Abstract
Research in proteomics has exploded in recent years with advances in mass spectrometry capabilities that have led to the characterization of numerous proteomes, including those from viruses, bacteria, and yeast. In comparison, analysis of the human proteome lags behind, partially due to the sheer number of proteins which must be studied, but also the complexity of networks and interactions these present. To specifically address the challenges of understanding the human proteome, we have developed HaloTag technology for protein isolation, particularly strong for isolation of multiprotein complexes and allowing more efficient capture of weak or transient interactions and/or proteins in low abundance. HaloTag is a genetically encoded protein fusion tag, designed for covalent, specific, and rapid immobilization or labelling of proteins with various ligands. Leveraging these properties, numerous applications for mammalian cells were developed to characterize protein function and here we present methodologies including: protein pull-downs used for discovery of novel interactions or functional assays, and cellular localization. We find significant advantages in the speed, specificity, and covalent capture of fusion proteins to surfaces for proteomic analysis as compared to other traditional non-covalent approaches. We demonstrate these and the broad utility of the technology using two important epigenetic proteins as examples, the human bromodomain protein BRD4, and histone deacetylase HDAC1. These examples demonstrate the power of this technology in enabling the discovery of novel interactions and characterizing cellular localization in eukaryotes, which will together further understanding of human functional proteomics.
Keywords: Cellular Biology, Issue 89, proteomics, HaloTag, protein interactions, mass spectrometry, bromodomain proteins, BRD4, histone deacetylase (HDAC), HDAC cellular assays, and confocal imaging
Introduction
Understanding of cellular function, response to stimuli, and changes in development and/or disease states is intricately tied to de-convolution of the proteome throughout these different dynamic states1,2. Much has been done to understand the proteome of lower organisms, however, given the number of proteins and all possible interactions, this has been very challenging in addressing the human proteome1,3-7. Advances in mass spectrometry have greatly enabled the ability to undertake these studies, and here we present an additional and significant advancement with the development of HaloTag technology for both efficient capture of protein complexes and functional characterization of human proteins from mammalian cells8-10. This technology has recently been shown in several studies to be a critical factor for discovery of important interactions, allowing for insight into novel protein function and understanding of disease11-13.
The HaloTag protein fusion was initially developed to address several challenges of traditional affinity tags and antibodies as related to protein capture, purification, and labelling8-10. Within nearly all methods for protein capture and purification, there is a non-covalent interaction step which is used for enrichment of a particular protein14. During this step, the protein and/or complex can disassociate due to diffusion, especially if the process requires hours instead of minutes to complete. To address this concern, the fusion protein was specifically engineered to rapidly and irreversibly interact with its ligands, which can be either particles, surfaces, or fluorophores9. Therefore once it is bound to its ligand, it remains bound, which is one of the most differentiating factor as compared to other affinity tags. Also demonstrated here, is the ability to address different biological questions with a single construct, which is possible by alternating ligands9 (Figure 1). For example, to interrogate protein interactions or function, surface ligands are used for capture of protein or complexes (Figure 1). In a separate experiment, the same fusion protein can be fluorescently labelled to study cellular localization, trafficking, or protein turnover (Figure 1). It is important to remember however that once a ligand is bound, whether it is a surface or a fluorophore, it is irreversible and therefore other ligands cannot then be subsequently bound in the same experiment.
Analysis of existing technologies to map protein interactions reveal the difficulty to efficiently isolate specific interactions, including those which are more transient or are in lower abundance1,6,7,14,15. Also, recent work by numerous groups shows the concern that within traditional isolation methods, particularly in the area of human proteomics, there are a significant number of protein contaminants which may mask signals from true interactors in mass spectrometry analysis16. The properties developed for the HaloTag protein and its co-developed resin address well these concerns. In the protocol for complex pulldowns (Figure 1), binding of complexes can be achieved in 15 min, both promoting complex capture and reducing levels of non-specific binding8. In addition, irreversibly binding allows for efficient capture of low abundance proteins and allows for the use of far fewer cells, again reducing background8. Once capture of complexes is established, the protocol includes mild wash conditions to maintain complex integrity. Elution of captured complexes can be performed by two methods and choosing which method is dictated by the goal of the downstream analysis. If the desire is to analyse or discover interacting proteins by Western blot or mass spectrometry, then it is recommended to elute complexes with denaturants such as SDS or urea (Figure 1). This is particularly advantageous for mass spectrometry as the protein originally fused to HaloTag, termed the bait protein, will remain behind bound to the resin and therefore will not dominate the population of peptides detected in the mass spectrometry. This also means however that the bait protein will likely not be visible in analysis by western blot, a significant difference as compared to other non-covalent affinity purifications or co-immunoprecipitations. If the goal is to purify a complex intact to be used for functional studies, elution can be performed using TEV (Tobacco Etch Virus) protease, which recognizes its cognate cleavage sequence encoded between the fusion protein and the bait protein (Figure 1). These samples could also be analysed by mass spectrometry, but as later shown, they will contain a significant amount of bait protein that may prevent detection of lower abundance, specific interactors.
Here we present pulldown and imaging protocols applied to two important therapeutical proteins, the bromodomain protein BRD4 and a histone deacetylase, HDAC117. We have shown with these examples, interaction with expected partners as determined by mass spectrometry, enzymatic activity after complex isolation for HDAC1, and proper cellular localization for both. Together, these results demonstrate the multifunctional nature and strength of the technology for characterization of eukaryotic protein interactions and function.
Protocol
Note: The following protocol can be used with any HaloTag fusion and in any cell line of choice stably or transiently transfected. For these examples, we will give protocols for transient transfection of HaloTag fusions in HEK293T or HeLa cells. For all experiments, we recommend use of the protein only control.
1. Fusion Protein Expression Test
- Obtain fusion protein construct:
- Obtain pre-made HaloTag fusion vector or construct using existing vectors. For more information about available constructs, please see Table of Specific Reagents.
- If making any clone, it is recommended to sequence verify the DNA.
- Testing expression of fusion protein and control:
- For each sample, prepare 250 μl of 1X SDS Loading Dye buffer (60 mM Tris HCL pH6.8, 0.75 mM bromophenol blue, 12.5% glycerol, 100 mM dithiothreitol, 0.5% SDS).
- For each fusion protein and control, plate 0.5 ml of HEK293T or HeLa cells in their appropriate media at a density of 2-4 x 105 cells/ml in a standard 24 well plate.
- Incubate for 18-24 hr at 37 °C and 5% CO2, then transfect as recommended.
- 24 hr post-transfection, add 0.5 μl of 5mM tetra-methyl rhodamine (TMR) ligand to the media of each well and gently mix plate.
- Incubate transfected cells containing ligand for 15 min at 37 °C and 5% CO2.
- Aspirate off media containing ligand and wash each well of cells gently with 1 ml RT PBS.
- Aspirate off PBS and perform a second wash with 1 ml RT PBS.
- Remove PBS and add 200 µl of 1X SDS Loading dye buffer directly to cells.
- Pipette lysate from plate into 1.5 ml eppendorf tube.
- Boil lysate for 5 min at 95 °C.
- Remove 5-10 μl of the reaction and load onto SDS-PAGE gel.
- Use a fluorescent detection scanner (TMR Excitation: 555 nm, Emission: 585 nm) to detect bands. Use fluorescent protein markers as standards.
- If a fluorescent detection scanner is unavailable, perform western blots using antibodies against either the bait protein or protein fusion tag to detect expression of the fusion protein.
2. Protein Pulldowns
- Preparation of transiently transfected cells:
- For each fusion or control prepare one 15 cm dish with 30 ml of cells at 3-4 x 105 cells/ml or 1-1.2 x 107 cells total per construct.
- Incubate for 18-24 hr at 37 °C and 5% CO2, then transfect construct as recommended.
- After 24-48 hr post-transfection, remove the media and gently wash the cell layer with 20-25 ml of ice cold PBS.
- Remove PBS wash, then add 25-30 ml of 4 °C chilled PBS and gently scrape cells off the plate.
- Collect cells into conical tubes and centrifuge for 5-10 min at 2,000 x g and 4 °C.
- Discard supernatant and place cell pellet at -80 °C for a minimum of 30 min or a maximum of 6 months.
- Equilibration of the HaloLink resin:
- For each fusion or control sample, prepare 12 ml of Resin Equilibration/Wash buffer (100 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.005% IGEPAL CA-630). Note: It is very important to make and use the Resin Equilibration Buffer within 12 hr as diluted IGEPAL CA-630 is not stable or effective beyond this time frame.
- Gently shake or mix resin to obtain a uniform suspension.
- For each pull-down experiment, dispense 200 µl of resin into a 1.5 ml microcentrifuge tubes.
- Centrifuge for 1 min at 800 x g (e.g., 3,000 rpm in a microcentrifuge), then carefully remove and discard the supernatant (ethanol) without disturbing resin at the bottom of the tube.
- Add 800 µl of Resin Equilibration/Wash buffer and mix thoroughly by inverting the tube several times.
- Centrifuge for 2 min at 800 x g, then carefully remove and discard the supernatant.
- Repeat steps 2.2.5 and 2.2.6 two more times for a total of 3 washes.
- Do not remove the final wash or supernatant until ready to add cellular lysates described below (Step 2.3.8) to prevent the resin from drying out.
- Binding and washing of fusion complexes:
- For each sample, prepare 500 µl of Mammalian Lysis Buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% Na Deoxycholate) and 1 ml of 1X TBS buffer (100 mM Tris-HCl pH 7.5 and 150 mM NaCl).
- Thaw the cell pellets and resuspend in 300 µl of Mammalian Lysis Buffer by pipetting up and down or briefly vortexing.
- Add 6 µl of 50X Protease Inhibitor Cocktail (800 µg/ml benzamidine HCl, 500 µg/ml phenanthroline, 500 µg/ml aprotinin, 500 µg/ml leupeptin, 500 µg/ml pepstatin A, 50mM PMSF). Note: Protease cocktails which include AEBSF cannot be used as these interfere with protein fusion tag binding.
- Add 3 µl RQ1 DNase and invert for 10 min at RT.
- Dounce with glass homogenizer 2.0 ml size; 25-30 strokes on ice, or pass cells through a 25 or 27 G needle 5-10 times to complete lysis. Note: Sonication is not recommended as complexes may fall apart and over-heating may affect the protein fusion tag activity.
- Centrifuge at 14,000 x g for 5 min at 4 °C to clear the lysate.
- Transfer clear lysate, approximately 300 µl total volume, to new tube and place on ice.
- Add to clear lysate an additional 700 µl of 1X TBS buffer and mix well by pipetting up and down.
- Take equilibrated resin tubes prepared in Step 2.2.8 and remove final wash/supernatant from resin without disturbing resin at the bottom of the tube.
- Add to each tube of resin, the 1 ml of diluted lysate.
- Incubate with mixing on a tube rotator (or gentle mixer) for 15 min at 22 °C. Note: Settling of resin during this time reduces binding efficiency. If binding at 4 °C is desired, do so by mixing for 1 hr.
- Centrifuge resin tubes for 2 min at 800 x g and discard supernatant.
- Add 1 ml of Resin Equilibration/Wash buffer made at Step 2.2.1 and mix thoroughly by inverting the resin tube by hand several times.
- Centrifuge resin tubes for 2 min at 800 x g and discard the wash.
- Repeat Steps 2.3.12 followed by 2.3.14 three additional times.
- Add 1 ml of Resin Equilibration/Wash buffer and incubate at 22 °C for 5 min with constant rotation.
- Centrifuge resin tubes for 2 min at 800 x g and discard the wash.
- Depending upon end application (see Introduction for further explanation), proceed to either Section 2.4 or 2.5.
- SDS elution for denaturing gels, western blots, or mass spectrometry:
- Resuspend the resin from each sample in 50 µl SDS Elution buffer (1% SDS and 50 mM Tris-HCl pH7.5)
- Shake tubes at RT for 30 min.
- Centrifuge for 2 min at 800 x g and transfer eluates to fresh tubes for analysis.
- For western blot or silver stain gel, load 5-10 µl on a SDS denaturing gel.
- For mass spectrometry, save 40 µl of each sample at -20 °C.
- TEV Protease elution for functional assays, western blots, or mass spectrometry:
- After removing the last wash, resuspend the resin in 50 µl ProTEV cleavage buffer and 30 units of TEV enzyme.
- Incubate at 25 °C with shaking for 1 hr.
- Centrifuge for 2 min at 800 x g.
- Transfer eluate to fresh tubes.
- Store sample at 4 °C for immediate use in functional assays.
3. Cellular Imaging of Fluorescently Labelled Fusion Proteins Using a Cytoplasmic and Nuclear Permeable Ligand
- Transfect, label, and image cells:
- In an 8 well chambered coverglass for each fusion protein or control, plate 400 μl of HeLa cells in their appropriate media in each well at a density of 1-2 x 105 cells/ml.
- Incubate for 18-24 hr at 37 °C and 5% CO2, then transfect as recommended.
- 18-24 hr post-transfection, dilute TMR ligand 1:200 in appropriate cellular media, then add 100 µl of this solution to each well and gently mix.
- Incubate transfected cells containing ligand for 15 min at 37 °C and 5% CO2.
- Aspirate off media containing ligand and replace with 500 μl of appropriate media lacking protein fusion tag ligand, which has been pre-warmed to 37 °C.
- Repeat twice for a total of three washes.
- Put cells back into incubator (37 °C and 5% CO2) for 30 min.
- Aspirate off media and replace with 500 μl of appropriate media, which has been pre-warmed to 37 °C.
- Image on a microscope using appropriate acquisition parameters (TMR Excitation: 555 nm, Emission: 585 nm).
Representative Results
When working with any new fusion protein, it is important to first test for expression of that protein after transfection and also validate that a protein of the proper molecular weight is being produced. As HaloTag fusion proteins can be fluorescently and covalently labelled with permeable or depending upon localization, impermeable ligands, it is possible to quickly determine expression by applying cellular lysates to denaturing gel electrophoresis followed by scanning on a fluorimager. Using the protocol described in Section 1.2, expression of Halo-BRD4 (189 kD) and the HaloTag alone control (Ctrl) is observed (34 kD, Figure 2A). As mentioned in the protocol, the expression of fusion proteins can also be detected using traditional Western blots with anti-HaloTag antibodies, or if they are available, antibodies to the bait protein. If possible, it is recommended to use the fluorescent ligand instead as it is more specific, faster, and easier than antibody detection, and also quantitative10.
After expression of the proper full-length fusion protein is verified, protein pulldowns can be performed. Shown in Figure 2B are the silver stained gels of biological replicates of Halo-BRD4 and Ctrl pulldowns eluted by SDS (Protocol Section 2.4) which demonstrate high reproducibility. The silver stain gels show a significant number of proteins are found to interact with the BRD4 protein and very low background in the control (Figure 2B). As mentioned in the Introduction, in this process of elution, the Halo-BRD4 will not be eluted from the resin as it remains covalently bound. Therefore, there is not a significant band at this molecular weight being detected in the silver stain (Figure 2B) or western blot (data not shown). To determine if these proteins are specific to BRD4, liquid chromatography mass spectrometry (LC-MS/MS) was performed on the complex mixture obtained after SDS elution. Shown in Figure 2C are spectral counts and normalized spectral abundance factor (NSAF) values for known interactors of BRD418-20 found in the Halo-BRD4 mass spectrometry analysis. The high abundance of components from pTEFb18,20 and also the BRD919 protein confirm specific capture of BRD4 complexes. As predicted by the silver stain gels (Figure 2B), numerous other proteins were also identified as potential interactors of BRD4 that were not observed in the control (data not shown). As these are previously unknown, they need to be independently verified by other methodologies to confirm whether the protein is a true interactor, and if so, if it is directly or indirectly associated with BRD4.
Isolated complexes can also be studied for activity; it is recommend to elute complexes using TEV protease (Protocol Section 2.5) so that they maintain functionality. In Figure 3A, a silver stain gel of Halo-HDAC1 pulldown complexes released from the resin using TEV protease is shown. As TEV protease will cleave in a linker region between the protein fusion tag and its fusion partner, significant amounts of the bait protein, in this case HDAC1, are observed (Figure 3A). To determine if this fraction contained HDAC1 activity, eluted TEV samples were tested in a luminescent HDAC assay, HDAC-Glo21. As shown in Figure 3B, HDAC1 pulldown samples showed high levels of HDAC1 activity (Column 1), which was specifically inhibited by a known HDAC inhibitor, SAHA22 (Column 2). As controls to further demonstrate specificity, no HDAC inhibition was observed with a related sirtuin family inhibitor, EX-52722 (Column 3) and no signal was detected using buffer alone without the HDAC1 pulldown sample added (Column 4).
A significant component of functional proteomics and understanding complexes, is also understanding protein localization and/or trafficking. As these same fusion constructs can be fluorescently labelled inside cells, we monitored their localization using confocal imaging. Following the protocol in Section 3, HeLa cells transiently transfected with Halo-BRD4 (Figure 4A) and Halo-HDAC1 (Figure 4B) were fluorescently labelled with the TMR ligand and imaged. As shown in Figures 4A and 4B, both localized to the nucleus as expected17. These data demonstrate that the presence of tag did not alter physiological cellular localization of its fusion partners.
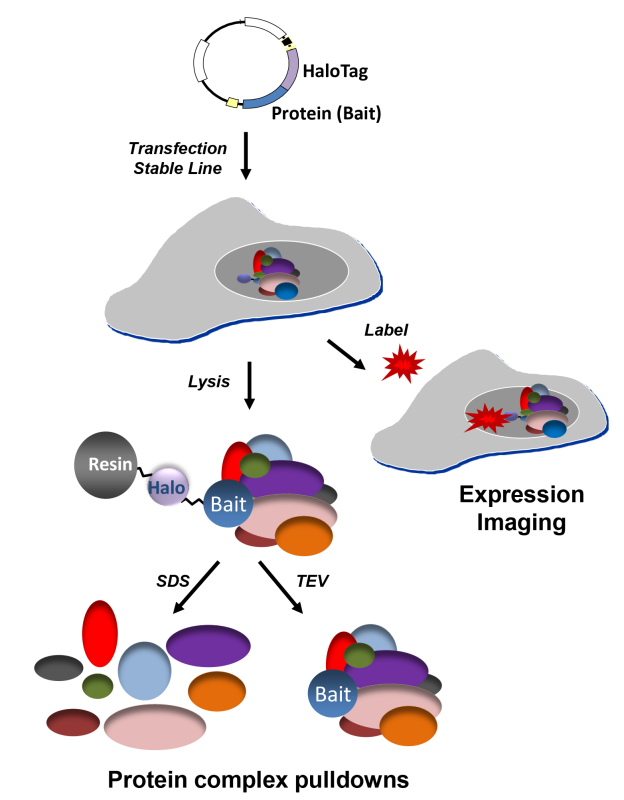 Figure 1.Schematic of protein pulldown and confocal imaging applications. Using a single construct several applications for understanding protein function in mammalian cells are possible. For all, a Halo fusion construct is either stably or transiently expressed in adherent or suspension mammalian cells. For protein complex pulldowns, cells are then lysed, complexes are covalently captured on resin, and eluted through either SDS elution (left pathway) or TEV cleavage (right pathway). SDS elution is recommended for proceeding to mass spectrometry analysis, while TEV cleavage is optimal for performing functional analysis. To characterize expression, cellular localization, trafficking events, or protein turnover, live cells expressing fusion proteins are fluorescently labelled and further analysed on SDS PAGE gels or using confocal imaging. Both cell permeable or impermeable fluorescent ligands are available depending upon the localization or presentation of the fusion protein within the cell.
Figure 1.Schematic of protein pulldown and confocal imaging applications. Using a single construct several applications for understanding protein function in mammalian cells are possible. For all, a Halo fusion construct is either stably or transiently expressed in adherent or suspension mammalian cells. For protein complex pulldowns, cells are then lysed, complexes are covalently captured on resin, and eluted through either SDS elution (left pathway) or TEV cleavage (right pathway). SDS elution is recommended for proceeding to mass spectrometry analysis, while TEV cleavage is optimal for performing functional analysis. To characterize expression, cellular localization, trafficking events, or protein turnover, live cells expressing fusion proteins are fluorescently labelled and further analysed on SDS PAGE gels or using confocal imaging. Both cell permeable or impermeable fluorescent ligands are available depending upon the localization or presentation of the fusion protein within the cell.
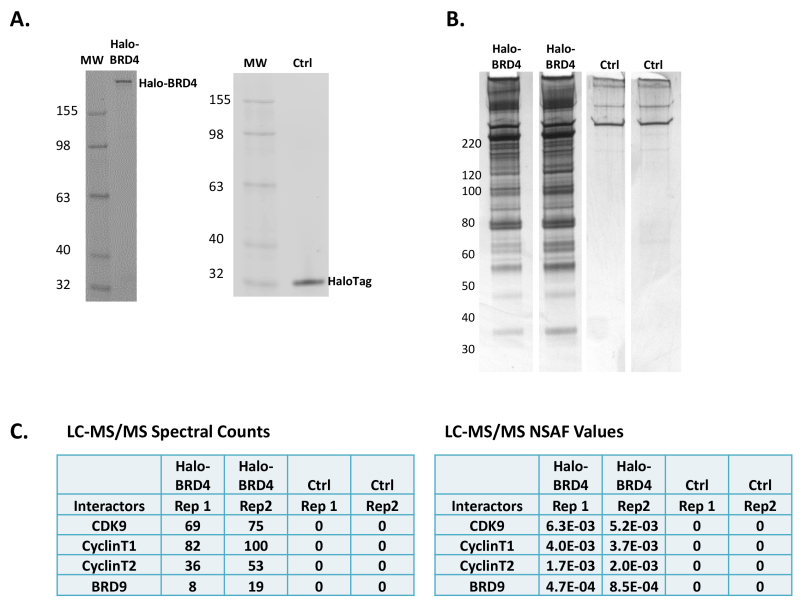 Figure 2. Halo-BRD4 protein expression, pulldown, and mass spectrometry analysis. (A) A SDS PAGE gels showing the expression of Halo-BRD4 fusion protein, 189 kD, and Halo protein fusion tag alone, 34 kD, control (Ctrl) within a HEK293T cellular lysate labelled with TMR ligand (Protocol Section 2.1). Gels were scanned with fluorimager for detection and a fluorescent molecular weight marker was used. (B) Silver stain gels of biological replicates for pulldowns of Halo-BRD4 and Ctrl samples eluted with SDS. Molecular weight sizes are indicated for the gel. (C) Spectral counts (left panel) and normalized spectral abundance factors (NSAF) values (right panel) which account for protein molecular weight of proteins identified in mass spectrometry analysis of biological replicates of Halo-BRD4 and Ctrl. Shown are proteins known to interact with BRD4, including components of pTEFb (CDK9 and Cyclin T)18,20, as well as BRD919. No peptides from these proteins were identified in the Ctrl.
Figure 2. Halo-BRD4 protein expression, pulldown, and mass spectrometry analysis. (A) A SDS PAGE gels showing the expression of Halo-BRD4 fusion protein, 189 kD, and Halo protein fusion tag alone, 34 kD, control (Ctrl) within a HEK293T cellular lysate labelled with TMR ligand (Protocol Section 2.1). Gels were scanned with fluorimager for detection and a fluorescent molecular weight marker was used. (B) Silver stain gels of biological replicates for pulldowns of Halo-BRD4 and Ctrl samples eluted with SDS. Molecular weight sizes are indicated for the gel. (C) Spectral counts (left panel) and normalized spectral abundance factors (NSAF) values (right panel) which account for protein molecular weight of proteins identified in mass spectrometry analysis of biological replicates of Halo-BRD4 and Ctrl. Shown are proteins known to interact with BRD4, including components of pTEFb (CDK9 and Cyclin T)18,20, as well as BRD919. No peptides from these proteins were identified in the Ctrl.
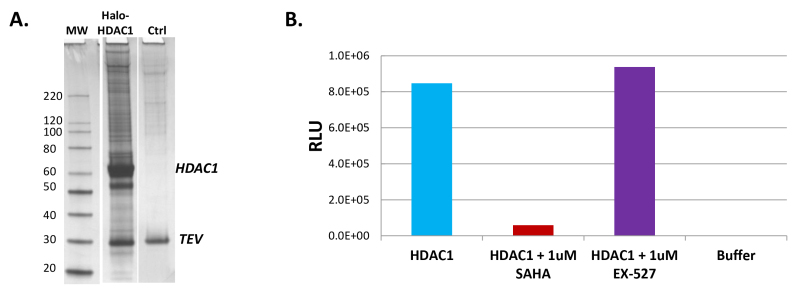 Figure 3. Halo-HDAC1 complex isolation and activity analysis. (A) Silver stain gels showing the isolation of Halo-HDAC1 complexes and background from the Ctrl after TEV cleavage (Protocol Section 2.5). The prominent HDAC1 band (55 kDa) and the TEV protease band are labelled. The free HDAC1 is generated by TEV cleavage within an optimized linker between the HaloTag and HDAC1 fusion sequence after capture on the resin (Figure 1). (B) Graph showing the activity of HDAC1 complex isolations samples in a luminescent histone deacetylase assay, HDAC-Glo21. Column 1 of the graph shows high levels of HDAC activity contained with the Halo-HDAC1 pulldown samples (HDAC1). Column 2 reveals this activity can be specifically decreased by addition of the HDAC inhibitor, SAHA22, to the HDAC1 pulldown samples. As controls, Column 3 demonstrates a deacetylase inhibitor specific to the sirtuin family, but not HDACs, EX-52722, does not inhibit HDAC1 activity and Column 4 shows no activity is observed using buffer alone.
Figure 3. Halo-HDAC1 complex isolation and activity analysis. (A) Silver stain gels showing the isolation of Halo-HDAC1 complexes and background from the Ctrl after TEV cleavage (Protocol Section 2.5). The prominent HDAC1 band (55 kDa) and the TEV protease band are labelled. The free HDAC1 is generated by TEV cleavage within an optimized linker between the HaloTag and HDAC1 fusion sequence after capture on the resin (Figure 1). (B) Graph showing the activity of HDAC1 complex isolations samples in a luminescent histone deacetylase assay, HDAC-Glo21. Column 1 of the graph shows high levels of HDAC activity contained with the Halo-HDAC1 pulldown samples (HDAC1). Column 2 reveals this activity can be specifically decreased by addition of the HDAC inhibitor, SAHA22, to the HDAC1 pulldown samples. As controls, Column 3 demonstrates a deacetylase inhibitor specific to the sirtuin family, but not HDACs, EX-52722, does not inhibit HDAC1 activity and Column 4 shows no activity is observed using buffer alone.
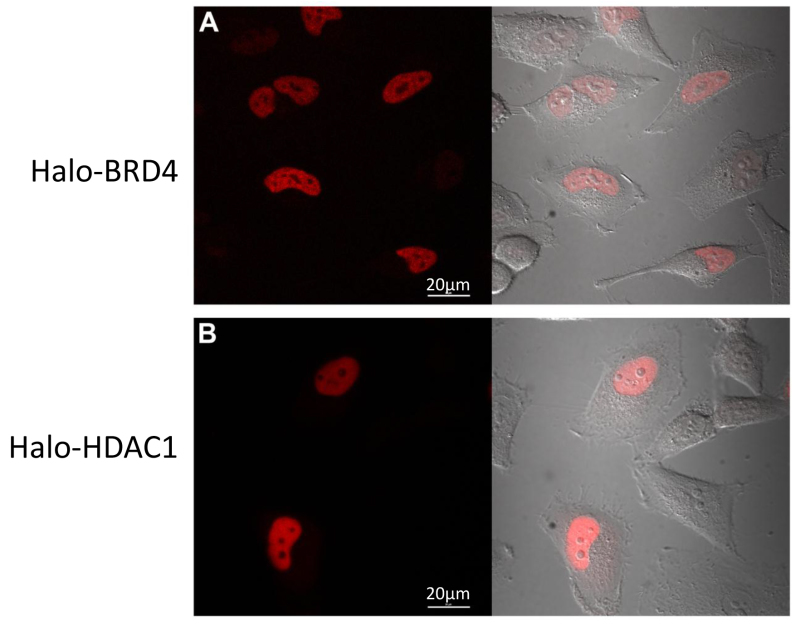 Figure 4. Halo-BRD4 and Halo-HDAC1 confocal imaging. Live cell confocal imaging of HeLa cells transfected with Halo-BRD4 (A) or Halo-HDAC1 (B) fluorescently labelled with TMR ligand. (A) Halo-BRD4 expression is restricted to the nucleus and (B) Halo-HDAC1 expression is predominantly nuclear. Left side of panels is fluorescent channel and right side is an overlay of the fluorescent channel with the DIC channel for each. Images were acquired on a confocal microscope equipped with a 37 °C + CO2 environmental chamber using appropriate filter sets. Scale bars = 20 µm.
Figure 4. Halo-BRD4 and Halo-HDAC1 confocal imaging. Live cell confocal imaging of HeLa cells transfected with Halo-BRD4 (A) or Halo-HDAC1 (B) fluorescently labelled with TMR ligand. (A) Halo-BRD4 expression is restricted to the nucleus and (B) Halo-HDAC1 expression is predominantly nuclear. Left side of panels is fluorescent channel and right side is an overlay of the fluorescent channel with the DIC channel for each. Images were acquired on a confocal microscope equipped with a 37 °C + CO2 environmental chamber using appropriate filter sets. Scale bars = 20 µm.
Discussion
Presented here are two fusion proteins, Halo-BRD4 and Halo-HDAC1, characterized in eukaryotic mammalian cells for expression, protein complex isolation and activity, and cellular localization. In working through these various protocols, there are several important steps for the success of each experiment. As is the case with any fusion protein, expression level and placement of the tag itself can be critical for maintaining physiology. It is therefore important to consider that N- and/or C-terminal fusions will need to be designed if prior knowledge or work with another tag has not been demonstrated for the particular protein of choice. Regarding expression, if the level is too high, dilution of DNA can be performed during transfection or use of vectors with weaker promoters are possible to achieve the level that is appropriate. Previous work has been performed showing efficient isolation of macromolecular complexes at endogenous levels of expression8, enabling work at very low levels of expression. If possible, cellular localization studies would also be able to provide insight as to the optimal tag placement position as well as relative expression level needed for proper physiology.
Once the fusion proteins are ready for pulldown experiments, to obtain maximum success with the protocol it is very important to follow the recommended time frames in the lysis, binding, and washing sections, as one of the biggest advantages is the speed of the complex isolation process. If any of these steps are lengthened in time, such as is required for an antibody-based capture method, there is a risk of complex disassociation or increased non-specific binding8. Similarly if the times are shortened during cellular lysis or binding, cells may not be completely lysed or efficiently captured respectively. If the number of washes is decreased or good mixing of the resin does not occur during the binding or washes, then background levels of non-specific proteins will be increased. Also, the means of lysis is very important as related to the binding efficiency to the resin. Attempts to sonicate the samples, remove the recommended detergent, add SDS or other strong detergent, and/or include the protease inhibitor AEBSF will result in reduced or loss of binding of fusion proteins and their complexes to the resin.
Existing methods for complex isolation from mammalian cells and human proteomic analysis struggle with the challenge of reducing background in analysis by mass spectrometry16. This has been so significant that a repository of contaminating proteins has been created by numerous mass spectrometry groups16. Background in mass spectrometry pulldown samples can be defined as anything which prevents identification of true interactors of the bait protein. As such, background can arise from contaminating non-specific proteins or also large concentrations of bait protein or antibodies used to precipitate bait proteins. Significant work was done in optimizing both the pulldown protocol presented here as well as the resin to minimize the level of the non-specific contaminating proteins in the pulldown process. This is evident in the silver stain gels and mass spectrometry analysis of the control (Figure 2). To address the high levels of bait protein or antibodies that can be equally detrimental as contaminants and with which other methodologies struggle, the pulldown with SDS elution is recommended (Protocol Section 2.4). Due to the covalent attachment to the resin, this process will leave the majority of the starting fusion protein covalently attached to the resin. A small percentage of bait is observed in the mass spectrometry analysis, which is believed to occur through hydrolysis, but it does not present a problem for detection of other weaker or transient interactions8.
As mentioned in the introduction, significant advances in proteomics have been enabled by significant advances in mass spectrometry1,7. Therefore, it is important to highlight important parameters of the choice of mass spectrometry analysis to deconvolute the mixture of proteins obtained in the pulldowns. Instrumentation must be robust and capable of routinely and efficiently analysing samples containing a small amount of protein, often less than <1 µg. Nanoscale chromatography involving 50-75 µm internal diameter HPLC columns with flow rates in the 100-300 nl/min range is typically employed for compatibility with the small sample sizes and to maximize the sensitivity of the mass spectrometer. To maximize the information acquired in a single analysis state of the art mass spectrometers capable of acquiring high resolution mass spectra on a time scale compatible with aforementioned nanoscale separations, ≥10 Hz, are typically employed. These instruments have attomolar levels of sensitivity and can routinely acquire data with sub ppm precursor and product ion mass error tolerances. These performance characteristics serve to increase the yield of the number of identified proteins and the confidence associated with those identifications.
With these data we demonstrate physiological cellular localization, proper protein:protein interactions along with potential for discovery of novel interactions, and isolation of active complexes all using a single construct. Indeed alternative technologies can be used for each of these different aspects, but likely not for all23,24. With the HaloTag technology, a multifunctional approach can be used, advancing proteomics studies and obtaining a more complete understanding of protein function in mammalian cells.
Disclosures
Publication fees for this article are sponsored by Promega Corporation. Danette L. Daniels, Jacqui Méndez, Hélène Benink, Andrew Niles, Nancy Murphy and Marjeta Urh are employees of Promega Corporation, the commercial owner by assignment of patents of the HaloTag technology and its applications. Michael Ford, Richard Jones, Ravi Amunugama, and David Allen are employees of MSBioworks, which provides mass spectrometry services described in this manuscript.
Acknowledgments
We thank Dr. Martin Rosenberg, Dr. Gary Tarpley, and Dr. Keith Wood for support of this work, and Dr. James Cali for critical reading of the manuscript. DLD, JM, HB, NM, AN, JC, and MU are employees of Promega Corporation. MF, RJ, RA, and DA are employees of MS Bioworks, LLC.
References
- Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- Coulombe B, et al. Interaction networks of the molecular machines that decode, replicate, and maintain the integrity of the human genome. Mol Cell Proteomics. 2004;3:851–856. doi: 10.1074/mcp.R400009-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Sardiu ME, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- Daniels DL, et al. Examining the complexity of human RNA polymerase complexes using HaloTag technology coupled to label free quantitative proteomics. J Proteome Res. 2012;11:564–575. doi: 10.1021/pr200459c. [DOI] [PubMed] [Google Scholar]
- Encell LP, et al. Development of a dehalogenase-based protein fusion tag capable of rapid, selective and covalent attachment to customizable ligands. Curr Chem Genomics. 2012;6:55–71. doi: 10.2174/1875397301206010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohana RF, et al. HaloTag-based purification of functional human kinases from mammalian cells. Protein Expr Purif. 2011;76:154–164. doi: 10.1016/j.pep.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Black JC, et al. KDM4A Lysine Demethylase Induces Site-Specific Copy Gain and Rereplication of Regions Amplified in Tumors. Cell. 2013;154:541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, et al. HIF1A Employs CDK8-Mediator to Stimulate RNAPII Elongation in Response to Hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Choudhary JS. Mapping multiprotein complexes by affinity purification and mass spectrometry. Curr Opin Biotechno. 2008;19:324–330. doi: 10.1016/j.copbio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Collins SR, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Halley F, et al. A bioluminogenic HDAC activity assay: validation and screening. J Biomol Screen. 2011;16:1227–1235. doi: 10.1177/1087057111416004. [DOI] [PubMed] [Google Scholar]
- Mack GS. To selectivity and beyond. Nat Biotechnol. 2010;28:1259–1266. doi: 10.1038/nbt.1724. [DOI] [PubMed] [Google Scholar]
- Daniels DL, Urh M. Isolation of intracellular protein--DNA complexes using HaloCHIP, an antibody-free alternative to chromatin immunoprecipitation. Methods Mol Biol. 2013;977:111–124. doi: 10.1007/978-1-62703-284-1_9. [DOI] [PubMed] [Google Scholar]
- Urh M, Rosenberg M. HaloTag a Platform Technology for Protein Analysis. Curr Chem Genomics. 2012;6:72–78. doi: 10.2174/1875397301206010072. [DOI] [PMC free article] [PubMed] [Google Scholar]


