Abstract
We aimed to design and develop a comprehensive mammography database system (CMDB) to collect clinical datasets for outcome assessment and development of decision support tools. A Health Insurance Portability and Accountability Act (HIPAA) compliant CMDB was created to store multi-relational datasets of demographic risk factors and mammogram results using the Breast Imaging Reporting and Data System (BI-RADS) lexicon. The CMDB collected both biopsy pathology outcomes, in a breast pathology lexicon compiled by extending BI-RADS, and our institutional breast cancer registry. The audit results derived from the CMDB were in accordance with Mammography Quality Standards Act (MQSA) audits and national benchmarks. The CMDB has managed the challenges of multi-level organization demanded by the complexity of mammography practice and lexicon development in pathology. We foresee that the CMDB will be useful for efficient quality assurance audits and development of decision support tools to improve breast cancer diagnosis. Our procedure of developing the CMDB provides a framework to build a detailed data repository for breast imaging quality control and research, which has the potential to augment existing resources.
Keywords: database, breast cancer, mammography, pathology lexicon, data repository
Introduction
Mammography, one of the few screening tests supported by high-quality randomized controlled trials demonstrating mortality benefit, plays a key role in early breast cancer diagnosis. Mammography helps decrease breast cancer deaths; however, it is not perfect and has nontrivial false-positive (recall for additional images or biopsy) and false-negative (missed cancer) rates. Thus, substantial effort is being invested to improve interpretive performance of mammography via decision support tools.1,2 Further advancement of these tools demands mammography databases store a large number of mammography reports for their training, testing, and evaluation. Another principle method of improving performance is to implement medical audits to ensure adequate mammography screening performance,3 which also demands mammography databases store mammography reports for comparison with existing benchmarks and guidelines. Therefore, mammography databases play an important role in improving breast cancer screening and diagnosis.
Substantial effort has been already expended to develop mammography databases. Most of publically available mammography databases are used to store mammography images for developing computer aided detection (CAD) systems, including the Mammographic Image Analysis Society (MIAS) Digital Mammogram Database,4 the Nijmegen Digital Mammogram Database,5 the Lawrence Livermore National Laboratories/University of California at San Francisco Database,6 the University of Chicago/University of North Carolina Database,7 INbreast Database,8 and the BancoWeb LAPIMO Database.9 The Digital Database for Screening Mammography (DDSM) stores both mammography images and mammography reports;10 however, it currently contains approximately 2,500 cases, which may be inadequate for training and testing decision support tools that would perform accurately in practice. In addition, cases in many of these databases are selected to represent the variety of observations, which do not emulate actual mammography practice because diagnoses are not accurately represented by expected, real-world prevalence. It is necessary to collect consecutive mammography reports for developing reliable decision support tools and implementing accurate medical audits. On the other hand, structured reporting systems are currently available to aid mammography facilities to maintain consistency, accuracy, and completeness of mammography reports. These systems have back-end databases to record important variable and generate reports for millions of mammograms collected from each facility; however, they use proprietary methods for storage and indexing of reports,11 instead of standardized, well-documented, and transparent methods, which makes data sharing and communication for implementing audits and developing decision support tools difficult.
The substantial opportunities for mammography facilities to benefit from database management strategies are in large part due to the efforts of the American College of Radiology (ACR). The ACR developed the Breast Imaging Reporting and Data System (BI-RADS) lexicon,12 which standardizes mammography reporting and provides a guide on mammography audits and performance measures.13 More recently, the ACR has launched the centralized National Mammography Database (NMD),14 which collects demographics and mammogram results in order to provide outcomes for practices to judge consistency and accuracy of mammogram interpretation as compared to national benchmarks and guidelines.15 National standards, like BI-RADS and the NMD, provide an excellent foundation for the development and use of a public database system in breast cancer research.16
Several specifications are necessary to fully realize the potential to enable database management systems for breast cancer research. In a typical clinical practice, biopsy results (pathology) are used as a ground truth/outcome measure, presenting several challenges. While mammogram results are summarized according to the BI-RADS lexicon, pathology outcomes are typically only available using a free-text reporting model rather than a structured lexicon, which largely precludes automated outcome matching processes and data sharing. This limitation motivated our use of a breast pathology lexicon to standardize pathology outcomes. Further complicating this issue is the fact that core biopsy pathology results can be incorrect in 5–15% of cases17–20 due to under-sampling. As a more accurate reference standard, breast cancer registry matching has been recommended,21 and as a result, many of the national benchmarks and guidelines are based on cancer registries. A database management practice ideally includes registries for accurate and complete evaluation of interpretive performance.
Moreover, resources like the NMD only focus on outcomes on the mammogram level, whereas radiologists often rely on decision support tools to estimate the risk of breast cancer on the abnormality level and primary care physicians are often more interested in results on the patient level. Accordingly, the NMD records descriptions of the single-most significant mammographic finding, but does not support collection of secondary or associated findings which may substantively influence care and result tracking.16 A well-designed entity-relationship model is preferred when developing a database that will take into account millions of mammogram results on the abnormality, mammogram, and patient levels.22
The purpose of this manuscript is to illustrate our design and development of a comprehensive mammography database system (CMDB) with an entity-relationship database model, which provides rich and reproducible outcomes (structured breast pathology and a cancer registry match), supports data collection and sharing on multiple levels relevant to care, and enables development of decision support tools for improving mammography practice.
Methods
The Institutional Review Board (IRB) of the University of Wisconsin Hospital and Clinics (UWHC) exempted this Health Insurance Portability and Accountability Act (HIPAA) compliant database design study from requiring informed consent.
Database design and implementation
Database tables for mammogram results
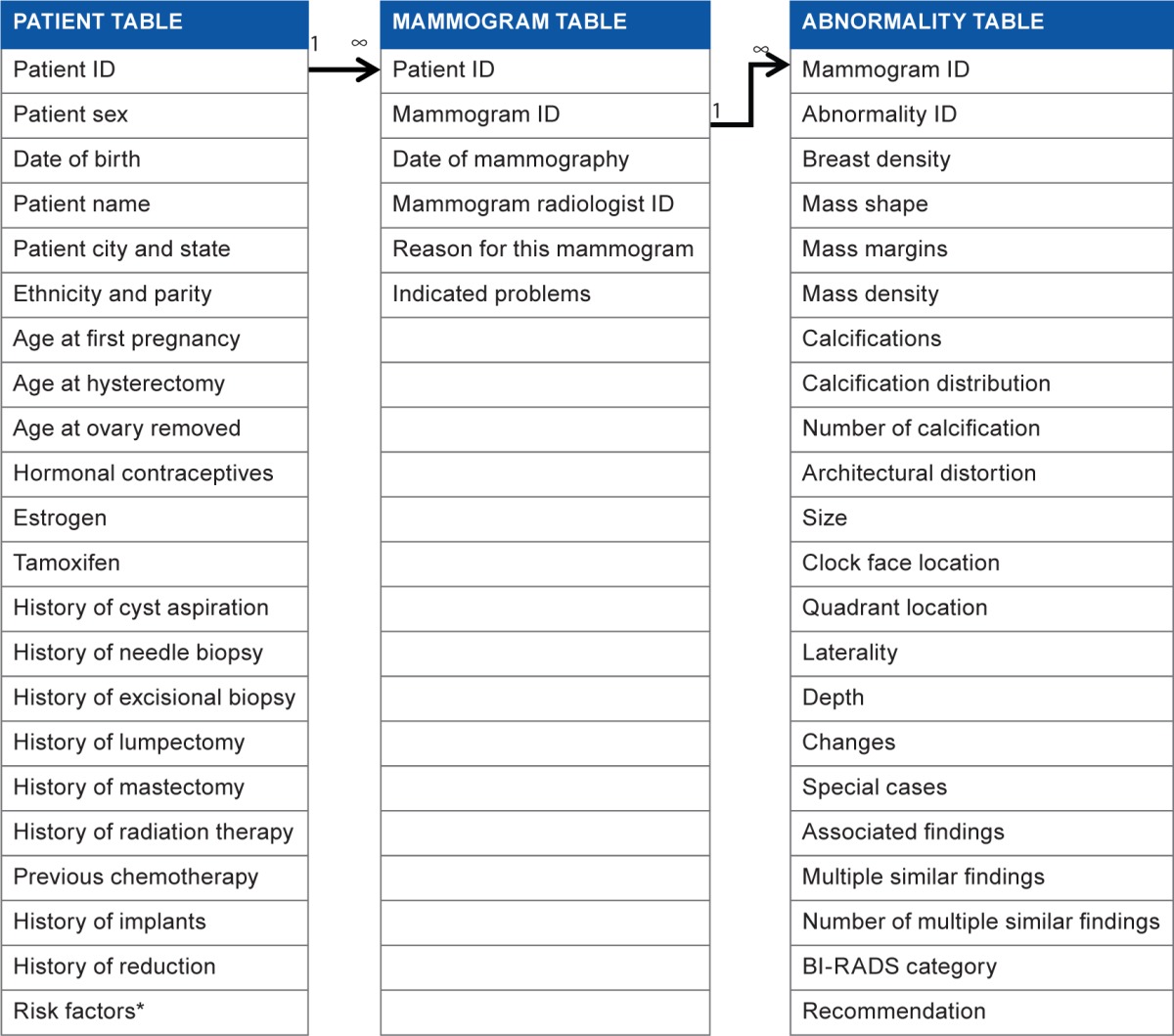
Based on BI-RADS lexicon, we use an entity-relationship database structure to manage mammography data on three levels of representation: patient, mammogram, and abnormality. In our CMDB, specifically, we use a patient table to store patient’s demographic information and use the patient ID to specify each entry uniquely (Table 1). We use a mammogram table to save mammogram data (date of mammography, ID of the radiologist, who interprets this mammogram, the reason for the mammogram, and indicated problems); we use a mammogram ID to specify each mammogram uniquely. Similarly, we use an abnormality table to record abnormalities of mammograms and use an abnormality ID to specify each abnormality entry. Unique IDs in each table assure that no duplicate entries exist.
Table 1.
Patient table, mammogram table, and abnormality table.
|
Database tables for image-guided biopsy pathology outcomes
To address the lack of standardization and uniformity in breast biopsy result-reporting, we designed and compiled a breast pathology lexicon including pathology description, pathology description abbreviation, pathology category (benign vs. high risk vs. malignant), and display order (an example was shown in Table 2). We created the terminology of breast diseases and their abbreviation by uniquely identifying each possible breast pathology diagnoses as described in BI-RADS third edition.23 Pathology category is specified by a consensus panel of radiologists and pathologists for each breast disease.
Table 2.
Pathology lexicon (an example).
| PATHOLOGYDESCRIPTION (PATHDX) | PATHDX ABBREVIATION | PATHOLOGY CATEGORY | DISPLAY ORDER |
|---|---|---|---|
| Mixed invasive ductal and DCIS | DCISDCNOS | malignant | 1 |
| Invasive ductal carcinoma (DCNOS) | DCNOS | malignant | 2 |
| Invasive lobular carcinoma (LC) | LC | malignant | 3 |
| Mixed invasive lobular and LCIS (LCLCIS) | LCLCIS | malignant | 4 |
| Intraductal carcinoma (DCIS) | DCIS | malignant | 5 |
| Invasive papillary carcinoma | PapCA | malignant | 6 |
| Medullary carcinoma | MedCA | malignant | 7 |
| Mucinous (colloid) carcinoma | CollCA | malignant | 8 |
| Tubular carcinoma | TubCA | malignant | 9 |
| Fibroadenoma | FA | benign | 10 |
| Papilloma | Pap | benign | 11 |
| Cyst | Cy | benign | 12 |
| …… | …… | …… | …… |
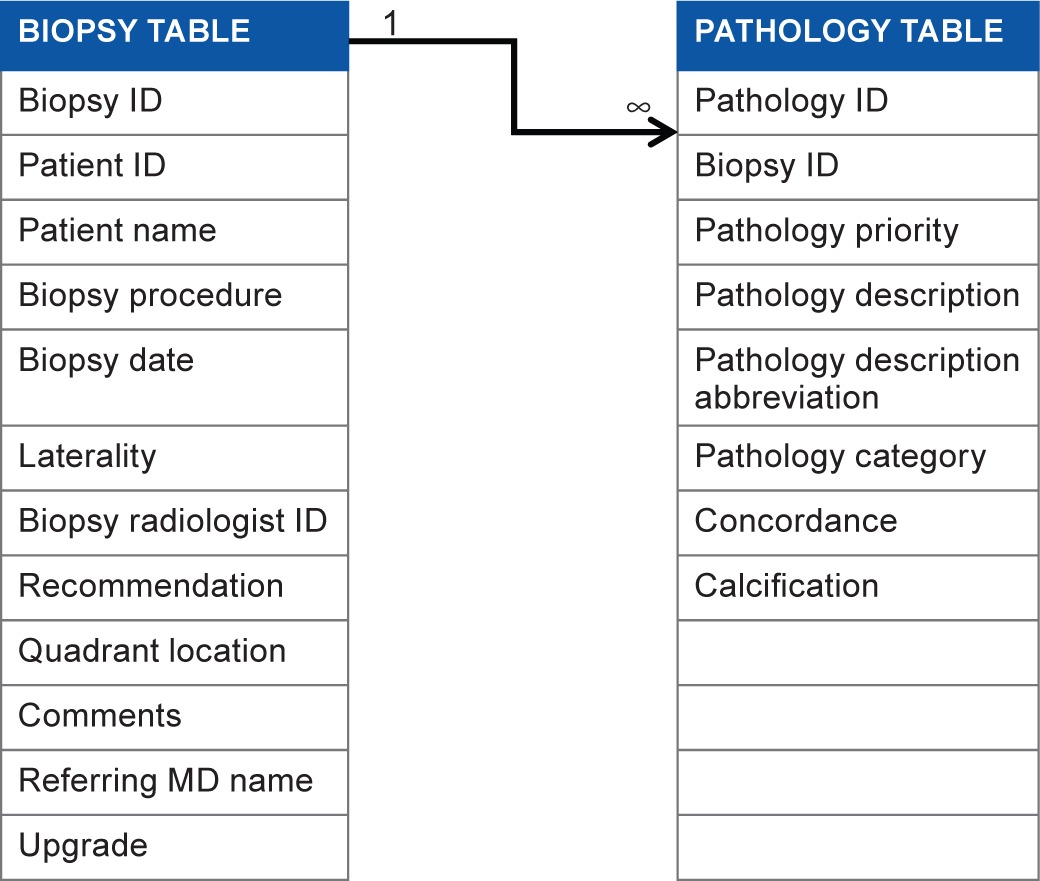
Once the breast pathology lexicon was compiled, we extended the entity-relationship structure of our CMDB to utilize these standard terms for results and outcome tracking. We manage biopsy pathology outcomes on two levels of representation; we use a biopsy table to record details from each biopsy and a pathology table to record pathologic observations by using breast pathology lexicon. We use a biopsy ID and a pathology ID to uniquely specify each record in two tables (Table 3).
Table 3.
Biopsy table and pathology table.
|
Database table for breast cancer registry outcomes
We use a registry table to store registry outcomes, which contains a registry ID field to uniquely specify each record obtained from our cancer center registry (Table 4).
Table 4.
Registry table.
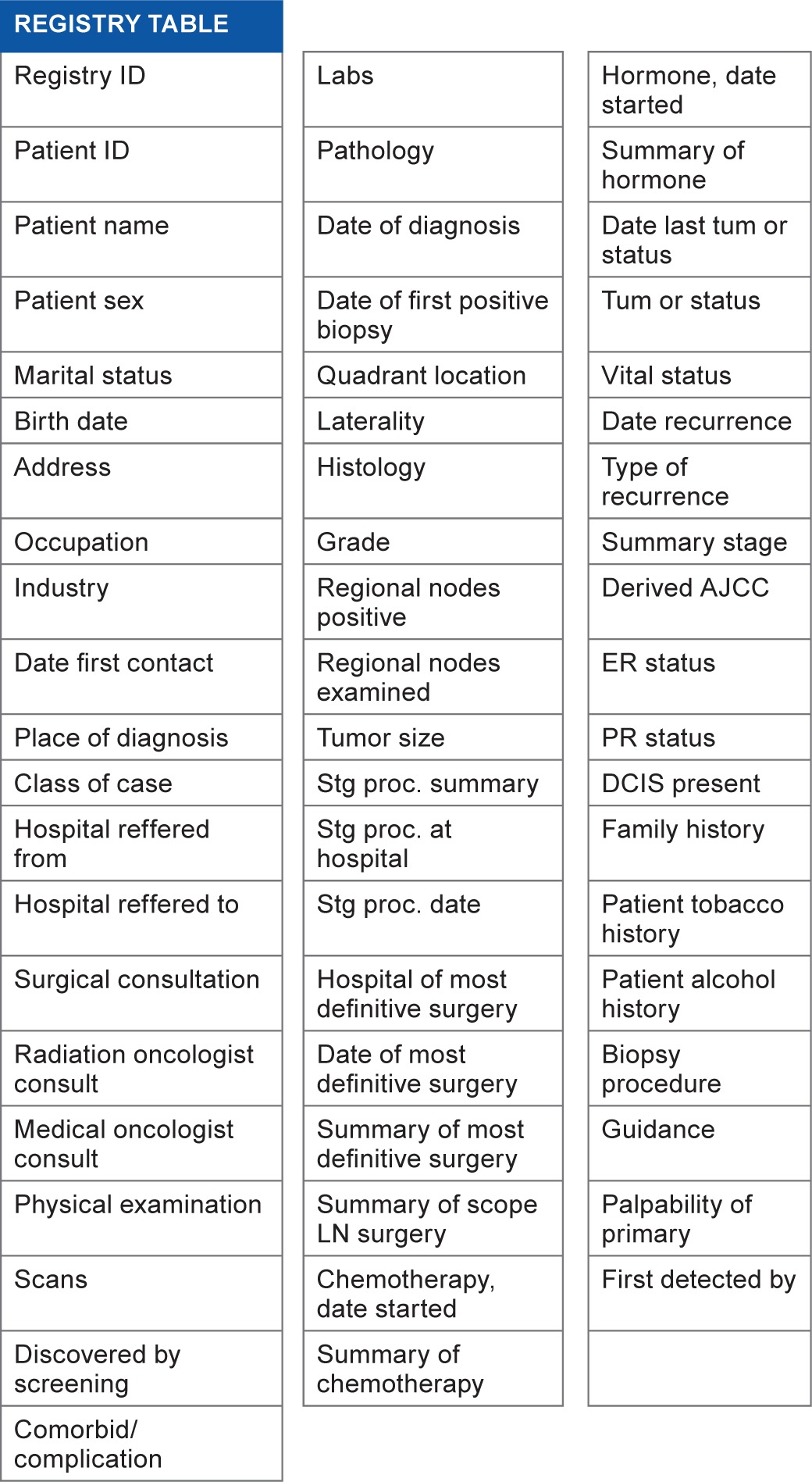
|
Database table linking
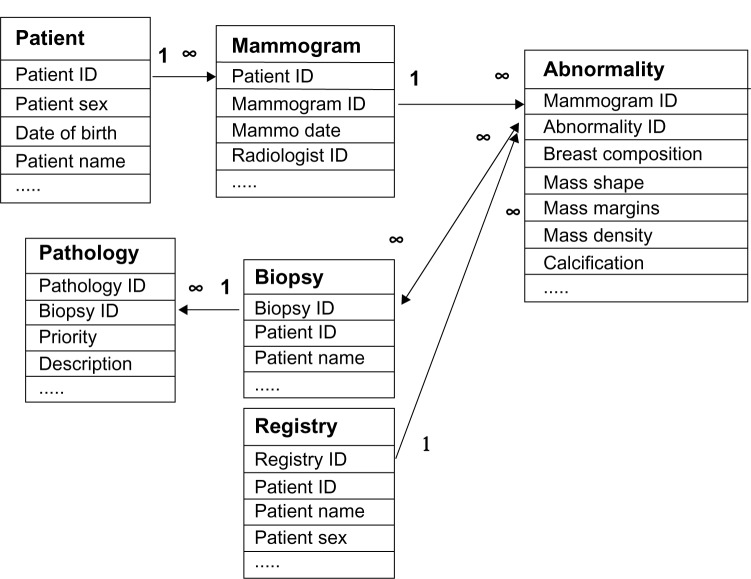
After tables for demographics and mammogram results were created, we established one-to-many relationships between the patient table and the mammogram table, and between the mammogram table and the abnormality table since a patient may have multiple mammograms over time, and radiologists may detect several abnormalities on each mammogram. These relationships are shown in the schema depicted in Figure 1. Based on these relationships, we can easily find the patient ID and the mammogram ID for each abnormality in the abnormality table.
Figure 1.
Sketched entity-relationship diagram for our comprehensive database management system. ∞ = associated multiple entries in a relationship.
For biopsy pathology outcomes, similarly, we established a one-to-many relationship between the biopsy table and the pathology table because multiple pathologic findings may exist in a single biopsy.
We use the following matching procedures to build up a many-to-many relationship between the abnormality table and the biopsy table. We assume a match exists between a biopsy and an abnormality if the mammographic abnormality and the biopsy occur in the same patient (patient ID matches), if the biopsy date is within 3 months of the mammography date, and if the biopsy is performed on the same location (laterality and quadrant) as the abnormality. When these three criteria are met, we create a link within our CMDB to identify that the biopsy is a match for that abnormality. We developed a Java computer program to find these biopsy matches automatically to the greatest degree possible, with limited oversight as necessary by radiologists.
The relationship between the abnormality table and the registry table is many-to-one. We assume a match exists between a registry record and an abnormality if the abnormality and the registry belong to the same patient, if the biopsy date of the registry entry is within 12 months of the mammography date,1,24 and if the biopsy location of the registry entry (laterality and quadrant) is the same as that of the abnormality. When these three criteria are met, we create a link within our CMDB to identify that the registry entry is a match for that abnormality. We developed another Java computer program to find these registry matches automatically.
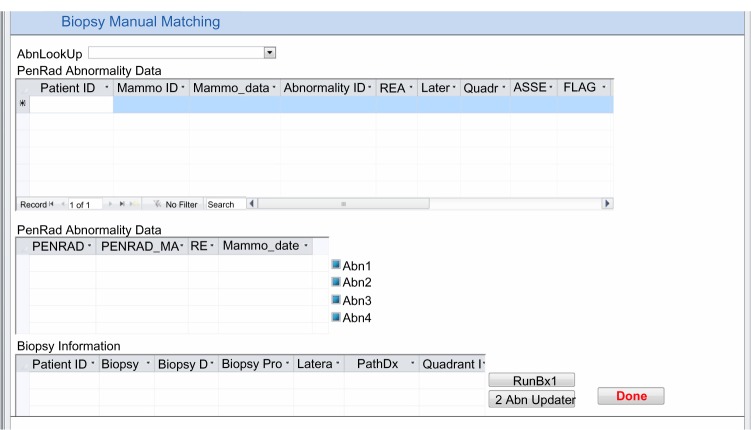
We have automatic processes for both biopsy and registry matching if there is a single abnormality, a single biopsy, and a single registry record depicting the same location. If there is more than one record, or if the location description is missing or inconsistent, these cases are automatically flagged by Java computer programs, and manual resolution is performed, using the electronic medical record if necessary (Figs. 2 and 3). The relationship among abnormalities, biopsy pathology outcomes, and cancer registry is demonstrated in Figure 1.
Figure 2.
Microsoft access form for manual matching between mammography results and biopsy outcomes.
Figure 3.
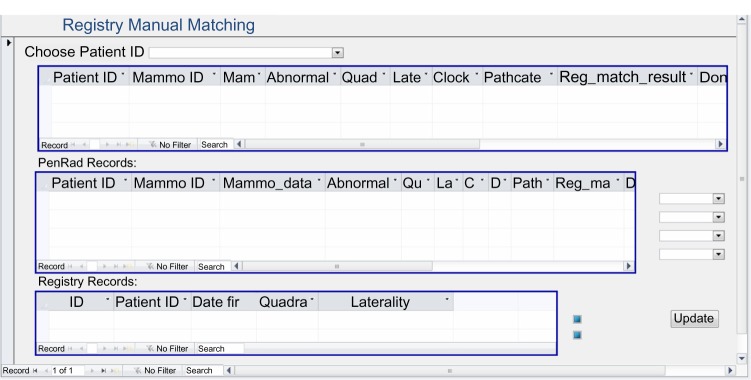
Microsoft access form for manual matching between mammography results and registry.
Clinical data collection
We collect demographics and mammogram results from our clinical structured reporting system (PenRad Technologies, Inc, Buffalo, MN) into the CMDB. In our practice, clinical data are captured in PenRad. Medical record numbers entered during patient scheduling from the radiology information system (RIS) are used as a unique patient identifier (patient ID). Technologists enter demographic risk factors including the patient’s birth date, race, family and personal history of breast cancer, use of hormone replacement therapy, reason for mammography, indicated problems (palpable abnormality, pain, skin, or nipple changes) into PenRad from patient intake sheets. During mammogram interpretation, radiologists use PenRad to record the presence of mammographic abnormalities and their characteristics according to BI-RADS. PenRad automatically assigns a unique mammogram ID for each mammogram and a unique abnormality ID for each abnormality found on each mammogram. Every 3 months, we query PenRad for demographics and mammogram results, prepare these data in NMD format, and upload these data into the appropriate tables in our CMDB according to the fields (feature variables) specified in each table.
We collect biopsy pathology in standardized lexicon and registry outcomes from distinct clinical sources. Specifically, we record pathology outcomes into our CMDB during a weekly Radiology/Pathology Consensus Conference in which each image-guided biopsy performed during the previous week is reviewed by the consensus panel of radiologists and pathologists (Fig. 4). We obtain breast cancer registry outcomes annually from our Comprehensive Cancer Center Registry.25
Figure 4.
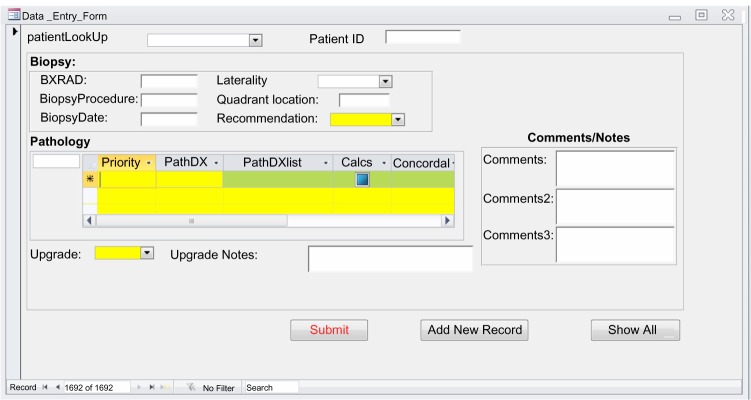
Microsoft access form for the input of biopsy pathology.
Database quality
Studies have confirmed that a series of factors or attributes affect database quality. We focus on three most important factors in health care: accuracy, completeness, and confidentiality.26 To ensure the accuracy of data entered in our CMDB, we check outcomes against Mammography Quality Standards Act (MQSA) audit requirements27,28 as well as national benchmarks. To ensure completeness, we include all abnormalities including secondary or associated ones in our CMDB. We also include both biopsy pathology outcomes and registry outcomes. To comply with HIPAA for confidentially, we store our system developed using Microsoft Access on a firewall-protected server supported by our hospital. All data collected into our system are protected from unauthorized access through established security measures. To develop decision support tools and implement QC audits, we will create a limited dataset29 by removing all direct identifiers under IRB approved protocols.
Outcome measurements and analysis
Our CMDB facilitates data collection and analysis on several levels of granularity (patient, mammogram, and abnormality) to better understand performance and quality in our practice. Important practice characteristics can be obtained on the patient level such as evaluating the prevalence and the incidence of breast cancer in our patient population.30 However, in this study, we focus on several important characteristics found on the mammographic finding level and the mammographic examination level. The analysis on the finding level provides an analysis that is truly representative of the decisions that a radiologist makes, ie, the decision to work up or biopsy a given abnormality. On the examination level, auditing by radiologists often occurs in routine clinical practice. For the sake of clarity in the analysis, we first make a distinction between a “finding” and an “examination.” A single mammographic finding (henceforth called “finding” or “mammographic finding”) is either a suspicious abnormality seen on a mammogram or a negative mammogram without specifications of any abnormalities. We use a mammographic examination (henceforth called “examination” or “mammographic examination”) to represent an overall characteristic of the entire mammogram. For analysis on the examination level in this study, we synthesized all findings on a mammogram to mammographic examination by choosing the most worrisome finding (using the BI-RADS categories ranked in the order of suspicion in routine clinic practice, 5 > 4 > 0 > 3 > 2 > 1).1,31
We collect registry outcomes for each finding. As prescribed in the literature,1 we consider a finding with a registry record of ductal carcinoma in situ or any invasive carcinoma as positive. Findings with other pathologic categories or without a matched registry are considered negative. We also determine registry outcomes for each examination by consolidating outcomes of its findings. In order to compare with the national performance benchmarks and guidelines,15 we evaluated interpretive performance of the radiologists by using mammographic examinations obtained from our CMDB. The analysis was implemented on screening and diagnostic mammography separately. Our clinical practice occasionally assigns BI-RADS category 0 to some diagnostic mammograms. When one or more mammograms follow an initial mammogram that is assessed as category 0, all mammograms up to and including the first mammogram with a non-zero assessment (within 180 days) are treated as a single observation.32
Results
General characteristics of the CMDB
From 1/1/2006 to 12/31/2011, our CMDB accumulated 28,029 patients (27,543 female and 486 male), 77,950 mammograms, and 22,074 abnormalities. On examination level, there were 64,244 screening mammographic examination and 13,706 diagnostic examinations collected from our CMDB for performance analysis. On finding level, there were 66,568 findings from screening mammography and 15,607 findings from diagnostic mammography.
For our breast cancer pathology lexicon, we cataloged 117 distinct pathologic diagnoses. Using this pathology lexicon, our CMDB recorded 3,595 biopsies associated with 4,011 pathology observations from 1/1/2006 to 12/31/2011.
Collection and analysis enabled on multiple levels relevant to care
We obtained audit data by analyzing performance on mammographic examinations from our CMDB, which were in accordance with national benchmarks and guidelines for screening and diagnostic mammography15 (Table 5). We reported age distribution and breast composition characteristics associated with mammographic examinations (Table 6). There were 82,175 mammographic findings (1,229 malignant and 80,946 benign) available for training, testing, and evaluation of decision support tools. Those breast cancers included 612 masses, 345 micro-calcifications, 118 false negatives without abnormality findings, and 154 findings categorized as “other.”
Table 5.
Abnormal interpretation for 64,244 screening mammographic examinations and 13,706 diagnostic mammographic examinations with registry outcomes.
| MEASUREMENT AND DATA | SCREENING EXAMINATIONS | SCREENING GUIDELINES15 | DIAGNOSTIC EXAMINATIONS | DIAGNOSTIC GUIDELINES15 |
|---|---|---|---|---|
| Recall rate (%) | 7.7 | 5–12 | 13.77 | 9–25 |
| No. of abnormality interpretations* | 4937 | 1888 | ||
| Total no. of examinations | 64,244 | 13,706 | ||
| PPV1, abnormal interpretations (%) | 6.7 | 3–8 | NA | |
| No. of cancers | 329 | NA | ||
| No. of abnormal interpretation* | 4937 | NA | ||
| PPV2, biopsy recommended (%) | 26.0 | 20–40 | 30.8 | 20–45 |
| No. of cancers | 258 | 581 | ||
| No. of abnormal interpretation+ | 994 | 1888 | ||
| PPV3, performed (%) | 29.4 | NA | 35.0 | 25–50 |
| No. of cancers | 229 | 513 | ||
| No. of abnormal interpretation++ | 778 | 1464 | ||
| Cancer detection rate (per 1000) | 5.9 | > = 2.5 | 49.7 | > = 30 |
| No. of cancers | 381 | 681 | ||
| Total no. of examinations | 64,244 | 13,706 |
Notes:
An abnormal interpretation was based on assignment of BI-RADS Category 0, 4, or 5 for screening examinations, BI-RADS Category 4 or 5 for diagnostic examinations.
A classification of biopsy recommendation was based on the assignment of BI-RADS Category 4 or 5 at the final assignment.
A classification of biopsy recommendation and performed was based on the assignment of BI-RADS Category 4 or 5 at the final assignment.
Table 6.
Distribution of age and breast composition for 77,950 mammographic examinations.
| CHARACTERISTIC | NUMBER OF EXAMINATIONS |
|---|---|
| Age (years) | |
| < = 45 | 14,355 (18.4%) |
| 46–50 | 11,792 (15.1%) |
| 51–55 | 12,918 (16.6%) |
| 56–60 | 12,573 (16.1%) |
| 61–65 | 10,021 (12.9%) |
| >65 | 16,291 (20.9%) |
| Breast composition | |
| Predominantly fatty | 7,945 (10.1%) |
| Scattered fibroglandular densities | 35,207 (45.2%) |
| Heterogeneously dense | 29,680 (38.1%) |
| Extremely dense | 5,037 (6.5%) |
| Missing | 81 (0.1%) |
Discussion
In this manuscript, we present a procedure of developing a standardized database system for mammography audit and breast cancer research in order to improve breast cancer diagnosis. We demonstrate that the CMDB integrates patient demographic risk factors, mammogram results, biopsy pathology results, and breast cancer registry outcomes to provide a foundation on which we can implement quality analysis and develop decision support tools. Important informatics concepts underlie this database design including a multiple-level database structure, which relies on a reliable entity-relationship model, heavy utilization of the already established BI-RADS lexicon, and nearly automated matching procedures.
The current version of the NMD export is by definition on the mammography level. Our structured reporting software package allows an NMD export on the abnormality level, which enables us to design the CMDB for developing decision support tools and evaluating our practice on this, more detailed, level of granularity. The procedure of developing our CMDB could be adopted in other mammography facilities. Different mammography facilities may choose different structured reporting systems to record mammography results. However, mammography results can be generated in the NMD format from all systems since the software vendors of the systems are required to comply with NMD requirements in order to enable the automatic upload of facility audit data directly to the ACR. Our CMDB was developed based on NMD specifications in order to make our methods portable, facilitating development, and implementation of similar mammography databases in other practices.
In addition, one of the most important characteristics of our CMDB is uniform description of breast pathology based on a breast pathology lexicon. Previous research has shown that variability in breast pathology reporting causes difficulty in comparing cancer data between different facilities and in developing/populating cancer registries.33 Recently, several organizations, including the Association of Directors of Anatomic and Surgical Pathology (ADASP) and the College of American Pathologists (CAP) Cancer Committee,34–36 have developed public guidelines for breast pathology reporting; however, these guidelines are clumsy to use and their completion is inefficient.37 Systematized Nomenclature of Medicine—Clinic Terms (SNOMED-CT) is considered to be the most comprehensive, multilingual clinical healthcare terminology in the world for systematically organizing collection of general medical terminology but it is not adequate to the needs of imaging related clinical practice.38,39 In our study, we compiled a compact pathology lexicon (based on the BI-RADS third edition pathology listing) for breast biopsy specifically, which includes pathology description, pathology description abbreviation, pathology category, and display order. This simple and compact lexicon can provide a first step in standardization of pathology reporting between radiologists and pathologists for improving communication and data analysis. Our breast pathology lexicon can be expanded and/or adapted to include more pathology diagnoses as new discoveries are made in breast pathology.
It is important to include rich outcomes in mammography database for breast cancer audit and research. Registry outcomes have been proven more accurate than biopsy pathology outcomes. They have been used to establish national benchmarks and guidelines15 and build decision support tools. However, they can be onerous, costly, and difficult to incorporate into a typical practice. Our CMDB defines a nearly automated method to match both biopsy pathology and registry outcomes to imaging findings, which allows us to find possible difference in performance measurement and evaluate the quality of mammography practice.
There are several advantages to parsing the mammographic datasets into separate patient, mammogram, and abnormality tables. First, this structure is a natural representation of clinical practice, enabling practices to understand this design schema, and to use this concept as a blueprint when designing their own mammography database systems. Second, this multiple-level structure avoids the problems (including redundancy and explosion in database size) associated with collapsing multiple levels of data into one single table. We used an abnormality table to record all abnormalities, which allow radiologists to take into account each individual finding with realization that each finding may eventually prove to be important in clinical decision making. Finally, this structure is extensible to include the data from other modalities used in breast imaging such as ultrasound and MRI. As breast cancer research and treatment advance, a myriad of data from other imaging modalities will be generated. Our CMDB provides a reliable solution to address the issue of mammography data management with a comprehensive database system.26
There are existing limitations to our CMDB, which we plan to address in future work. First, during data linking, if matches do not fulfill all the criteria for automatic matching, they are flagged for manual resolution of near matches, which is both labor-intensive and possibly error-prone. One possible solution is to learn the previous matches and find matching patterns for near matches automatically.40 Another potential solution of reducing the burden of matching is to annotate abnormality findings at the time of interpretation as is done elegantly in software packages like annotation and image markup (AIM).41 Second, we collect datasets from different sources and update them periodically. In the future, we plan to develop a processing pipeline to organize these steps. We intend to build a standardized application programming interface (API) to coordinate the steps for quality control and efficiency purpose. Third, we developed our CMDB by using Microsoft Office Access; therefore, it works only on Windows operating system, which limits the usage of our CMDB. A planned improvement is to migrate our CMDB to MySQL or Oracle in order to handle large data and enable cross-platform interoperability. Finally, in the future, we will implement our CMDB client using programming languages that allow multiple users to access the data most easily.
Conclusion
The proposed mammography database management system provides efficient storage and management of complex, multi-relational datasets to improve breast cancer diagnosis. Our CMDB has managed the challenges of the multi-level structure demanded by the complexity of breast imaging practice. By capitalizing on BI-RADS lexicon widely used in mammography practice, we have demonstrated the utility of analyzing data on different levels of granularity, which has the potential for improving diagnostic performance. Overall, our CMDB exemplifies data collection and management of mammography practice with a multi-relational structure and automated matching processes for enhancing data integrity, evaluation, and utilization in breast cancer diagnosis.
Footnotes
Author Contributions
DLR and ESB conceived and designed the experiments. YW and ESB analyzed the data. YW wrote the first draft of the manuscript. RWW, ME, and ESB contributed to the writing of the manuscript. All authors agree with manuscript results and conclusions. YW and ESB jointly developed the structure and arguments for the paper. All authors made critical revisions and approved final version. All authors reviewed and approved the final manuscript.
ACADEMIC EDITOR: JT Efird, Editor in Chief
FUNDING: This work was supported by the National Institutes of Health (grants K07-CA114181, R01-CA127379, R01-LM010921, and R01-CA165229).
COMPETING INTERESTS: YW discloses grants from Hologic, Inc, outside the published work. Other authors disclose no competing interests.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants. Provenance: the authors were invited to submit this paper.
REFERENCES
- 1.Burnside ES, Davis J, Chhatwal J, et al. Probabilistic computer model developed from clinical data in national mammography database format to classify mammographic findings. Radiology. 2009;251(3):663–72. doi: 10.1148/radiol.2513081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chhatwal J, Alagoz O, Lindstrom MJ, Kahn CE, Jr, Shaffer KA, Burnside ES. A logistic regression model based on the national mammography database format to aid breast cancer diagnosis. AJR Am J Roentgenol. 2009;192(4):1117–27. doi: 10.2214/AJR.07.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohlich RE, Sickles EA, Burnside ES, Dee KE. Interpreting data from audits when screening and diagnostic mammography outcomes are combined. AJR Am J Roentgenol. 2002;178(3):681–6. doi: 10.2214/ajr.178.3.1780681. [DOI] [PubMed] [Google Scholar]
- 4.Suckling J, Parker J, Dance DR, et al. The mammographic image analysis society digital mammogram database; Paper presented at: 2nd International Workshop on Digital Mammography; Amsterdam: Excerpta Medica; 1994. [Google Scholar]
- 5.Karssemeijer N. Adaptive noise equalization and recognition of microcalcifications in mammography. Int J Pattern Recognit Artif Intell. 1993;7:1357–76. [Google Scholar]
- 6.Mascio LN, Frankel SD, Hernandez JM, Logan CM.Building the LLNL/UCSF digital mammogram library with image groundtruth.Paper presented at: 3rd International Workshop on Digital Mammography1996Chicago, IL [Google Scholar]
- 7.Nishikawa R, Wolverton D, Schmidt RA, Pisano ED, Hemminger BM, Moody J. A common database of mammograms for research in digital mammography; Paper presented at: 3rd International Workshop on Digital Mammography; Chicago, IL: 1996. [Google Scholar]
- 8.Moreira IC, Amaral I, Domingues I, Cardoso A, Cardoso MJ, Cardoso JS. INbreast: toward a full-field digital mammographic database. Acad Radiol. 2012;19(2):236–48. doi: 10.1016/j.acra.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Matheus BR, Schiabel H. Online mammographic images database for development and comparison of CAD schemes. J Digit Imaging. 2011;24(3):500–6. doi: 10.1007/s10278-010-9297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heath M, Bowyer K, Kopans D, Moore R, Kegelmeyer WP. The digital database for screening mammography; Paper presented at: 5th International Workshop on Digital Mammography; Toronto: 2000. [Google Scholar]
- 11.Burnside ES, Sickles EA, Bassett LW, et al. The ACR BI-RADS experience: learning from history. J Am Coll Radiol. 2009;6(12):851–60. doi: 10.1016/j.jacr.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.American College of Radiology . Breast Imaging Reporting and Data System (BI-RADS) Atlas. Reston, VA: ACR; 2003. [Google Scholar]
- 13.D’Orsi C, Kopans D. Mammography interpretation: the BI-RADS method. Am Fam Physician. 1997;55:1548–50. [PubMed] [Google Scholar]
- 14.Osuch JR, Anthony M, Bassett LW, et al. A proposal for a national mammography database: content, purpose, and value. AJR Am J Roentgenol. 1995;164:1329–34. doi: 10.2214/ajr.164.6.7754870. [DOI] [PubMed] [Google Scholar]
- 15.American College of Radiology . Breast Imaging Reporting and Data System (BI-RADS) Atlas. 5th ed. Reston, VA: ACR; 2013. [Google Scholar]
- 16.Reiner B. Medical imaging data reconciliation, part 4: reconciliation of radiology reports and clinical outcomes data. J Am Coll Radiol. 2011;8(12):858–62. doi: 10.1016/j.jacr.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Berg W, Hruban R, Kumar D, Singh H, Brem R, Gatewood O. Lessons from mammographic-histopathologic correlation of large-core needle breast biopsy. Radiographics. 1996;16(5):1111–30. doi: 10.1148/radiographics.16.5.8888394. [DOI] [PubMed] [Google Scholar]
- 18.Zuiani C, Mazzarella F, Londero V, Linda A, Puglisi F, Bazzocchi M. Stereotactic vacuum-assisted breast biopsy: results, follow-up and correlation with radiological suspicion. Radiol Med. 2007;112(2):304–17. doi: 10.1007/s11547-007-0143-7. [DOI] [PubMed] [Google Scholar]
- 19.Liberman L. Percutaneous imaging-guided core breast biopsy: state of the art at the millennium. AJR Am J Roentgenol. 2000;174:1191–9. doi: 10.2214/ajr.174.5.1741191. [DOI] [PubMed] [Google Scholar]
- 20.Liberman L, Drotman M, Morris EA, et al. Imaging-histologic discordance at percutaneous breast biopsy. Cancer. 2000;89(12):2538–46. doi: 10.1002/1097-0142(20001215)89:12<2538::aid-cncr4>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 21.Woods R, Lin Y, Wu Y, Burnside ES. Breast cancer detection rate differs based on biopsy proven or registry matched cancers; Radiological Society of North America Annual Meeting; Chicago, IL: 2010. [Google Scholar]
- 22.Chen P. The entity-relationship model: toward a unified view of data. ACM Trans Database Syst. 1976;1:9–36. [Google Scholar]
- 23.American College of Radiology . Breast Imaging Reporting and Data System (BI-RADS®) Atlas. 3rd ed. Reston, VA: ACR; 1998. [Google Scholar]
- 24.Houssami N, Irwig L, Simpson J, McKessar M, Blome S, Noakes J. The influence of clinical information on the accuracy of diagnostic mammography. Breast Cancer Res Treat. 2004;85:223–8. doi: 10.1023/B:BREA.0000025416.66632.84. [DOI] [PubMed] [Google Scholar]
- 25.Foote M. Wisconsin Cancer Reporting System: a population-based registry. Wis Med J. 1999;98(4):17–8. [PubMed] [Google Scholar]
- 26.Marcus DS, Erickson BJ, Pan T. Whitepapers on imaging infrastructure for research, Part 2: data management practices. J Digit Imaging. 2012;25:566–9. doi: 10.1007/s10278-012-9502-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Food and Drug Administration, Mammography facilities: requirements for accrediting bodies and quality standards and certification requirements - interim rules. Fed Regist. 1993;58:67558–72. [PubMed] [Google Scholar]
- 28.The Food and Drug Administration, State certification of mammography facilities. Fed Regist. 2002;67:5446–69. [PubMed] [Google Scholar]
- 29.Pan T, Erickson BJ, Marcus DS. Whitepapers on imaging infrastructure for research, Part 3: security and privacy. J Digit Imaging. 2012;25:692–702. doi: 10.1007/s10278-012-9493-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capocaccia R, Verdecchia A, Micheli A, Sant M, Gatta G, Berrino F. Breast cancer incidence and prevalence estimated from survival and mortality. Cancer Causes Control. 1990;1(1):23–9. doi: 10.1007/BF00053180. [DOI] [PubMed] [Google Scholar]
- 31.D’Orsi C, Newell M. BI-RADS decorded: detailed guidance on potentially confusing issues. Radiol Clin North Am. 2007;45:751–63. doi: 10.1016/j.rcl.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Sickles E, Miglioretti D, Ballard-Barbash R, et al. Performance benchmarks for diagnostic mammography. Radiology. 2005;235(3):775–90. doi: 10.1148/radiol.2353040738. [DOI] [PubMed] [Google Scholar]
- 33.Apple S. Variability in gross and microscopic pathology reporting in excisional biopsies of breast cancer tissue. Breast J. 2006;12(2):145–9. doi: 10.1111/j.1075-122X.2006.00222.x. [DOI] [PubMed] [Google Scholar]
- 34.Association of Directors of Anatomic and Surgical Pathology. Standardization of the surgical pathology report. Am J Surg Pathol. 1992;16:84–6. [Google Scholar]
- 35.Ruby S, Henson D. Practice protocols for surgical pathology A communication from the Cancer Committee of the College of American Pathologists. Arch Pathol Lab Med. 1994;118(2):120–1. [PubMed] [Google Scholar]
- 36.Lester S, Bose S, Chen Y, et al. Protocol for the examination of specimens from patients with ductal carcinoma in situ of the breast. Arch Pathol Lab Med. 2009;133(1):15–25. doi: 10.5858/133.1.15. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt RA. Synopses, systems, and synergism. Am J Clin Pathol. 2007;127:845–7. doi: 10.1309/0R7MP6QJHA38CV81. [DOI] [PubMed] [Google Scholar]
- 38.Sinha U, Yaghmai A, Thompson L, et al. Evaluation of SNOMED 3.5 in representing concepts in chest radiology reports: integration of a SNOMED mapper with a radiology reporting workstation; Paper presented at: Proceedings of the AMIA Symposium; Los Angeles, CA: 2000. [PMC free article] [PubMed] [Google Scholar]
- 39.Bell DS, Greenes RA. Evaluation of UltraSTAR: performance of a collaborative structured data entry system; Paper presented at: Proceedings of the Annual Symposium on Computer Application in Medical Care; Washington, DC: 1994. [PMC free article] [PubMed] [Google Scholar]
- 40.Doan A, Domingo P, Levy A. Learning source descriptions for data integration; Proceedings of the 3rd International Workshop on the Web and Databases; Dallas, TX: 2000. [Google Scholar]
- 41.Rubin D. Finding the meaning in images: annotation and image markup. Philos Psychiatry Psychol. 2012;18(4):311–8. [Google Scholar]