Abstract
Nuclear hormone receptors of the NR4A subgroup have been implicated in cancer, atherosclerosis, and metabolic disease. However, little is known about the role of these receptors in kidney health or disease. Nr4a1-deficient rats (Nr4a1−/−) developed on a genetic background susceptible to kidney injury (fawn-hooded hypertensive rat [FHH]) were evaluated for BP, proteinuria, renal function, and metabolic parameters from 4 to 24 weeks-of-age. By week 24, Nr4a1−/− rats exhibited significantly higher proteinuria (approximately 4-fold) and decreased GFR compared with FHH controls. The severity of tubular atrophy, tubular casts, and interstitial fibrosis increased significantly in Nr4a1−/− rats and was accompanied by a large increase in immune cell infiltration, predominantly macrophages and to a lesser extent T cells and B cells. Global transcriptome and network analyses at weeks 8, 16, and 24 identified several proinflammatory genes and pathways differentially regulated between strains. Bone marrow crosstransplantation studies demonstrated that kidney injury in Nr4a1−/− rats was almost completely rescued by bone marrow transplanted from FHH controls. In vitro, macrophages isolated from Nr4a1−/− rats demonstrated increased immune activation compared with FHH-derived macrophages. In summary, the loss of Nr4a1 in immune cells appears to cause the increased kidney injury and reduced renal function observed in the Nr4a1−/− model.
The incidence of CKD has increased over the last 2 decades.1 Current estimates suggest that approximately 10% of adults in the United States have CKD.2 Considering only federal Medicare spending, the direct economic effect of CKD is staggering. The cost to treat CKD (predialysis) in 2010 was $41 billion, in addition to ESRD (dialysis and transplantation) costs estimated at $32.9 billion.3 A significant problem is that current treatment options are not ideal and only serve to slow the progression of CKD. Thus, the identification of genes and/or biologic pathways that mediate progressive kidney disease is critical to both developing new and/or improved treatments for CKD and curbing the associated financial burden.
Nuclear hormone receptors are transcription factors that play a role in a variety of biologic processes and are induced in multiple tissues by diverse stimuli, such as β-adrenoceptor agonists, cold, fatty acids, glucose, insulin, and cholesterol.4 The nuclear receptor (NR) 4A subgroup of nuclear hormone receptors is composed of three members: Nr4a1 (Nur77), Nr4a2 (Nurr1), and Nr4a3 (Nor1). In contrast with other NRs, the activity of NR4A receptor family is ligand independent and regulated at the level of gene expression and protein stability.5 The NR4A subgroup has been linked to cancer,6 obesity,7 diabetes,8 and vascular disease.9 A significant role for NR4A1 has been established in chronic inflammatory diseases such as atherosclerosis, psoriasis, and rheumatoid arthritis.10 Although inflammation is a key factor in the pathogenesis of a variety of kidney diseases, little is known about the role that NR4A receptors may play in kidney health or disease.
This study aimed to explore the role of Nr4a1 in the context of a genetic model (fawn-hooded hypertensive [FHH] rat) that is sensitive to cardiovascular and kidney disease. We performed a longitudinal study of BP, renal injury, proteinuria, and renal function in Nr4a1−/− and FHH control animals from 4 to 24 weeks-of-age. BP between strains was similar, but kidneys from Nr4a1−/− rats demonstrated prominent tubulointerstitial injury and inflammation, suggesting that the mechanism of injury was immune mediated. Bone marrow crosstransplantation and in vitro studies using macrophages isolated from both strains suggest that loss of Nr4a1 in immune cells such as macrophages is the likely cause of kidney injury and renal dysfunction in Nr4a1−/− rats. In summary, these findings suggest a novel and previously unknown role for Nr4a1 in the pathogenesis of kidney disease.
Results
Association of Nr4a1 Mutation with Proteinuria by Linkage Analysis
To confirm and validate that the point mutation (TAC [tyrosine] to TAA [stop]) in Nr4a1−/− animals was associated with proteinuria (renal injury), a genetic analysis was performed using an F2 (FHH×Nr4a1−/−) segregating population (n=175 animals) (Supplemental Figure 1). Several parameters including body weight, kidney weight, proteinuria, fluid intake, and urine output were measured at 8 and 12 weeks-of-age. At both time points, animals with the A/A genotype (which results in a premature stop codon) exhibited significantly higher proteinuria compared with the wild-type C/C genotype (Supplemental Figure 1). The linkage analysis also demonstrated that animals having the A/A genotype were significantly smaller (233±2.1 g), while having significantly larger kidneys (10.5±0.1 mg/g body wt) compared with the wild-type G/G genotype (249±2.1 g and 9.5±0.1 mg/g body wt, respectively; P<0.01). In summary, linkage analysis provided conclusive evidence that the Nr4a1 locus (point mutation) is linked to increased proteinuria and associated renal measures.
Temporal Changes in Renal, Cardiovascular, and Metabolic Traits
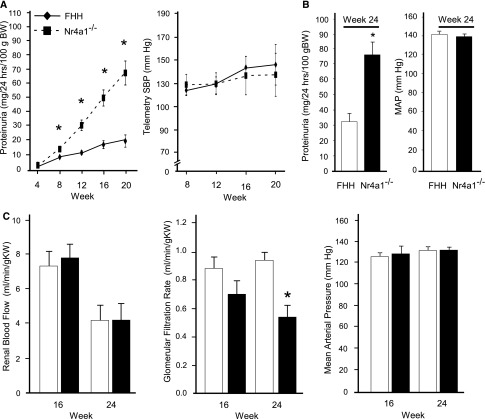
FHH and Nr4a1−/− rats were evaluated for changes in BP, proteinuria, and renal function from 4 to 24 weeks-of-age (Figure 1, A and B). Consistent with the linkage analysis, Nr4a1−/− rats demonstrated significantly higher albuminuria and proteinuria compared with FHH rats starting at week 8 (Figure 1B, Table 1). By week 24, Nr4a1−/− rats exhibited approximately 3-fold higher proteinuria compared with controls. A similar increase in telemetry-measured BP was observed from weeks 8 to 20 between strains. BP measured at week 24 (by indwelling catheter) also demonstrated similar BP between strains (Figure 1B). Likewise, no significant difference in cardiac hypertrophy was observed throughout the time course (Table 1). Renal hemodynamics were evaluated at weeks 16 and 24 to more comprehensively evaluate renal function. No significant difference in renal blood flow (RBF) or GFR was observed between strains at week 16 (Figure 1C). By week 24, a significant reduction in GFR was observed in Nr4a1−/− rats compared with FHH rats, which was not accompanied by changes in BP. As expected, impairment of RBF autoregulation was observed in the FHH rats because RBF increased when renal perfusion pressure varied from 90 to 140 mmHg (Supplemental Figure 2). A similar impairment in RBF autoregulation was also observed in the Nr4a1−/− rats. Significant increases in kidney weight and BUN and a significant decrease in creatinine clearance was observed in Nr4a1−/− (compared with FHH) at week 8, 16, and 24 (Table 1). Several other metabolic parameters, including elevated cholesterol and triglycerides was observed in the Nr4a1−/− as well as significant lipid deposits in the liver (Table 1, Supplemental Figure 3).
Figure 1.
Temporal evaluation of BP, proteinuria, and renal hemodynamics in FHH and Nr4a1−/− rats. (A) Proteinuria and telemetry-measured BP are assessed in the FHH and Nr4a1−/− rats from 4 to 20 weeks-of-age (n=28 per group at week 4 to n=6 at week 20; 6 rats are euthanized at weeks 8, 12, and 16). (B) Proteinuria and MAP at week 24 (n=6–8 per group; separate group from A). BP is measured in conscious animals by indwelling animals. (C) RBF, GFR, and MAP at weeks 16 and 24 for animals (n=6–8 per group). *P<0.05 compared with FHH (same time point). Mean values±SEM are presented. SBP, systolic BP; BW, body weight; MAP, mean arterial pressure; KW, kidney weight.
Table 1.
Summary of body weight, organ weights, blood, and kidney function parameters in FHH and Nr4a1−/− rats at weeks 8, 16, and 24
| Trait | Week 8 | Week 16 | Week 24 | |||
|---|---|---|---|---|---|---|
| FHH Rats | Nr4a1−/− Rats | FHH Rats | Nr4a1−/− Rats | FHH Rats | Nr4a1−/− Rats | |
| Body weight (g) | 207±3.1 | 190±2.6a | 352±5.8 | 328±5.2a | 429±9.5 | 352±14.0a |
| Kidney weight (mg/g) | 9.2±0.27 | 10.3±0.14a | 9.2±0.27 | 10.3±0.14a | 8.1±0.18 | 11.0±0.53a |
| Heart weight (mg/g) | 3.7±0.23 | 3.9±0.8 | 3.7±0.23 | 3.9±0.8 | 3.1±0.06 | 3.4±0.13 |
| Liver weight (mg/g) | 35.6±0.73 | 42.7±1.56a | ||||
| Drinking volume (ml) | 48±4.0 | 38±3.4 | 36±6.2 | 28±4.4 | 20±5.0 | 22±2.4 |
| Urine volume (ml) | 47±3.4 | 37±2.1 | 39±5.3 | 31±3.7 | 24±3.7 | 24±2.1 |
| Albuminuria (mg/24 h per 100 g body wt) | 0.6±0.09 | 8±2.3.5a | 12±6.1 | 32±5.5a | 17±3.6 | 52±5.8a |
| Creatinine clearance (ml/min per g kidney weight) | 1.2±0.07 | 1.0±0.12 | 0.9±0.06 | 0.7±0.06 | 1.2±0.2 | 0.6±0.2a |
| BUN (mg/dl) | 14±1.5 | 14±0.6 | 14±0.6 | 17±0.6a | 14±1.6 | 23±1.6a |
| Cholesterol | 70±4.7 | 71±2.1 | 84±7.9 | 122±8.7a | 102±9.7 | 283±29.4a |
| Triglycerides | 32±1.4 | 30±3.0 | 37±5.3 | 82±12.6a | 50±4.4 | 192±43.3a |
P<0.05.
Characterization of Glomerular, Tubular, and Vascular Renal Injury
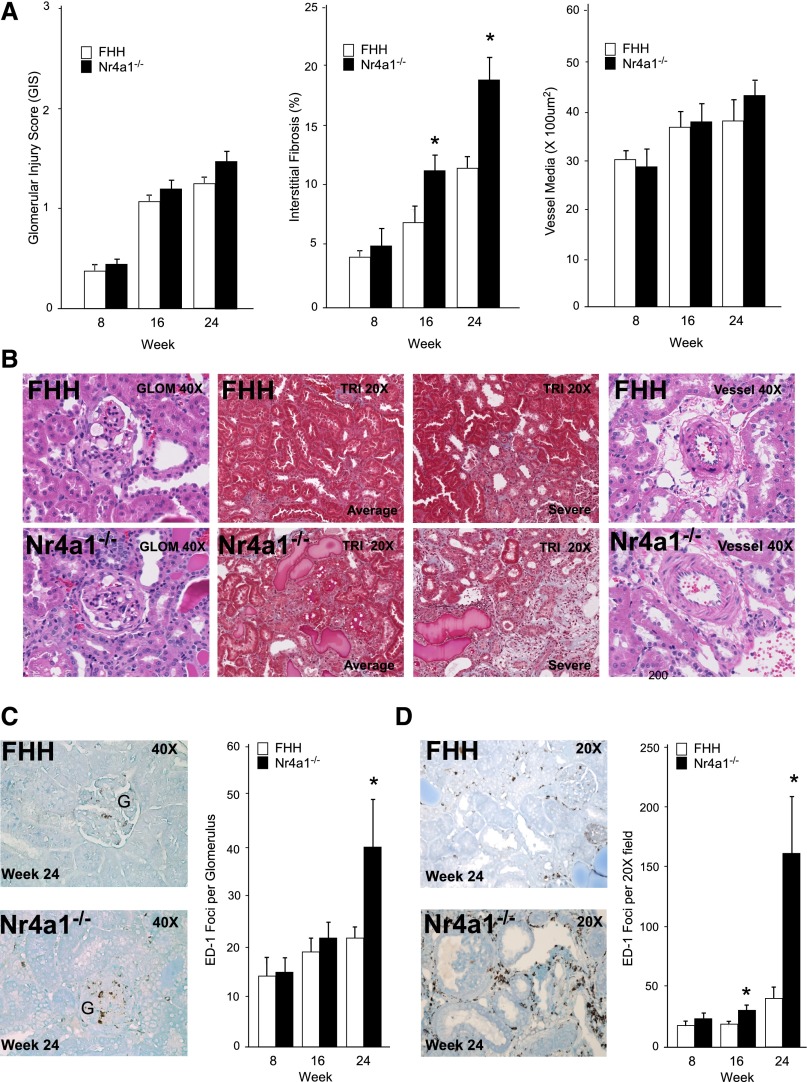
The severity of kidney injury in FHH and Nr4a1−/− rats was assessed at weeks 8, 16, and 24. Moderate increases in mesangial matrix expansion and glomerulosclerosis were observed in both stains with progressive injury from weeks 8 to 24. However, there was no significant difference in the severity of glomerular injury between strains (Figure 2, A and B). Electron microscopy analysis showed that glomeruli exhibited a similar degree of mesangial expansion and podocyte foot process effacement between strains (Supplemental Figure 4). The severity of tubular injury by evaluation of tubular atrophy, dilation, and protein casts was significantly increased from weeks 16 to 24 in Nr4a1−/− rats compared with FHH rats (Supplemental Figure 5). Consistent with tubular injury, the degree of interstitial fibrosis was significantly increased in Nr4a1−/− rats (approximately 2-fold) compared with controls starting at week 16 (Figure 2, A and B). Electron microscopy analysis demonstrated enhanced matrix formation between adjacent tubules and cell degeneration in proximal tubules cells of Nr4a1−/− rats (Supplemental Figure 4). No difference in renal arteriole morphology was detected between strains from 8 to 24 weeks-of-age.
Figure 2.
Temporal evaluation of renal injury in FHH and Nr4a1−/− rats at weeks 8, 16, and 24. (A) Glomerular, tubulointerstitial, and vascular injury. Each glomerulus is scored on a scale from 0 (normal) to 4 (global sclerosis). A minimum of 20 glomeruli are scored for each kidney section (n=6 per group/time). For tubulointerstitial injury (fibrosis), a minimum of 20 random images per kidney section are studied (n=6 animals per group). Vascular injury is assessed by measurement of vessel wall thickening. Vessel wall thickening (vessel media, micrometers squared) is calculated by measuring the outer circumference of the vessel minus the inner circumference of the lumen (20 random images per animal, n=6 animals per group). (B) Representative images of kidney sections from FHH and Nr4a1−/− rats at week 24. (C) Quantification of macrophage (ED-1–positive cells) infiltration in glomeruli (weeks 8, 16, and 24) and representative immunohistochemistry images at week 24. (D) Macrophage infiltration in tubulointerstitial region (weeks 8, 16, and 24) and representative immunohistochemistry images at week 24. *P<0.05 compared with FHH.
Increased macrophage infiltration was observed in kidney from Nr4a1−/− rats starting at week 16 in the tubulointerstitial region and by week 24 was 3- to 4-fold higher than FHH rats (Figure 2D). No significant difference in glomerular macrophage infiltration was observed until week 24. Kidneys from Nr4a1−/− rats also demonstrated a significant increase in T- and B-cell infiltration by week 24 (CD43; 34.2±7.4 versus 16.3±2.2 foci per ×20 field; CD22: 9.3±2.04 versus 4.4±1.2, respectively; P<0.05), but to a lesser extent compared with macrophage infiltration. Gene expression markers of macrophages (CD68 and SOCS3) and tubular injury genes (Kim-1 and Ngal) were upregulated in the Nr4a1−/− rats (Supplemental Figure 5). Taken together, these findings indicate that loss of Nr4a1 exacerbates macrophage infiltration predominantly in the tubulointerstitial region and culminates in significant injury and renal function decline.
Time Course Transcriptome Analyses
Gene expression profiling was performed at weeks 8, 16, and 24 using kidney from FHH and Nr4a1−/− rats. A two-way ANOVA was performed to identify genes that were significantly different between strains (Nr4a1−/− and FHH) and over time (weeks 8, 16, and 240; i.e., strain [n=277], time [n=975], and interaction [n=635]) using fold change ≥±1.2 and a P<0.01. Principal component analysis identified distinct differences between sample groups and gene expression changes over time. At week 8, gene expression patterns were similar between Nr4a1−/− and FHH kidneys. However, at weeks 16–24, Nr4a1−/− gene expression differences were distinct from FHH rats and each other (Supplemental Figure 6), whereas FHH gene expression differences were similar from weeks 16 and 24. Ingenuity Pathway Analysis identified inflammatory response as the top biologic function associated with gene expression differences. Identified genes were associated with key upstream regulators of inflammation (e.g., LPS, IFN-ɣ, IL-1β, IL-6, and TNF-α). Important inflammatory genes observed to be differentially expressed (by microarray) between Nr4a1−/− and FHH were validated by real-time PCR, including Ccr2, Tgf-β1, Tgf-β2, Ifn-ɣ, Tnf-α, Il-1β, and Il-6 (Supplemental Figure 5).
Bone Marrow Cross-Transplantation Studies between Nr4a1−/− and FHH Rats
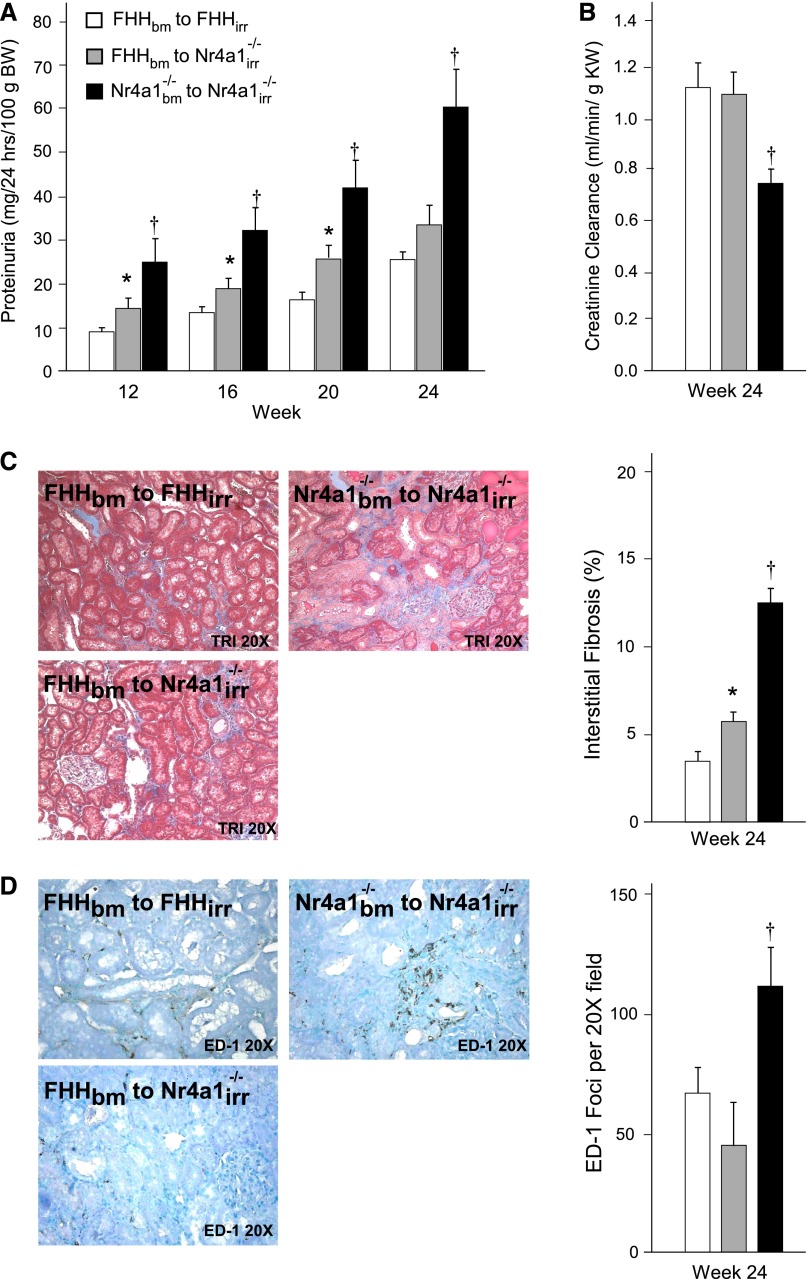
On the basis of the specificity of the histologic injury, increase in immune cell infiltration, and inflammatory gene expression differences in kidney from Nr4a1−/− rats (compared with FHH rats), we aimed to determine whether loss of Nr4a1 promotes kidney injury via a mechanism involving immune cells. Bone marrow (bm) crosstransplantation studies were performed in three groups of irradiated (irr) animals: (1) FHHbm into FHHirr (FHH control), (2) FHHbm into Nr4a1irr−/− (experimental), and (3) Nr4a1bm−/− into Nr4a1irr−/− (Nr4a1−/− control). The transplant of bone marrow from FHH into Nr4a1 was confirmed by the identification of FHH genotype in Nr4a1 animals (Supplemental Figure 7). FHHbm into Nr4a1irr−/− animals showed a significant attenuation of proteinuria starting as early as week 12 (4 weeks after transplantation) and continued through week 24 compared with the Nr4a1bm−/− into Nr4a1irr−/− (control) animals (Figure 3A). Creatine clearance was similar between FHH controls compared with FHHbm into Nr4a1irr−/− animals, but both were significantly higher compared with Nr4a1−/− control animals (Figure 3B). Improved renal injury and renal function in FHHbm into Nr4a1irr−/− animals was corroborated by reduced tubular injury (Supplemental Figure 7), interstitial fibrosis, and tubulointerstitial macrophage infiltration (Figure 3, C and D) compared with Nr4a1−/− controls. FHHbm into Nr4a1irr−/− animals demonstrated a significant reduction of serum cholesterol and triglycerides compared with Nr4a1−/− controls (Supplemental Figure 7).
Figure 3.
Bone marrow crosstransplantation studies in FHH and Nr4a1−/− rats. (A and B) Proteinuria and renal function in FHH or Nr4a1−/− rats undergoing reciprocal bone marrow transplantation. Recipient animals are irradiated (irr) at week 8, provided bone marrow (bm) isolated from donor animals, and evaluated for proteinuria and/or renal function markers at weeks 12, 16, 20, and 24. (C) Morphometric analysis of tubulointerstitial injury (fibrosis) and representative Masson’s trichrome images. (D) Quantification of tubulointerstitial macrophage infiltration and immunohistochemistry kidney images from FHH and Nr4a1−/− bone marrow transplant animals at week 24. *P<0.05 versus control groups; †P<0.05 versus other groups. BW, body weight; KW, kidney weight.
Thioglycolate-Elicited and Bone Marrow–Derived Macrophage Isolation and Cell Culture
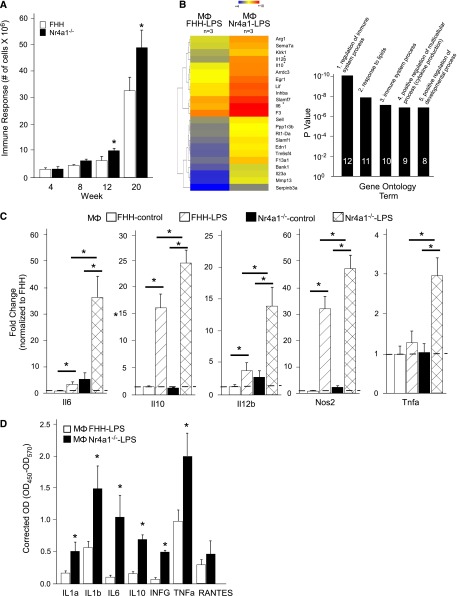
Thioglycolate-elicited immune cells were isolated from FHH and Nr4a1−/− rats at weeks 4, 8, 12, and 20. At weeks 4–8, there was no difference in immune response (i.e., number of cells isolated from intraperitoneal cavity). However, at weeks 12–20, there was a more significant cellular response in Nr4a1−/− rats compared with FHH controls (Figure 4A). Gene expression changes in important genes that characterize M1 or M2 polarized macrophages were studied at each time point (Supplemental Figure 8A). Macrophages isolated from Nr4a1−/− rats demonstrated downregulation of Il10, Il12b, and Tnf-α and upregulation of Nos2 from weeks 4 to 8, but by week 12 or 20 only Nos2 was significantly changed (increased in Nr4a1−/− rats). Kruppel-like factor 4 was significantly downregulated and suppressor of cytokine signaling 3 (Socs3) was upregulated.
Figure 4.
Thioglycolate-elicited macrophages and primary cell culture from FHH and Nr4a1−/− rats. (A) Quantitation of intraperitoneal cell infiltration at weeks 4, 8, 12, and 20. (B) Hierarchical clustering and gene ontology of differentially expressed genes between LPS-treated cultured macrophages (Mɸ) from FHH and Nr4a1−/− rats at week 20. Mɸ FHH-LPS is set as the control. The heat map colors denoted how genes are downregulated or upregulated within each group and across groups (FHH versus Nr4a1−/−). Genes denoted in blue are downregulated and genes denoted in red are upregulated. (C) Quantitative real-time PCR confirmation of important inflammatory genes identified by microarray on untreated and LPS-treated macrophages from each group. (D) Measurement of secreted inflammatory chemokines/cytokines by ELISA. n=6 for animal macrophage isolation. n=3–4 independent samples for primary culture studies. *P<0.05 versus FHH. RANTES, regulated upon activation, normal T cell expressed and secreted.
Macrophages isolated at week 20 were selected for additional study because this time frame (weeks 16–24) demonstrated the most significant renal injury based on proteinuria, histology, and decline in kidney function. Macrophages isolated from both groups were cultured with or without LPS. Whole genome microarray analysis of LPS-treated macrophages from Nr4a1−/− rats demonstrated a more significant upregulation of proinflammatory immune response genes, including Il-6, Il-12b, Il23, Tnfa, and Nos2 that typical characterize M1 polarized macrophages, and to a lesser extent several anti-inflammatory genes (Il-10 and Arg1) compared with controls (Figure 4, B and C). Quantitative real-time PCR and evaluation of secreted cytokines and chemokines (measured by ELISA) confirmed the microarray findings that LPS-treated macrophages isolated from Nr4a1−/− rats exhibited more robust proinflammatory immune activation compared with those isolated from FHH controls (Figure 4, C and D). Similar to the thioglycolate-elicited macrophages, Nr4a1−/− macrophages derived from bone marrow demonstrated a significant upregulation of Il-6, Il-12b, Tnfa, and Nos2 compared with FHH macrophages upon stimulation with LPS (Supplemental Figure 8B).
Discussion
In this study, we aimed to determine the role of Nr4a1 in the progression of kidney injury using the FHH rat, which is known to develop hypertension, proteinuria, and glomerular disease.11–13 Time course evaluation of renal injury and renal function in FHH rats deficient in Nr4a1 (Nr4a1−/−) demonstrated that loss of Nr4a1 leads to early onset of kidney injury and progressive decline in kidney function compared with wild-type FHH rats and this effect is independent of changes in systemic or renal hemodynamics. Histologically, kidneys from FHH and Nr4a1−/− rats exhibited a similar degree of glomerular and vascular injury that became more severe over the course of study. By contrast, tubulointerstitial injury was significantly increased in Nr4a1−/− rats and was characterized by macrophage infiltration and upregulation of inflammatory pathways, suggesting a role of immune cell infiltration in the pathogenesis of kidney disease in the Nr4a1−/− model. In support of these findings, transplantation of bone marrow from wild-type FHH into Nr4a1−/− rats resulted in an almost complete rescue of proteinuria and renal function phenotypes by week 24. In vitro studies using macrophages isolated from Nr4a1−/− rats demonstrated significant upregulation of proinflammatory immune response genes and secretion of chemokines/cytokines upon stimulation with LPS compared with controls. In total, these data suggest that the loss of Nr4a1 in cells of hematopoietic origin (e.g., lymphocytes) is the primary mechanism of the kidney injury in the Nr4a1−/− rats.
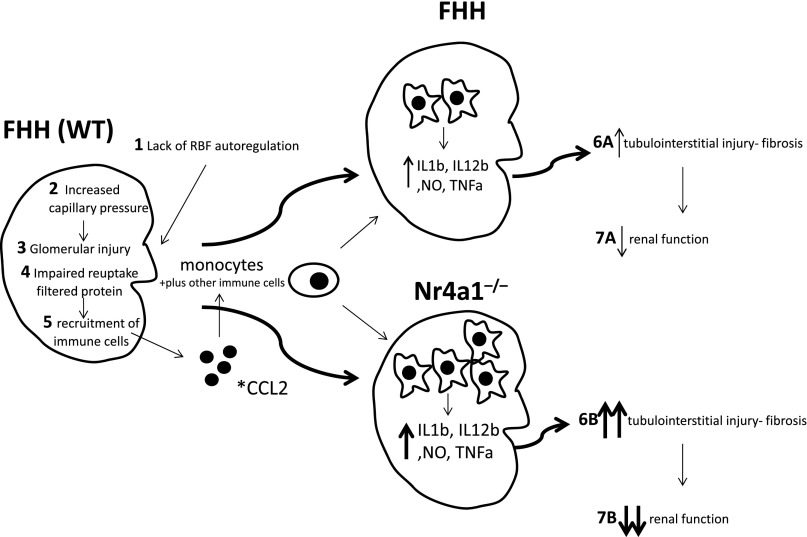
The enhanced kidney injury observed in Nr4a1−/− rats is likely caused, in part, by the FHH genetic background, which is permissive for the development of kidney injury. The mechanism of kidney injury in the FHH rats (summarized in Figure 5) is proposed to occur primarily through lack of RBF autoregulation that results in transmission of elevated systemic BP to the glomerulus, promoting increased capillary pressure and glomerular injury.14,15 The glomerular injury results in an increase in filtered protein that in combination with tubular defects impairs reuptake of filtered protein16 and leads to additional tubular injury and recruitment of inflammatory cells (monocytes). Thus, the FHH genetic background provides an environment in which loss of Nr4a1 enhances inflammatory processes that are already underway, leading to more injury in the kidney and culminating in an additional loss of renal function (Figure 5).
Figure 5.
Proposed pathophysiologic mechanism of enhanced immune kidney injury in Nr4a1−/− compared with FHH rats. Wild-type FHH animals exhibit a genetic predisposition to renal injury via the following: (1) lack of RBF autoregulation, (2) increased capillary pressure, (3) glomerular injury, (4) protein overload and tubular defects, and (5) recruitment of inflammatory cells. This mechanism, including lack of autoregulation, tubular defects, and inflammation, promotes the progressive injury and reduced renal function (6 and 7) observed in the FHH. Loss of Nr4a1 on the FHH genetic background provides an environment in which enhanced immune activation (shift toward M1 macrophages and possibly other immune cells) exacerbates and/or accelerates inflammatory processes that lead to more injury, culminating in an additional reduction in renal function compared with FHH. *CCL2 also known or monocyte chemoattractant protein-1 (MCP-1) was the top differentially expressed gene and upregulated in kidney from Nr4a1−/− compared with FHH rats (Supplemental Figure 6). WT, wild type; NO, nitric oxide.
Kidney injury is a complex pathophysiologic process that involves a number of different types of immune cells and processes. In particular, monocytes/macrophages play a pivotal role in regulating the function of the immune system and their dysfunction can lead to chronic inflammation.17 Macrophages are typically classified into two distinct types: M1, which represent a proinflammatory state, and M2 (with additional subcategories M2a, b, and c), which exhibit anti-inflammatory functions. A disruption in the balance between the two can lead to the sustained activation of M1 and predispose to chronic inflammatory states.17 Our data demonstrate that loss of Nr4a1 leads to a robust macrophage response in kidneys from Nr4a1−/− rats, which corresponds with progressive proteinuria and renal function decline. Although, we did not specifically assess the degree of M1 or M2 macrophages in this study, the gene expression profiles of unstimulated isolated macrophages at multiple time points suggest differential regulation of Il10, Il12b, Tnf-α, and Nos2 between FHH- and Nr4a1−/−-derived macrophages. In addition, downregulation of Kruppel-like factor 4 and upregulation of Socs3, both observed in Nr4a1−/− unstimulated macrophages, were previously associated with M1 macrophage activation.18,19
LPS-stimulated macrophages from Nr4a1−/− rats (either thioglycolate or bone marrow derived) demonstrate a significant and robust M1 macrophage response characterized by increased Il6, Il12b, Tnf-α, and Nos2 expression and protein levels compared with FHH rats. Contrary to these findings, LPS-stimulated macrophages also resulted in increased Il10 expression, an anti-inflammatory cytokine. This was unexpected, but may be a consequence of a mixed population that includes M2 polarized macrophages. In total, in vivo studies demonstrate significant macrophage infiltration that correlates with kidney injury and in vitro studies demonstrate that loss of Nr4a1 may result in an imbalance in M1 and M2 macrophages, with M1 macrophages demonstrating enhanced immune responses when stimulated.
NR4A receptors were first studied in the brain and early studies suggested a role in apoptosis and proliferation.20 Subsequently, in vitro and in vivo studies in metabolic tissues, including skeletal muscle, adipose, and liver cells, identified an association between the NR4A subgroup and specific aspects of lipid, carbohydrate, and energy homeostasis.21 For example, Nr4a1 (Nur77)−/− mice were shown to exhibit insulin resistance (muscle and liver), altered glucose metabolism, and increased lipid levels that resulted in hepatic steatosis.22 Increased lipid levels and hepatic steatosis were also observed in this study, suggesting that the observed metabolic changes in the Nr4a1−/− model are likely independent from the observed decline in kidney function.
The role of Nr4a1 (Nur77) in vascular disease, namely atherosclerosis, has been extensively investigated.4,9 Like CKD, atherosclerosis involves a significant inflammatory component that is characterized by infiltration of monocytes (in response to endothelial activation) as well as differentiation into macrophages that release cytokines/growth factors that aggravate inflammation and lead to activation of smooth muscle cells that proliferate into the lesion.23 A recent study found that mice transplanted with Nr4a1-deficient bone marrow exhibited a significant increase in atherosclerosis compared with wild-type bone marrow transplanted controls.24 These animal studies as well as cell culture experiments (exposure to LPS) demonstrated that macrophages deficient in Nr4a1 leads to a proinflammatory phenotype (polarization toward M1) and exacerbates the development of atherosclerotic lesions.25 In addition, in vitro studies that performed knockdown of Nr4a1 found enhanced macrophage activation demonstrated by increased inflammatory cytokine expression.26 Macrophages stimulated with LPS or INF-γ assume an M1 proinflammatory phenotype characterized by a high expression level of Nos2, a high capacity to present antigen, and production of proinflammatory cytokines such as TNF-α and IL-1β. Conversely, overexpression of Nr4a1 in human THP-1 and U937 macrophages was observed to lead to decreased inflammatory cytokines after stimulation with LPS (IL-1β, IL-6, IL-8, macrophage inflammatory protein-1α, macrophage inflammatory protein-1β, and monocyte chemoattractant protein-1).26 Our bone marrow crosstransplantation studies, in vitro macrophage assessment, and proposed mechanism of kidney injury (Figure 5) are consistent with the conclusions reached by atherosclerosis experiments and in vitro macrophage studies (i.e., that loss of Nr4a1 in macrophages likely plays a prominent role in observed pathology).
To our knowledge, this is the first study to investigate the potential role of Nr4a1 in the context of CKD. There is a single report that describes the loss of Nr4a1 (in mice) in promoting epithelial apoptosis and inflammation after ischemia-reperfusion injury.27 In this case, loss of Nr4a1 in the kidney itself appears to play a significant role in the observed injury and although we cannot eliminate a possible role of loss of Nr4a1 in the kidney itself, an immune-mediated mechanism appears to be the predominant mechanism in our CKD model. This apparent pleiotropic role of Nr4a1 is consistent with functional studies demonstrating both a pro- and anti-inflammatory role for Nr4a1 that is dependent on acute versus chronic expression and cell type.21 Previous work on the genetic basis of GN identified Jund, another transcription factor, with macrophage activation and GN susceptibility in the WKY rat.28 This study provides additional evidence that transcription factors may play a vital role in macrophage activation status and may promote chronic inflammatory responses.
In summary, our data suggest that loss of Nr4a1 in cells of hematopoietic origin (e.g., macrophages) promotes increased kidney injury and reduced renal function observed in the Nr4a1−/− model. However, the ability of Nr4a1-deficient macrophages to exacerbate injury in the kidney is likely dependent on the predisposition of the genetic background (FHH) to exhibit established renal injury and inflammation. This work establishes that Nr4a1, through immune activation, can have a significant role in promoting kidney injury and future studies aimed at enhancing Nr4a1 expression or function could provide a novel therapeutic strategy to treat kidney disease.
Concise Methods
Animals and Study Design
FHH and Nr4a1−/− rats were obtained from the Medical College of Wisconsin. Nr4a1−/− rats were originally generated using an ENU mutagenesis strategy (N-ethyl-N-nitrosourea) on the genetic background of the FHH rat through the PhysGen Program (http://pga.mcw.edu) at the Medical College of Wisconsin. All experiments were approved by the institutional animal care and use committees at the Medical College of Wisconsin and the University of Mississippi Medical Center.
Protocol 1: Association of Genetic Mutation with Proteinuria
An F2 segregating population was developed to validate that the genetic mutation (i.e., knockout) was associated with renal injury (proteinuria).29 Male FHH animals were crossed to Nr4a1−/− female rats to produce F1(FHH×Nr4a1−/−) animals. F1 animals were intercrossed to produce the F2(FHH×Nr4a1−/−) (n=175). Proteinuria was measured at weeks 8 and 12 after 24 hours urine collection in metabolism cages as previously described.30 Nr4a1−/− were significantly smaller in size compared with FHH controls and a significant correlation was observed between proteinuria and body weight. Thus, it was necessary to normalize proteinuria for differences in body weight and it is reported as mg/24 hours per 100 g body wt.
Protocol 2: Temporal Changes in Renal and Cardiovascular Traits
Age-matched male Nr4a1−/− rats (n=28) and FHH rats (n=28) were studied for telemetry-measured BP (n=8), proteinuria, renal function, and renal injury at 4, 8, 12, 16, and 20 weeks-of-age. At each time point, six animals per group were euthanized to measure body, heart, kidney, and liver weights and the remainder of animals were aged to the next time point. Kidney, heart, and liver samples were processed for histologic examination and serum samples were obtained from cardiac puncture to measure blood parameters as previously described.30,31 A separate group of Nr4a1−/− rats (n=11) and FHH rats (n=11) were grown to week 24 for additional measurements.
Protocol 3: BP and Renal Hemodynamics
Age-matched male Nr4a1−/− and FHH rats were raised until week 16 (n=4–6 per group) and week 24 (n=6 per group). Mean arterial pressure, RBF, autoregulation, and GFR were measured as previously described.30
Protocol 4: Bone Marrow Transplantation Studies
Bone marrow transplant studies were performed on FHH and Nr4a1−/− animals. Donor animals were euthanized at week 8 with an overdose of pentobarbital. Femurs were removed from donor rats, either FHH (n=6) and Nr4a1−/− (n=6). Bone marrow was flushed from the femur using RPMI cell culture media containing 10% FBS (Sigma-Aldrich), 100 U/ml penicillin, and 100 g/ml streptomycin (Invitrogen). Cells were centrifuged under low-speed (250×g), washed, and resuspended in fresh medium at 5×107 cells/ml. Recipient animals were irradiated with approximately 6 Gy at 2.56 Gy/min (approximately 2.00 minutes) using γ rays from a cesium source irradiator (JL Shephard). Recipient animals were placed under anesthesia using 2%–3% isoflurane and were administered an intravenous injection of 0.2 ml of bone marrow cell preparation by tail vein within 2 hours of irradiation. Recipient animals were divided into the following four groups: (1) FHHbm to FHHirr (control; n=8), (2) FHHbm to Nr4a1irr−/− (n=7), (3) Nr4a1bm−/− to Nr4a1irr−/− (control; n=8), and (4) Nr4a1−/− bm to FHHirr (n=8). Rats were allowed to recover for 4 weeks to reconstitute their bone marrow, at which time the animals were evaluated (week 12) for measures of renal injury (proteinuria) and again at weeks 16, 20, and 24. DNA isolated from the spleen was genotyped for presence of the wild-type or mutation in exon 1 (Nr4a1) to confirm that bone marrow was successfully transplanted. FHHbm to Nr4a1irr−/− animals were found to be heterozygous (i.e., FHH and Nr4a1 genotype). However, we were unable to confirm the successful transplant of Nr4a1bm−/− to FHHirr, suggesting that the loss of Nr4a1 may have an effect on the ability of the Nr4a1-derived bone marrow cells to establish themselves in the context of wild-type hematopoietic cells. Thus, the Nr4a1bm−/− to FHHirr group was not included in the study.
Protocol 5: Macrophage Isolation and Cell Culture
Male FHH and Nr4a1−/− rats (n=6 per group/each time) were given an intraperitoneal injection of 4% thioglycolate (Sigma-Aldrich) solution in PBS (Thermo Fisher Scientific) at weeks 4, 8, 12, and 20 as previously described by others.32 At day 4 postinjection, animals were euthanized under 2%–3% isoflurane gas. PBS containing 3% FBS (Invitrogen) was injected into the peritoneal cavity. The abdomen was massaged briefly to release any adherent cells and a small incision was created in the lower portion of the abdomen and the fluid was removed using a sterile transfer pipet. Cells were collected by centrifugation, resuspended in sterile media, and counted using a cytometer (USA Scientific). For cell culture, approximately 4–6×106 cells (week 20) were placed in sterile, nontissue culture 100 mm×15 mm petri dishes with DMEM/F12 media (Invitrogen) supplemented with 10% FBS. Cells were incubated at 5% CO2/37°C in an incubator for 2 hours to allow macrophages to adhere. Nonadherent B and T cells were washed away using sterile media after the incubation. Cells were either (1) treated with 1 µg/ml LPS (Escherichia coli 055:B5; Sigma-Aldrich) in DMEM/F12/10% FBS solution for 18 hours or (2) untreated in DMEM/F12/10% FBS solution for 18 hours (control). After incubation, the supernatant was removed and stored in 96-well plates at −80°C for later analysis of cytokine/chemokine production using an ELISA (Qiagen). Bone marrow–derived macrophages were derived from FHH and Nr4a1−/− rats as previously described by others.28 Bone marrow was isolated from animals similar to protocol 4, and total bone marrow cells were counted, plated, and cultured for 5 days using DMEM (Gibco), heat inactivated FBS (Invitrogen), macrophage colony-stimulating factor, (ProSpec Bio), and penicillin/streptomycin (Invitrogen) in a 37°C/5% CO2 environment. Adherent cells were then treated with DMEM/F12-10 containing LPS (E. coli 055:B5; Sigma-Aldrich) or DMEM/F12-10 media alone for 4 hours. Macrophages were removed using sterile PBS and a cell scraper and were processed for RNA. RNA was used for gene expression using the Affymetrix GeneAtlas system or real-time PCR.
Histology and Immunohistochemistry
Kidneys were fixed in 10% buffered formalin, embedded in paraffin, cut into 4-μm sections and stained with hematoxylin and eosin and Masson’s trichrome (n=5–6 per group under each protocol/time point). Samples processed for electron microscopy were processed as previously described.33 Kidney sections were evaluated by light microscopy for the presence of glomerular injury (n=20 glomeruli per kidney section) including the degree of glomerulosclerosis and mesangial expansion as previously described.31,33 Interstitial fibrosis was determined by evaluation of slides stained with Masson’s trichrome to quantify the percent fibrosis (blue staining) compared with the background. Tubular injury was assessed by degree of tubular atrophy, vacuolization, dilation, and/or protein casts on a scale from 0 (normal) to 4 (global sclerosis) as previously described.34 Vascular injury was assessed by wall thickening and cellular proliferation. Vessel wall thickening (vessel media, in micrometers squared) was calculated by measuring the outer circumference of the vessel minus the inner circumference of the lumen (20 random images at ×40 per rat).
Macrophage, T-cell, and B-cell infiltration was assessed by immunohistochemistry on unstained sections using primary antibodies directed at CD68/ED-1, CD22, and CD43 (Santa Cruz Biotechnology) and detected by 3,3′-diaminobenzidine (Ultravision LPValue Detection System; Thermo Fisher Scientific).30 Slides were counterstained with methyl green (n=4 sections per group; 15–20 images). Images were captured using a Nikon 55i microscope with a DS-Fi1 5-Meg Color C digital camera (Nikon, Melville, NY) and were analyzed using Nis-Elements image analysis software (version 3.03; Nikon).
Molecular Methods
Genotyping and Sequencing
Genomic DNA was obtained by tail biopsy and prepared using Wizard SV 96 Genomic DNA kit (Promega, San Luis Obispo, CA). Genotyping was done using standard PCR-ethidium bromide-gel electrophoresis. The point mutation in exon 1 (Nr4a1) created an AseI restriction site in the Nr4a1−/− that was used for genotyping segregating population (Supplemental Figure 1). Primers were designed to amplify across the point mutation in exon 1 (Nr4a1-F, 5′-TCGTCCTCGGCCACCTCTCC-3′; Nr4a1-R, 5′-GCCTTCATAAGTCTGGCTGGGGG-3′). DNA sequencing was performed on a CEQ8000 using the DTCS Quick Start Kit (Beckman Coulter, Brea, CA). Sequencing reads were assessed for quality and aligned to the BN reference sequence using the DNASTAR Lasergene v7.2 software package.
Microarray and Real-Time PCR
Whole genome transcript analysis was performed using Affymetrix platforms (kidney tissue collected under protocol 1) from FHH and Nr4a1−/− rats at weeks 8 (n=3 per group), 16 (n=3), and 24(n=3). RNA was isolated using TRIzol and Invitrogen PureLink Kit (Life Technologies) and evaluated for quality and integrity (Bio-Rad Experion System) as previously described.35 Kidney RNA time course samples were processed per the manufacturer’s directions for specific application (GeneChip 1.0 ST) using Affymetrix equipment (Scanner 3000 7G System) or primary macrophage cultures (GeneAtlas). Hybridized chips were automatically washed, stained, and scanned at the University of Mississippi Medical Center Institutional Molecular and Genomics Core using Affymetrix equipment. Data obtained from these gene expression studies are deposited in the Gene Expression Omnibus database (accession number GSE54714; http://www.ncbi.nlm.nih.gov/geo).
Analysis of microarray data was performed using software provided by Affymetrix (Affymetrix Expression Console Software) and the commercially available GeneSifter software platform (http://www.genesifter.net) and/or Array Star 5 (DNASTAR Inc., Madison, WI). In brief, differentially expressed genes evaluated by a t test using two methods: (1) family-wise error rate procedure, using P<0.05 and a fold change ≥±1.2; and/or (2) Benjamani and Hochberg false discovery rate, which corrects for multiple comparison, using P<0.05 and a fold change ≥±1.2. Two-way ANOVA using a P<0.01 or lower was performed to identify strain×time interaction effects between groups. Gene networks and functional analysis were evaluated using Ingenuity Pathways Analysis (http://www.ingenuity.com; Ingenuity Systems). Gene expression differences were confirmed using SYBR Green dye chemistry on Bio-Rad CFX96 (n=6–8 per group). RNA was reverse-transcribed to cDNA using an iScript cDNA Synthesis Kit and real-time PCR (Bio-Rad) was performed using SsoFast EvaGreen Supermix (Bio-Rad). Predesigned PrimeTime quantitative PCR primers were purchased from IDT to assess gene expression levels of chemokines, cytokines, and growth factors.
Statistical Analyses
Linkage study associations between genotype and phenotype were assessed for the Nr4a1 locus using ANOVA within the SPSS program (Chicago, IL). An independent t test or one-ANOVA followed by Bonferroni correction for multiple comparisons was used for described animal experiments. A P<0.05 was considered statistically significant. All data are presented as the mean±SEM.
Disclosures
None.
Supplementary Material
Acknowledgments
M.R.G is supported by the National Institutes of Health National Heart, Lung, and Blood Institute (HL094446), the Robert M. Hearin Foundation, and the Research Center of Excellence in Pediatric Nephrology (RCEPN) at the Medical College of Wisconsin. K.R.R is supported by funds from RCEPN and the Clinical and Translational Science Institute at the Medical College of Wisconsin. The work performed through the University of Mississippi Medical Center Molecular and Genomics Facility is supported, in part, by funds from the National Institutes of Health National Institute of General Medical Sciences, including the Mississippi IDeA Networks of Biomedical Research Excellence (P20GM103476), Center for Psychiatric Neuroscience–Center of Biomedical Research Excellence (P30GM103328) and Obesity, Cardiorenal, and Metabolic Diseases–Center of Biomedical Research Excellence (P20GM104357). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013070786/-/DCSupplemental.
References
- 1.US Renal Data Service : USRDS 2011 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda, MD, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2011 [Google Scholar]
- 2.Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS: Prevalence of chronic kidney disease in the United States. JAMA 298: 2038–2047, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Trivedi H: Cost implications of caring for chronic kidney disease: Are interventions cost-effective? Adv Chronic Kidney Dis 17: 265–270, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Pearen MA, Muscat GEO: Minireview: Nuclear hormone receptor 4A signaling: Implications for metabolic disease. Mol Endocrinol 24: 1891–1903, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-González J, Badimon L: The NR4A subfamily of nuclear receptors: New early genes regulated by growth factors in vascular cells. Cardiovasc Res 65: 609–618, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Mohan HM, Aherne CM, Rogers AC, Baird AW, Winter DC, Murphy EP: Molecular pathways: The role of NR4A orphan nuclear receptors in cancer. Clin Cancer Res 18: 3223–3228, 2012 [DOI] [PubMed] [Google Scholar]
- 7.Kanzleiter T, Wilks D, Preston E, Ye J, Frangioudakis G, Cooney GJ: Regulation of the nuclear hormone receptor nur77 in muscle: Influence of exercise-activated pathways in vitro and obesity in vivo. Biochim Biophys Acta 1792: 777–782, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Fu Y, Luo L, Luo N, Zhu X, Garvey WT: NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: Potential role in insulin resistance. J Biol Chem 282: 31525–31533, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Bruemmer D: NR4A orphan nuclear receptors: Transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol 30: 1535–1541, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMorrow JP, Murphy EP: Inflammation: A role for NR4A orphan nuclear receptors? Biochem Soc Trans 39: 688–693, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Kuijpers MH, Gruys E: Spontaneous hypertension and hypertensive renal disease in the fawn-hooded rat. Br J Exp Pathol 65: 181–190, 1984 [PMC free article] [PubMed] [Google Scholar]
- 12.Simons JL, Provoost AP, Anderson S, Troy JL, Rennke HG, Sandstrom DJ, Brenner BM: Pathogenesis of glomerular injury in the fawn-hooded rat: Early glomerular capillary hypertension predicts glomerular sclerosis. J Am Soc Nephrol 3: 1775–1782, 1993 [DOI] [PubMed] [Google Scholar]
- 13.Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, Cowley AW, Jr, Jacob HJ: Chromosomal mapping of the genetic basis of hypertension and renal disease in FHH rats. Am J Physiol Renal Physiol 293: F1905–F1914, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Van Dokkum RPE, Alonso-Galicia M, Provoost AP, Jacob HJ, Roman RJ: Impaired autoregulation of renal blood flow in the fawn-hooded rat. Am J Physiol 276: R189–R196, 1999 [DOI] [PubMed] [Google Scholar]
- 15.van Dokkum RPE, Sun C-W, Provoost AP, Jacob HJ, Roman RJ: Altered renal hemodynamics and impaired myogenic responses in the fawn-hooded rat. Am J Physiol 276: R855–R863, 1999 [DOI] [PubMed] [Google Scholar]
- 16.Rangel-Filho A, Lazar J, Moreno C, Geurts A, Jacob HJ: Rab38 modulates proteinuria in model of hypertension-associated renal disease. J Am Soc Nephrol 24: 283–292, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray PJ, Wynn TA: Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 11: 723–737, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, Flask CA, Kim J, Doreian BW, Lu KQ, Kaestner KH, Hamik A, Clément K, Jain MK: Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest 121: 2736–2749, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold CE, Whyte CS, Gordon P, Barker RN, Rees AJ, Wilson HM: A critical role for SOCS3 in promoting M1 macrophage activation and function in vitro and in vivo. Immunology 141: 96–110, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson MA, Milbrandt J: The NGFI-B gene, a transcriptionally inducible member of the steroid receptor gene superfamily: Genomic structure and expression in rat brain after seizure induction. Mol Cell Biol 9: 4213–4219, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxwell MA, Muscat GE: The NR4A subgroup: Immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal 4: e002, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao LC, Wroblewski K, Zhang Z, Pei L, Vergnes L, Ilkayeva OR, Ding SY, Reue K, Watt MJ, Newgard CB, Pilch PF, Hevener AL, Tontonoz P: Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes 58: 2788–2796, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woollard KJ, Geissmann F: Monocytes in atherosclerosis: Subsets and functions. Nat Rev Cardiol 7: 77–86, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamers AAJ, Vos M, Rassam F, Marinković G, Kurakula K, van Gorp PJ, de Winther MP, Gijbels MJ, de Waard V, de Vries CJ: Bone marrow-specific deficiency of nuclear receptor Nur77 enhances atherosclerosis. Circ Res 110: 428–438, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Hanna RN, Shaked I, Hubbeling HG, Punt JA, Wu R, Herrley E, Zaugg C, Pei H, Geissmann F, Ley K, Hedrick CC: NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ Res 110: 416–427, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonta PI, van Tiel CM, Vos M, Pols TW, van Thienen JV, Ferreira V, Arkenbout EK, Seppen J, Spek CA, van der Poll T, Pannekoek H, de Vries CJ: Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler Thromb Vasc Biol 26: 2288–2294, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Balasubramanian S, Jansen M, Valerius MT, Humphreys BD, Strom TB: Orphan nuclear receptor Nur77 promotes acute kidney injury and renal epithelial apoptosis. J Am Soc Nephrol 23: 674–686, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behmoaras J, Bhangal G, Smith J, McDonald K, Mutch B, Lai PC, Domin J, Game L, Salama A, Foxwell BM, Pusey CD, Cook HT, Aitman TJ: Jund is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nat Genet 40: 553–559, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrett MR, Dene H, Rapp JP: Time-course genetic analysis of albuminuria in Dahl salt-sensitive rats on low-salt diet. J Am Soc Nephrol 14: 1175–1187, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Williams JM, Johnson AC, Stelloh C, Dreisbach AW, Franceschini N, Regner KR, Townsend RR, Roman RJ, Garrett MR: Genetic variants in Arhgef11 are associated with kidney injury in the Dahl salt-sensitive rat. Hypertension 60: 1157–1168, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regner KR, Harmon AC, Williams JM, Stelloh C, Johnson AC, Kyle PB, Lerch-Gaggl A, White SM, Garrett MR: Increased susceptibility to kidney injury by transfer of genomic segment from SHR onto Dahl S genetic background. Physiol Genomics 44: 629–637, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray A, Dittel BN: Isolation of mouse peritoneal cavity cells. J Vis Exp 35: 1488, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrett MR, Gunning WT, Radecki T, Richard A: Dissection of a genetic locus influencing renal function in the rat and its concordance with kidney disease loci on human chromosome 1q21. Physiol Genomics 30: 322–334, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Packard M, Saad Y, Gunning WT, Gupta S, Shapiro J, Garrett MR: Investigating the effect of genetic background on proteinuria and renal injury using two hypertensive strains. Am J Physiol Renal Physiol 296: F839–F846, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng H, Garrett MR, Dene H, Rapp JP: Localization of a blood pressure QTL to a 2.4-cM interval on rat chromosome 9 using congenic strains. Genomics 81: 210–220, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.