRNAs encoded in the mt genomes from P. acuta isolates A and B. Only three tRNA genes are identical between isolates A and B. Two graphical representations are shown for all tRNA genes that differ in sequence between isolates A and B. Typically such differences occurred in the loops, not the stems. Three letter codes identify the amino acid anticodon specificity. Irregular tRNAs are Gly, Ser (AGN) and Ser (UCN).
The 11.49% overall intraspecific divergence at nt level of complete mt genomes of P. acuta A and B exceeds that of two strains of B. glabrata (18 of 13,670 nt or 0.13%; uncorrected p-distance). This divergence is comparable to interspecific difference from total mt genome sequences among additional species within either the genus Aplysia or the genus Biomphalaria; however, it did not exceed the interspecific sequence differences from species within the genera Onchidella and Siphonaria. Regardless of the high intraspecific divergence, P. acuta is distinct from other genera. A direct comparison of the cox1 gene sequences from P. acuta isolate A compared with B. glabrata (representing the sister taxon), yielded over 20% sequence divergence between genera.
Mitochondrial gene order: P. acuta vs Panpulmonata
The mt gene order of P. acuta is novel compared to the rather standard gene order that has been recorded from other panpulmonates (Fig. 5). Despite the rearrangements evident from P. acuta, the coding directionality on the positive or negative strand is identical for gene homologues of all the panpulmonates. In addition, several groups of genes that occur adjacent in the mt genome of P. acuta were designated as gene clusters because identical groups of genes are present (in different order) in the mt genomes of other panpulmonates. The rearranged mt gene order of P. acuta may have resulted from processes that have retained several gene clusters (see Supplementary Material Table S5 for a scenario involving segmental duplications and deletions of gene duplicates that may explain the origin of the rearranged gene order in the mt genome of P. acuta).
Figure 5.
Linear alignment of mt gene order of Physella acuta compared with other panpulmonate gastropods. Phylogenetic relationships shown are based on analysis of 18S, 28S, 16S and COI sequences (Jörger et al., 2010). Protein-encoding and rRNA genes are colour coded to emphasize patterns and gene rearrangements. Single letters designate tRNA genes. The genes encoded on the negative strand (underlined) are the same for all species shown. Bold lines under the alignment delineate clusters of genes with the same internal order for the majority of the panpulmonates. Note that gene rearrangements among the panpulmonates are modest and rarely affect protein-encoding gene orders, with the exception of P. acuta. Alignment is not to scale.
Rate of mutation of mitochondrial genome of P. acuta
The ML tree of the equivalent amino acids predicted from equivalent codons of protein-encoding genes of the mt genomes of P. acuta and other selected gastropods is similar to published phylogenies (Fig. 6) (Grande et al., 2008; Klussman-Kolb et al., 2008; Jörger et al., 2010; Dayrat et al., 2011; White et al., 2011). NJ and MP analyses (not shown) yielded similar results. Briefly, the NJ tree showed the hygrophilid species B. glabrata and P. acuta as adjacent branches (low support), while the MP and ML analyses both showed B. glabrata as sister group to P. acuta (low support). The long branch lengths for P. acuta relative to most other clades (ML), especially close phylogenetic neighbours, is indicative of a higher substitution rate in the mt genomes of P. acuta.
Figure 6.
Phylogenetic analysis of selected gastropods. Representative gastropods with fully characterized mt genomes were selected to compare protein-encoding genes with those of Physella acuta. Optimized alignments of amino acid sequences of all 13 protein-encoding genes were concatenated for ML analysis (1,000 bootstrap replicates). Longer branch lengths indicate increased mutation rates of amino acid sequences across the mt genome. Note the relatively long branch of the Physella clade.
The relative rate analysis showed a highly significant acceleration in both nt (not shown) and amino acid substitutions in the mt genomes of P. acuta relative to B. glabrata (isolate A χ2 = 38.01, isolate B χ2 = 30.82, P < 0.000001 for each).
The dN/dS ratios for the terminal branches (Table 2) from the ML tree across the protein-encoding genes identified a significant increase of amino acid substitutions in P. acuta (0.091) as compared to other gastropods (0.019). Increased dN/dS values for individual genes were recorded for cox2, nad1, nad2, nad4, nad5, and (isolate B only) nad6, but not all were significant (see Table 2). The remaining protein-encoding genes had equivalent dN/dS ratios relative to other gastropods. Note that the cox1 of isolate A was the only gene with a lower dN/dS ratio as compared to other gastropods. Gene relocations resulting from putative gene rearrangements did not appear to associate with altered dN/dS ratios of particular genes of P. acuta as compared to other gastropods (Table 2).
Table 2.
Nonsynonymous per synonymous (dN/dS) substitution ratios, comparing Physella acuta with other gastropods (see Material and Methods).
| Gene sequence | dN/dS ratio |
|
|---|---|---|
| Other gastropods | P. acuta A/B | |
| Concatenated gene set* | 0.019 | 0.091 |
| cox1 | 0.011 | 0.006/0.011 |
| nad6 | 0.101 | 0.101/0.239 |
| nad5* | 0.016 | 0.091/0.074 |
| nad1* | 0.026 | 0.122 |
| cox2 | 0.042 | 0.076 |
| nad4L | 0.049 | |
| atp6 | 0.030 | |
| cox3 | 0.022 | |
| nad2* | 0.033 | 0.216 |
| cytb | 0.035 | |
| atp8 | n.d. | |
| nad3 | 0.082 | |
| nad4 | 0.059 | 0.139 |
The dN/dS ratios were calculated for individual and concatenated mt protein-encoding gene sequences with the exception of atp8. Single ratios indicate no difference between other gastropods and P. acuta isolates. Different ratios from isolates of P. acuta are separated by a slash; n.d. indicates not done. Genes with significantly different (P < 0.001) dN/dS ratio between the P. acuta isolates and the other gastropods are indicated by an asterisk.
DISCUSSION
The characterization of the mt genome of P. acuta revealed (1) considerable intraspecific differences in length and sequence composition, (2) a novel gene order that is unique among panpulmonates and (3) elevated substitution rates in protein-encoding genes compared with mt genomes of other gastropods.
The sequence data (ITS1, ITS2, 16S and COI) obtained from the physid snails collected from Stubblefield Lake identified isolate A and B as the same species, Physella acuta. The isolate-specific differences between the sequences that were analysed fell within the ranges of considerable intraspecific divergence that are routinely recorded from phylogenetic studies that employ such genes of other snail species (Thomaz, Guiller & Clarke, 1996; Stothard & Rollinson, 1997; DeJong et al., 2001; Dillon & Frankis, 2004; Armbruster & Bernhard, 2000; Nekola, Coles & Bergthorsson, 2009; Albrechts et al., 2009; Wethington, Wise & Dillon, 2009).
For P. acuta, the levels of intraspecific divergence were different for the nuclear ITS sequences (>98% identity) vs the mitochondrial 16S and COI sequences (95.25% identity). Differences in 16S and COI gene sequences between the two isolates did not exceed the 6% difference suggested to delineate separate species of Physella (Wethington & Lydeard, 2007). Additionally, phylogenetic reconstruction placed isolates A and B within the P. acuta clade, but as separate genetic lineages (Fig. 1). The characterization of the complete mt genomes revealed additional extensive differences in sequence and length that further increased the mt nucleotide divergence between P. acuta A and B to 11.49%. Based upon the limited number of reports available for such comparison of complete mt genomes, this level exceeds the intraspecific divergence of B. glabrata (DeJong et al., 2004) and it is more within the range of interspecific divergence within the genera Biomphalaria and Aplysia (DeJong et al., 2004; Knudsen et al., 2006; Jannotti-Passos et al., 2010; Medina et al., 2011). The observation in one gastropod species of minimal intraspecific differences in nuclear sequences combined with elevated divergence of mt sequences is not novel. Additional to P. acuta, another instance was reported for the slug Arion subfuscus (Stylommatophora) with mean pairwise sequence divergence of 21% for 16S and 0.3% for ITS1 (Pinceel, Jordaens & Backeljau, 2005). Dramatic intraspecific differences occur in some bivalve molluscs where doubly uniparental inheritance (DUI) of maternally (F genome) and paternally (M genome) transmitted mitochondrial genomes differ in size, gene order and sequence (Doucet-Beaupré et al., 2010). However, this does not apply here; P. acuta belongs to a different molluscan class and is a simultaneous hermaphrodite. Thomaz et al. (1996) proposed that intraspecific variance of mt genomes may stem from (1) rapid mt evolution, (2) sequence divergence in previously isolated populations, (3) selection acting to generate and maintain variability and (4) unusually structured or large populations. Thus, it is not unexpected that intraspecific divergence has developed in the mt genome of a globally invasive species with complex genetic population structures that are capable of reproduction by selfing such as P. acuta (Escobar, Nicot & David, 2008; Albrecht et al., 2009). The occurrence of variant mt genomes in P. acuta may result from putative mitochondrial introgression (Ballard & Whitlock, 2004), but more data are needed to test this hypothesis. These considerations and the findings in this study suggest that it may be informative for molecular sequence-based taxonomic identification of snails to employ combined analyses of sequences encoded by both mitochondrial and nuclear genomes.
A standard ancestral gene pattern has been postulated for molluscan mt genomes (Ki et al., 2010), but frequent and extensive rearrangements have led to highly diverse patterns of gene order in mt genomes across the phylogeny of molluscs (Boore et al., 2004; Grande et al., 2008). The mt genomes of Panpulmonata, however, display a relatively standard gene order with modest variations in the relative positions of tRNA genes and only rarely of protein-encoding genes (Kurabayashi & Ueshima, 2000; Knudsen et al., 2006; Grande et al., 2008). In light of the apparent standard gene pattern it was surprising that the gene order of the mt genome of P. acuta differed radically from that of phylogenetically close relatives within the Panpulmonata (Fig. 5). It remains unclear what mechanisms underlie the rearrangements of the mt genomes in this group (Grande et al., 2008; White et al., 2011), but the analysis of the mt gene order of P. acuta relative to the standard panpulmonate genome favours a combination of segmental duplication and selective deletion of supernumerary genes (Kurabayashi & Ueshima 2000; Knudsen et al., 2006; Grande et al., 2008) (Fig. 5; Supplementary Material Table S5). Despite extensive gene rearrangements, the mt genome of P. acuta still reflects the common mt gene order shared by many Panpulmonata. The directionality of gene homologues is the same and complements of genes that are encoded on either the H and L strands are identical. Several clusters of genes with the same relative internal positions as seen in other panpulmonates were identified in the divergent gene pattern of the mt genome of P. acuta. As proposed for other panpulmonates (Grande et al., 2008; White et al., 2011), the location of the POR of P. acuta is predicted in the intergenic region between cox3 and trnI (Fig. 3), within a gene cluster that remained undisturbed during the rearrangements. This suggests that rearrangements involved groups of genes (segments of the mt genome) rather than individual genes.
Additional differences in the mt genome of P. acuta vs other panpulmonates are the longer branch lengths (Fig. 6), accelerated amino acid substitutions (relative rate test) and increased substitution rates (Table 2). These are indications that the mt genome of P. acuta is evolving faster than those of several other gastropods. Nevertheless, phylogenetic analysis performed with concatenated protein-encoding gene sequences place P. acuta in the clade Hygrophila (Fig. 6). This is in agreement with other phylogenetic trees based on mt and nuclear DNA sequences (Grande et al., 2008; Klussman-Kolb et al., 2008; Jörger et al., 2010; Dayrat et al., 2011; White et al., 2011). A number of processes may account for increased branch lengths and the increased dN/dS rates: (1) increased substitution rates that create a spectrum of mutations which may generate increased amino acid replacements, (2) relaxation of selection which could allow for an increase in the number of substitution sites, (3) mechanistic flaws in replication and/or mismatch DNA repair, (4) population effects such as bottlenecking or reproduction, especially because P. acuta is a simultaneous hermaphrodite that is capable of selfing (Neiman et al., 2010). Finally, increased substitutions rates may be explained by genome rearrangements via genome duplication and selective loss of genes.
In summary, two isolates of P. acuta that appear side by side in the Stubblefield Lake in New Mexico have highly similar ITS1 and ITS2 sequences, yet display high mt sequence divergence and differ considerably in length of their mt genomes. Few studies provide the entire mt genome from multiple individuals of the same species among Gastropoda, but none of these match the intraspecific sequence divergence of entire mt genome sequences as seen within P. acuta. The physid snails have an mt gene order that is strikingly different from the relatively conserved pattern previously described from within panpulmonates and phylogenetic analysis indicates overall elevated substitution rates, yet phylogenetic placement of P. acuta remains within Hygrophila (Panpulmonata). The mt genomes from P. acuta may be used in future studies of topics such as intra- and interspecific sequence divergence, genome evolution and establishing phylogeny aided by gene rearrangements. We conclude that White et al. (2011) correctly assumed that with increased mt genomes being sequenced, there would be increased detection of gene rearrangements. Also, Boore (1999) validly cautioned against interpretation of phylogenetic relationships solely based on mt gene rearrangements within Mollusca due to the phylum's myriad of gene rearrangements, which are not connected with any type of molecular clock. It appears that P. acuta provides an intriguing example of the diversity of mt genomes within Mollusca.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at Journal of Molluscan Studies online.
ACKNOWLEDGEMENTS
Dr Sara V. Brant (CETI, University of New Mexico UNM) provided the field collected physid snails. Dr Jeffrey C. Nekola (UNM) provided helpful discussion and contributed primers. J.R.N. was supported in part by PREP (NIH R25 GM075149). C.M.A. acknowledges support from NIH grant number P20GM103452. U.B. was supported by NSF grant number DEB-0952342. Sequencing was performed at the Molecular Biology Facility and supported in part by NIH grant number P20GM103452 from the National Institute of General Medical Sciences.
REFERENCES
- ALBRECHT C., KROLL O., MORENO-TERRAZAS E., WIEKE T. Invasion of ancient Lake Titicaca by globally invasive Physa acuta (Gastropoda: Pulmonata: Hygrophila) Biological Invasions. 2009;11:1821–1826. [Google Scholar]
- ALTSCHUL S.F., MADDEN T.L., SCHÄFFER A.A., ZHANG J., ZHANG Z., MILLER W., LIPMAN D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARMBRUSTER G.F.J., BERNHARD D. Taxonomic significance of ribosomal ITS-1 sequence markers in self-fertilizing land snails of Cochlicopa (Stylommatophora, Cochilcopidae) Mitteilungen aus dem Museum für Naturkunde in Berlin, Zoologische Reihe. 2000;76:11–18. [Google Scholar]
- ARMBRUSTER G.F.J., KORTE A. Genomic nucleotide variation in the ITS1 rDNA spacer of land snails. Journal of Molluscan Studies. 2006;72:211–213. [Google Scholar]
- BALLARD J.W.O., WHITLOCK M.C. The incomplete natural history of mitochondria. Molecular Ecology. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- BANDYOPADHYAY P.K., STEVENSON B.J., OWNBY J.-P., CADY M.T., WATKINS M., OLIVERA B.M. The mitochondrial genome of Conus textile, coxI-coxII intergenic sequences and conoidean evolution. Molecular Phylogenetics and Evolution. 2008;46:215–223. doi: 10.1016/j.ympev.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOORE J.L. Animal mitochondrial genomes. Nucleic Acids Research. 1999;27:1767–1280. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOORE J.L., MEDINA M., ROSENBERG L.A. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Molecular Biology and Evolution. 2004;21:492–1503. doi: 10.1093/molbev/msh090. [DOI] [PubMed] [Google Scholar]
- BOUSSET L., HENRY P.Y., SOURROUILLE P., JARNE P. Population biology of the invasive freshwater snail Physa acuta approached through genetic markers, ecological characterization and demography. Molecular Ecology. 2004;13:2023–2036. doi: 10.1111/j.1365-294X.2004.02200.x. [DOI] [PubMed] [Google Scholar]
- BURCH J.B. North American freshwater snails. Hamburg, MI: Malacological Publications; 1989. [Google Scholar]
- CASTRO L.R., COLGAN D.J. The phylogenetic position of Neritimorpha based on the mitochondrial genome of Nerita melanotragus (Mollusca: Gastropoda) Molecular Phylogenetics and Evolution. 2010;57:918–923. doi: 10.1016/j.ympev.2010.08.030. [DOI] [PubMed] [Google Scholar]
- CORREA A.C., ESCOBAR J.S., DURAND P., RENAUD F., DAVID P., JARNE P., POINTIER J.-P., HURTREZ-BOUSSÈS S. Bridging gaps in the molecular phylogeny of the Lymnaeidae (Gastropoda: Pulmonata), vectors of Fascioliasis. BMC Evolutionary Biology. 2010;10:381. doi: 10.1186/1471-2148-10-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAYRAT B., CONRAD M., BALAYAN S., WHITE T.R., ALBRECHT C., GOLDING R., GOMES S.R., HARASEWYCH M.G., MARTINS A.M. Phylogenetic relationships and evolution of pulmonate gastropods (Mollusca): new insights from increased taxon sampling. Molecular Phylogenetics and Evolution. 2011;59:425–437. doi: 10.1016/j.ympev.2011.02.014. [DOI] [PubMed] [Google Scholar]
- DE RIJK P., WUYTS J., DE WACHTER R. RnaViz2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- DEJONG R.J., EMERY A.M., ADEMA C.M. The mitochondrial genome of Biomphalaria glabrata (Gastropoda: Basommatophora), intermediate host of Schistosoma mansoni. Journal of Parasitology. 2004;90:991–996. doi: 10.1645/GE-284R. [DOI] [PubMed] [Google Scholar]
- DEJONG R.J., MORGAN J.A.T., PARAENSE W.L., POINTIER J.-P., AMARISTA M., AYEH-KUMI P.F.K., BABIKER A., BARBOSA C.S., BRÉMOND P., CANESE A.P., PEREIRA DE SOUZA C., DOMINGUEZ C., FILE S., GUTIERREZ A., INCANI R.N., KAWANO T., KAZIBWE F., KPIKPI J., LWAMBO N.J.S., MIMPFOUNDI R., NJIOKOU F., PODA J.N., SENE M., VELÁSQUEZ L.E., YONG M., ADEMA C.M., HOFKIN B.V., MKOJI G.M., LOKER E.S. Evolutionary relationships and biogeography of Biomphalaria (Gastropoda: Planorbidae) with implications regarding its role as host of the human Bloodfluke, Schistosoma mansoni. Molecular Biology and Evolution. 2001;18:222–239. doi: 10.1093/oxfordjournals.molbev.a003769. [DOI] [PubMed] [Google Scholar]
- DILLON R.T., JR., FRANKIS R.C. High levels of mitochondrial DNA sequence divergence in isolated populations of freshwater snails of the genus Goniobasis Lea, 1862. American Malacological Bulletin. 2004;19:69–77. [Google Scholar]
- DILLON R.T., JR., WETHINGTON A.R., LYDEARD C. The evolution of reproductive isolation in a simultaneous hermaphrodite, the freshwater snail Physa. BMC Evolutionary Biology. 2011;11:144. doi: 10.1186/1471-2148-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOUCET-BEAUPRÉ H., BRETON S., CHAPMAN E.G., BLIER P.U., BOGAN A.E., STEWART D.T., HOEH W.R. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evolutionary Biology. 2010;10:50. doi: 10.1186/1471-2148-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRAPARNAUD J.P.R. Histoire naturelle des mollusques terrestres et fluviatiles de la France. Montpellier, Paris: 1805. VII Physe Physa. [Google Scholar]
- ESCOBAR J.S., NICOT A., DAVID P. The different sources of variation in inbreeding depression, heterosis and outbreeding depression in a metapopulation of Physa acuta. Genetics. 2008;180:1593–1608. doi: 10.1534/genetics.108.092718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FELDMEYER B., HOFFMEIER K., PFENNINGER M. The complete mitochondrial genome of Radix balthica (Pulmonata, Basommatophora), obtained by low coverage shotgun next generation sequencing. Molecular Phylogenetics and Evolution. 2010;57:1329–1333. doi: 10.1016/j.ympev.2010.09.012. [DOI] [PubMed] [Google Scholar]
- FOLMER O., BLACK M., HOEH W., LUTZ R., VRIJENHOEK R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;5:294–299. [PubMed] [Google Scholar]
- GRANDE C., TEMPLADO J., ZARDOYA R. Evolution of gastropod mitochondrial genome arrangements. BMC Evolutionary Biology. 2008;8:61. doi: 10.1186/1471-2148-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALL T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- HATZOGLOU E., RODAKIS G.C., LECANIDOU R.L. Complete sequence and gene organization of the mitochondrial genome of the land snail Albinaria coerulea. Genetics. 1995;140:1353–1366. doi: 10.1093/genetics/140.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANNOTTI-PASSOS L.K., RUIZ J.C., CALDEIRA R.L., MURTA S.M., COELHO P.M., CARVALHO O.S. Phylogenetic analysis of Biomphalaria tenagophila (Orbigny, 1935) (Mollusca: Gastropoda) Memórias do Instituto Oswaldo Cruz. 2010;105:504–551. doi: 10.1590/s0074-02762010000400027. [DOI] [PubMed] [Google Scholar]
- JEX A.R., HALL R.S., LITTLEWOOD D.R.J., GASSER R.B. An integrated pipeline for next-generation sequencing and annotation of mitochondrial genomes. Nucleic Acids Research. 2010;38:522–533. doi: 10.1093/nar/gkp883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JÖRGER K.M., STÖGER I., KANO Y., FUKUDA H., KNEBELSBERGER T., SCHRÖDL M. On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BMC Evolutionary Biology. 2010;10:323. doi: 10.1186/1471-2148-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAYAL E., ROURE B., PHILIPPE H., COLLINS A.G., LAVROV D.V. Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evolutionary Biology. 2013;13:5. doi: 10.1186/1471-2148-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEFFORD B.J., NUGEGODA D. No evidence for a critical salinity threshold for growth and reproduction in the freshwater snail Physa acuta. Environmental Pollution. 2005;134:377–383. doi: 10.1016/j.envpol.2004.09.018. [DOI] [PubMed] [Google Scholar]
- KI J.S., LEE Y.M., JUNG S.O., HORIGUCHI T., CHO H.S., LEE J.S. Mitochondrial genome of Thais clavigera (Mollusca: Gastropoda): affirmation of the conserved, ancestral gene pattern within the molluscs. Molecular Phylogenetics and Evolution. 2010;54:1016–1020. doi: 10.1016/j.ympev.2009.12.003. [DOI] [PubMed] [Google Scholar]
- KILPERT F., PODSIADLOWSKI L. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genomics. 2006;7:241. doi: 10.1186/1471-2164-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLUSSMANN-KOLB A., DINAPOLI A., KUHN K., STREIT B., ALBRECHT C. From sea to land and beyond – new insights into the evolution of euthyneuran Gastropoda (Mollusca) BMC Evolutionary Biology. 2008;8:57. doi: 10.1186/1471-2148-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KNUDSEN B., KOHN A.B., NAHIR B., MCFADDEN C.S., MOROZ L.L. Complete DNA sequence of the mitochondrial genome of the sea-slug, Aplysia californica: conservation of the gene order in Euthyneura. Molecular Phylogenetics and Evolution. 2006;38:459–469. doi: 10.1016/j.ympev.2005.08.017. [DOI] [PubMed] [Google Scholar]
- KOSAKOVSKY-POND S.L., FROST S.D.W. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Molecular Biology and Evolution. 2005;22:478–485. doi: 10.1093/molbev/msi031. [DOI] [PubMed] [Google Scholar]
- KURABAYASHI A., UESHIMA R. Complete sequence of the mitochondrial DNA of the primitive opisthobranch gastropod Pupa strigosa: systematic implication of the genome organization. Molecular Biology and Evolution. 2000;17:266–277. doi: 10.1093/oxfordjournals.molbev.a026306. [DOI] [PubMed] [Google Scholar]
- LEE Y.S., LEE S.G., KANG S.W., JEONG J.E., BAEK M.K., CHOI S.H., CHAE S.H., JO Y.H., HAN Y.S., PARK H.S. Expressed sequence tag analysis of Physa acuta: a freshwater pulmonate in Korea. Journal of Shellfish Research. 2011;30:127–132. [Google Scholar]
- LIU G.H., WANG S.Y., HUANG W.Y., ZHAO G.H., WEI S.J., SONG H.Q., XU M.J., LIN R.Q., ZHOU D.H., ZHU X.Q. The complete mitochondrial genome of Galba pervia (Gastropoda: Mollusca), an intermediate host snail of Fasciola spp. PLoS ONE. 2012;7:e42172. doi: 10.1371/journal.pone.0042172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MADSEN H., FRANDSEN F. The spread of freshwater snails including those of medical and veterinary importance. Acta Tropica. 1989;46:139–146. doi: 10.1016/0001-706x(89)90030-2. [DOI] [PubMed] [Google Scholar]
- MAS-COMA S., VALERO M.A., BARGUES M.D. Chapter 2. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Advances in Parasitology. 2009;69:41–146. doi: 10.1016/S0065-308X(09)69002-3. [DOI] [PubMed] [Google Scholar]
- MAYNARD B.T., KERR L.J., MCKIERNAN J.M., JANSEN E.S., HANNA P.J. Mitochondrial DNA sequence and gene organization in Australian blacklip abalone Haliotis rubra (Leach) Marine Biotechnology. 2005;7:645–658. doi: 10.1007/s10126-005-0013-z. [DOI] [PubMed] [Google Scholar]
- MEDINA M., LAL S., VALLÈS Y., TAKAOKA T., DAYRAT B., BOORE J., GOSLINER T. Crawling through time: transition of snails to slugs dating back to the Paleozoic, based on mitochondrial phylogenomics. Marine Genomics. 2011;4:51–59. doi: 10.1016/j.margen.2010.12.006. [DOI] [PubMed] [Google Scholar]
- MORGAN J.A., DEJONG R.J., JUNG Y., KHALLAAYOUNE K., KOCK S., MKOJI G.M., LOKER E.S. A phylogeny of planorbid snails, with implications for the evolution of Schistosoma parasites. Molecular Phylogenetics and Evolution. 2002;25:477–488. doi: 10.1016/s1055-7903(02)00280-4. [DOI] [PubMed] [Google Scholar]
- NEI M., KUMAR S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- NEIMAN M., HEHMAN G., MILLER J.T., LOGSDON J.M., JR., TAYLOR D.R. Accelerated mutation accumulation in asexual lineages of a freshwater snail. Molecular Biology and Evolution. 2010;27:954–963. doi: 10.1093/molbev/msp300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEKOLA J.C., COLES B.F., BERGTHORSSON U. Evolutionary pattern and process within the Vertigo gouldii (Mollusca: Pulmonata, Pupillidae) group of minute North American land snails. Molecular Phylogenetics and Evolution. 2009;53:1010–1024. doi: 10.1016/j.ympev.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALUMBI S., MARTIN A., ROMANO S., MCMILLAN W.O., STICE L., GRABOWSKI G. The simple fool's guide to PCR, version 2.0. Department of Zoology and Kewalo Marine Laboratory, University of Hawaii; 1991. [Google Scholar]
- PAPADOPOULOS J.S., AGARWALA R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- PARAENSE W.L., POINTIER J.P. Physa acuta Draparnaud, 1805 (Gastropoda: Physidae): a study of topotypic specimens. Memórias do Insitituto Oswaldo Cruz. 2003;98:513–517. doi: 10.1590/s0074-02762003000400016. [DOI] [PubMed] [Google Scholar]
- PINCEEL J., JORDAENS K., BACKELJAU T. Extreme mtDNA divergences in a terrestrial slug (Gastropoda, Pulmonata, Arionidae): accelerated evolution, allopatric divergence and secondary contact. Journal of Evolutionary Biology. 2005;18:1264–1280. doi: 10.1111/j.1420-9101.2005.00932.x. [DOI] [PubMed] [Google Scholar]
- RAWLINGS T.A., MACINNIS M.J., BIELER R., BOORE J.L., COLLINS T.M. Sessile snails, dynamic genomes: gene rearrangements within the mitochondrial genomes of a family of caenogastropod molluscs. BMC Genetics. 2010;11:440. doi: 10.1186/1471-2164-11-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROKAS A., HOLLAND P.W.H. Rare genomic changes as a tool for phylogenetics. Trends in Ecology & Evolution. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- ROZEN S., SKALETSKY H.J. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S.A., editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- ROZENDAAL J.A. Freshwater snails. In: Rozendaal J.A., editor. Vector control – methods for use by individuals and communities. WHO; 1997. pp. 337–356. , Geneva, Switzerland. [Google Scholar]
- RUTHERFORD K., PARKHILL J., CROOK J., HORSNELL T., RICE P., RAJANDREAM M.A., BARRELL B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- SÁNCHEZ-ARGÜELLO P., FERNÁNDEZ C., TARAZONA J.V. Assessing the effects of fluoxetine on Physa acuta (Gastropoda, Pulmonata) and Chironomus riparius (Insecta, Diptera) using a two-species water-sediment test. Science of the Total Environment. 2009;407:1937–1946. doi: 10.1016/j.scitotenv.2008.12.004. [DOI] [PubMed] [Google Scholar]
- SIMISON W.B., LINDBERG D.R., BOORE J.L. Rolling circle amplification of metazoan mitochondrial genomes. Molecular Phylogenetics and Evolution. 2006;39:562–567. doi: 10.1016/j.ympev.2005.11.006. [DOI] [PubMed] [Google Scholar]
- STOTHARD J.R., ROLLINSON D. Partial DNA sequences from the mitochondrial cytochrome oxidase subunit I (COI) gene can differentiate the intermediate snail hosts Bulinus globosus and B. nasutus (Gatropoda: Planorbidae) Journal of Natural History. 1997;31:727–737. [Google Scholar]
- STOTHARD P., WISHART D.S. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- TAJIMA F. Simple methods for testing molecular clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAMURA K., PETERSON D., PETERSON N., STECHER G., NEI M., KUMAR S. MEGA5: molecular Evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERRETT J.A., MILES S., THOMAS R.H. Complete DNA sequence of the mitochondrial genome of Cepaea nemoralis (Gastropoda: Pulmonata) Journal of Molecular Evolution. 1996;42:160–168. doi: 10.1007/BF02198842. [DOI] [PubMed] [Google Scholar]
- THOMAZ D., GUILLER A., CLARKE B. Extreme divergence of mitochondrial DNA within species of pulmonate land snails. Proceedings of the Royal Society of London B. 1996;263:363–368. doi: 10.1098/rspb.1996.0056. [DOI] [PubMed] [Google Scholar]
- VALLES Y., BOORE J.L. Lophotrochozoan mitochondrial genomes. Integrative and Comparative Biology. 2006;46:544–547. doi: 10.1093/icb/icj056. [DOI] [PubMed] [Google Scholar]
- WADE C.M., MORDAN P.B. Evolution within the gastropod molluscs; using the ribosomal RNA gene-cluster as an indicator of phylogenetic relationships. Journal of Molluscan Studies. 2000;66:565–570. [Google Scholar]
- WETHINGTON A.R., LYDEARD C. A molecular phylogeny of Physidae (Gastropoda: Basommatophora) based on mitochondrial DNA sequences. Journal of Molluscan Studies. 2007;73:241–257. [Google Scholar]
- WETHINGTON A.R., WISE J., DILLON R.T. Genetic and morphological characterization of the Physidae of South Carolina (Gastropoda: Pulmonata: Basommatophora), with description of a new species. Nautilus. 2009;123:282–292. [Google Scholar]
- WHITE T.R., CONRAD M.M., TSENG R., BALAYAN S., GOLDING R., MARTINS A.M., DAYRAT B.A. Ten new complete mitochondrial genomes of pulmonates (Mollusca: Gastropoda) and their impact on phylogenetic relationships. BMC Evolutionary Biology. 2011;11:295. doi: 10.1186/1471-2148-11-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINNEPENNINCKX B., BACKELJAU T., DE WACHTER R. Extraction of high molecular weight DNA from molluscs. Trends in Genetics. 1993;12:407. doi: 10.1016/0168-9525(93)90102-n. [DOI] [PubMed] [Google Scholar]
- XIA X. DNA replication and strand asymmetry in prokaryotic and mitochondrial genomes. Current Genomics. 2012;13:16–27. doi: 10.2174/138920212799034776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMAZAKI N., UESHIMA R., TERRETT J.A., TOKOBORI S.I., KAIFU M., SEGAWA R., KOBAYASHI T., NUMACHI K.I., UEDA T., NISHIKAWA K., WATANABE K., THOMAS R.H. Evolution of pulmonate gastropod mitochondrial genomes: comparisons of gene organizations of Euhadra, Cepaea and Albinaria and implication of unusual tRNA secondary structures. Genetics. 1997;145:749–758. doi: 10.1093/genetics/145.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]