Abstract
Glioblastoma multiforme (GBM) is the most common malignant brain tumor. GBM samples are classified into subtypes based on their transcriptomic and epigenetic profiles. Despite numerous studies to better characterize GBM biology, a comprehensive study to identify GBM subtype- specific master regulators, gene regulatory networks, and pathways is missing. Here, we used FastMEDUSA to compute master regulators and gene regulatory networks for each GBM subtype. We also ran Gene Set Enrichment Analysis and Ingenuity Pathway Analysis on GBM expression dataset from The Cancer Genome Atlas Project to compute GBM- and GBM subtype-specific pathways. Our analysis was able to recover some of the known master regulators and pathways in GBM as well as some putative novel regulators and pathways, which will aide in our understanding of the unique biology of GBM subtypes.
Keywords: glioblastoma multiforme, GBM, pathways, classical, mesenchymal, neural, proneural, master regulators, gene regulatory networks, GBM subtypes
Introduction
Glioblastoma multiforme (GBM) is the most lethal form of brain cancer with a median survival of 14 months.1,2 There have been numerous studies to generate high-throughput datasets to better understand and characterize these tumors at the genomic, genetic, and epigenetic levels.1,3–8 Among these studies, The Cancer Genome Atlas Project (TCGA) has generated a vast amount of genomic data for about 500 GBM samples.5,7
GBM samples are classified into molecular subtypes based on their transcriptomic and epigenetic profiles. Verhaak et al classified GBM samples based on gene expression into four subtypes, namely classical, mesenchymal, neural, and proneural.7 GBM samples are further classified into two major subtypes based on their DNA methylation profiles, namely glioma-CpG island methylator phenotype (G-CIMP) positive and G-CIMP negative.8 A majority of the G-CIMP positive samples are also proneurals,8 which allows splitting proneural group into proneural G-CIMP positive and proneural G-CIMP negative subtypes (hereafter, proneural+ and proneural−, respectively).
Various studies have characterized GBM subtypes based on individual mutations, genetic alterations, and pathways associated with each subtype. Specifically, in mesenchymal subtype, genes in the tumor necrosis factor superfamily pathway and NF-kB pathway, such as TRADD, RELB, and TNFRSF1A are highly expressed.7 In classical samples, genes in Notch (NOTCH3, JAG1, and LFNG) and Sonic hedgehog (SMO, GAS1, and GLI2) signaling pathways are highly expressed.7 Also, EGFR amplification is common in both neural and classical samples.7 In proneural+ samples, IDH1 mutation is common.9
Several studies have evaluated the master regulators of GBM subtypes. Carro et al reported CEBP and STAT3 as the master regulators of the mesenchymal subtype.10 Bhat et al also reported mesenchymal-specific regulators such as MAFB, HCLS1, TAZ, and YAP.11
Despite these findings, there is still no study that gives a comprehensive report of GBM- and GBM subtype-specific master regulators, regulatory networks and pathways. In this study, we have two main objectives: (i) to compute GBM-and GBM subtype-specific master regulators and regulatory networks of GBM at the transcriptional level, (ii) to compute GBM- and GBM subtype-specific pathways. We used two different pathway enrichment algorithms and looked at the overlapping significantly enriched pathways for each GBM subtype. We also used FastMEDUSA to compute putative master regulators and regulatory networks of each subtype. Our analysis identified some existing master regulators and putative novel regulators of each GBM subtype. We also identified several GBM- and GBM subtype-specific pathways. Our results provide testable hypotheses to further investigate potential therapeutic targets for GBM subtypes.
Materials and Methods
Preprocessing of expression dataset
We downloaded the TCGA exon expression level 3 dataset of 500 GBM and 10 normal brain tissue samples at TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga, accessed in March 2012). We obtained the molecular subtype and G-CIMP calls of these samples from the data freeze package by TCGA analysis working group (https://wiki.nci.nih.gov/download/attachments/39921481/dataFreeze_9_3_2011.tar.bz2?version=1&modificationDate=1315077027000). We filtered out 61 samples that were reference samples or samples without an assigned subtype. We filtered out about 600 low-signal genes whose log-transformed signal intensity was less than 4. We also detected samples that were either outliers within their subtype or had similar expression profiles to the samples of another subtype. To detect these samples, we computed pairwise Pearson correlation between the expression profiles of all samples and removed 87 samples, which had r < 0.87 for at least 20% of the samples within their subtypes. After sample filtering, we were left with 101 classical, 95 mesenchymal, 57 neural, 57 proneural−, 31 proneural+, and 10 normal samples.
Analysis of gene expression dataset
We performed batch effect control based on two covariates, namely batch ID, which was assigned by TCGA analysis center, and institution ID that provided the tissue. We used Partek Genomics Suite version 6.5 (Copyright © 2010 Partek Inc., St. Louis, MO, USA) to check and remove batch effect. We applied one-way analysis of variance (ANOVA) in Partek to compute differentially expressed genes (DEGs) between each subtype and normal samples with Benjamini–Hochberg False Discovery Rate (FDR) ≤0.0512 and fold change ≥1.5.
Computing significant pathways and IPA networks
We utilized QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity) and Gene Set Enrichment Analysis (GSEA)13 tools to compute GBM- and GBM subtype-specific significant pathways. GSEA is a computational method to determine whether an a priori defined set of genes with an interpretable function shows statistically significant differences between two biological states.13 We ran GSEA using GBM expression dataset to identify upregulated and downregulated expression signatures associated with individual subtypes. The source of the gene sets used in this study was the c2.all.v3.0.symbols.gmt collection from the Broad Institute Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/collections.jsp#C2), which contains canonical pathways; gene sets derived from experimental data with chemical and genetic perturbation, as well as gene sets derived from the reactome pathway database (www.reactome.org). All comparisons of the five subtypes were performed by mapping all gene sets with size ranging from 10 to 500 in MsigDB v3.0 c2 curated database to the ranked gene expression profiles. The enrichment scores were calculated by walking down the ordered list, and the statistical significance of nominal P values of the enrichment scores was estimated using Kolmogorov–Smirnov statistics by constructing a cumulative null distribution with 1000 permutations. To optimize the thresholds for selecting significant gene sets in our study, three random datasets, such as one generated from real data, two from simulated data, 15 from random gene sets, were generated and six methods of multiple comparisons correction including Benjamini–Hochberg, Stepdown FDR, Bonfferoni, q value, familywise error rate, and GSEA FDR were compared for aiding us to make decisions.
To analyze data in IPA, we uploaded the DEGs with fold changes for each subtype as input. After running the core analysis, we selected canonical pathways that were statistically significant (FDR ≤0.05). Then, we computed the overlap between significant pathways computed by GSEA and IPA. Computing common pathways between IPA and GSEA outputs was not straightforward since pathway nomenclature is different in these two systems and gene lists in proprietary IPA pathways are not readily available. Thus, we developed a pipeline to compare the results. First, we exported the list of DEGs that were present in each significant IPA pathway (IPA does not allow to export all genes in pathways but does allow to export DEGs). We also obtained a list of all genes in significant GSEA pathways. We first converted all gene names to HUGO gene symbols, then compared gene sets of each GSEA and IPA pathway pairwise. We selected pathway matches whose overlap comprised 50% of genes in GSEA and 25% of DEGs in IPA pathways for manual verification. During manual verification, we eliminated false positive matches.
To further annotate overall relationship between some genes based on literature, we also built IPA networks using these genes. Given a set of input genes, IPA uses a heuristic algorithm to construct networks iteratively while optimizing the interconnectivity and number of input genes in the network under a constraint of a maximal network size, which is 35 by default. IPA first builds networks from input genes iteratively based on their connectivity, then merges these networks and grows them to the maximum size by adding other genes in its global network iteratively.
Computing significant master regulators and gene regulatory networks
We utilized FastMEDUSA14 to compute significant transcription factors (TFs) and TF–gene interactions that were common and unique to each GBM subtype. FastMEDUSA is a machine learning algorithm that builds a predictive model based on the expression states of genes and motif presence in their promoter regions.14 FastMEDUSA incorporates discretized gene expression, promoter sequence data, and a list of candidate TFs as input. We obtained the list of candidate TFs from Genomatix software (Genomatix, www.genomatix.de). We obtained 1000 bases upstream of 5′ UTR of genes from University of California, Santa Cruz (UCSC) Genome Browser database.15 For the genes that did not exist in UCSC genome browser, we obtained their promoter sequences from Genomatix and Biomart.16
To discretize gene expression data, we computed fold change of expression signal of a gene in a sample to the gene’s median expression across normal samples. We called a gene in a sample upregulated if its fold change is ≥t, downregulated if it is ≤−t, and baseline otherwise. We discretized expression data for different choices of t and decided to use t = 1.2 for genes and t = 1.1 for TFs as it allowed a clear distinction of subtypes (Supplementary Fig. 1). We used a lower threshold for TFs as they are known to be expressed in lower levels than other genes.17 For FastMEDUSA analysis, we ignored genes that were consistently up or down across all subtypes, because FastMEDUSA could build a predictive model on genes that have high variation among experimental conditions.
We first ran FastMEDUSA with cross-validation mode to determine a number of optimal boosting steps. We set the number of boosting iterations to 1300 as it was optimal based on the error ratio curve that was obtained to predict the expression state of the test set (Supplementary Fig. 2). Then, we ran FastMEDUSA five times on the entire data for 1300 iterations and selected top significant TFs as following: for each FastMEDUSA run, we computed the significance score of each TF by computing the prediction score of genes of interest on the original model and on the model after removing that TF and all subtrees rooted by that TF as described before.18 TFs were ranked by their maximum prediction score, which was computed by adding up all scores of nodes containing the TF, and top 30 TFs were selected. Among the top 30 TFs, the ones with a ratio of their prediction score to their maximum score higher than 0.35 were chosen as the top TF list of that FastMEDUSA run. Finally, TFs that were in the top list in three out of five FastMEDUSA runs were selected as the final significant TF list.
Assuming that the majority of TFs would be positively correlated with their downstream target, we computed prediction score of upregulated (downregulated) TFs by choosing upregulated (downregulated) DEGs as target genes of interest. In order to compute gene regulatory networks, we computed the same score for each TF–gene pair in each subtype and filtered out the low-scored pairs and plot the remaining TF–gene pairs in Cytoscape.19 We used NIH Biowulf cluster to run FastMEDUSA in parallel. We used 200–300 cores and got the results for each run in about 13 hours.
Survival analysis
To further explore potential master regulators of GBM subtypes, we used REMBDRANDT (https://caintegrator.nci.nih.gov/rembrandt/home.do) to check survival of GBM patients based on the expression status of these TFs. TFs that stratify patients in low and high survival groups based on their expression could be potential biomarkers. We chose a twofold change threshold to determine the expression category of genes and chose the maximum intensity probe to determine the expression of the gene.
Gene ontology (GO) enrichment analysis
We used DAVID’s functional annotation chart20 pipeline to compute the GO term enrichment of genes in gene regulatory networks computed by FastMEDUSA. We chose GOTERM_ BP_FAT, GOTERM_CC_FAT, and GOTERM_MF_FAT categories in DAVID, which refers to biological process, cellular component, and molecular function, respectively. We applied Benjamini Hochberg FDR ≤0.05 as threshold to select significant GO terms.
Results
Removing batch effect and computing DEGs
We computed the source of variation in the expression data by establishing a three-way ANOVA model, where subtype, institution ID, and batch ID were the covariates. We found out that 4.94% of the variation was due to the batch ID, and the source of variation due to institution ID was negligible (Supplementary Fig. 3). We removed batch effect that was due to the batch ID by using the batch effect removal module in Partek Genomics Suite. We applied one-way ANOVA to compute DEGs for each subtype with respect to normal samples using Partek (FDR ≤0.05 and fold change = 1.5). Figure 1 shows the Venn diagram of upregulated and downregulated genes for each subtype.
Figure 1.
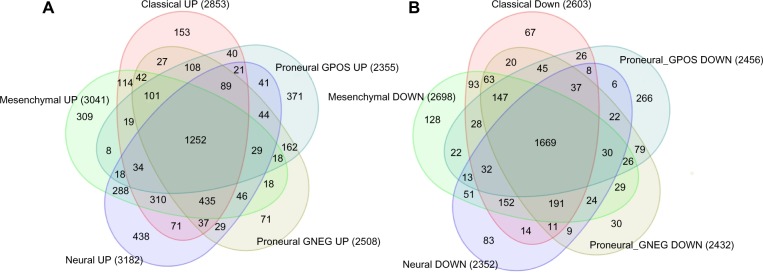
Venn diagram for (A) upregulated and (B) downregulated DEGs for each subtype with respect to normal samples. proneural GPOS: proneural+, proneural GNEG: proneural−.
Subtype-specific master regulators computed by Fast-MEDUSA
In order to run FastMEDUSA, we first excluded DEGs that were differentially expressed in all subtypes. For the remaining DEGs, we obtained 1000 bases upstream of their 5′ UTR from UCSC Genome Browser database. For about 30 genes whose promoter sequences were not available in UCSC Genome Browser database, we obtained their promoter sequences from Genomatix and BioMart. In total, we had promoter sequences of 5141 out of 5167 DEGs. Among these DEGs, 499 of them were used as candidate TFs based on Genomatix annotation.
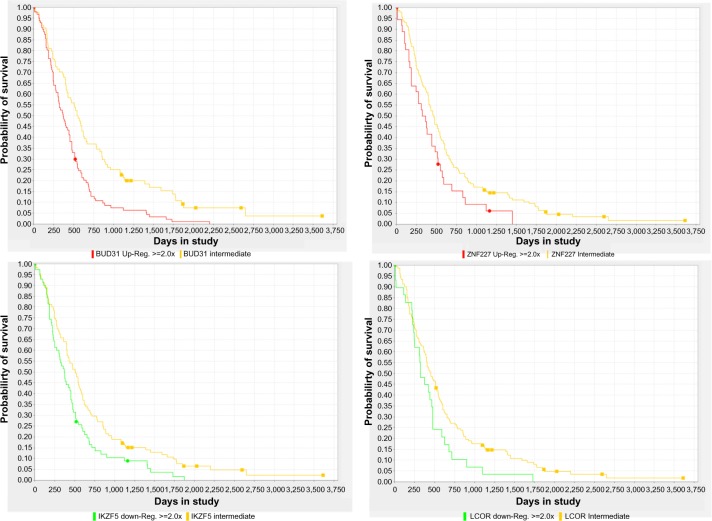
We ran FastMEDUSA five times with a unique random seed each time and obtained five different models. We post-processed these models to compute significant TFs (ie, master regulators) (see Materials and Methods section). The lists of significant upregulated and downregulated master regulators for each subtype are shown in Tables 1 and 2, respectively. Among these TFs, some of them are known previously to have a role in GBM such as CEBPD,21 RUNX1,21 LEF1,22 HES6,23 ASCL1,24 EBF1,25,26 SP100,27 and AEBP1.28 To check the effects of these TFs on survival, we computed survival plots of these TFs based on GBM gene expression data in REMBRANDT database and found out that some of these TFs have significant survival difference based on their expression status (Fig. 2 and Supplementary Fig. 4).
Table 1.
List of upregulated master regulators for each GBM subtype.
| CLASSICAL | NEURAL | MESENCHYMAL | PRONEURAL− | PRONEURAL+ |
|---|---|---|---|---|
| AEBP1 | BUD31 | AEBP1 | BUD31 | CTDSP1 |
| BUD31 | CEBPB | BUD31 | EN2 | DMTF1 |
| CEBPD | EN2 | EN2 | HES6 | EBF1 |
| EN2 | FOXJ1 | HEYL | HEYL | HES6 |
| FOXJ1 | HES6 | HNF4G | HOXD11 | PHC1 |
| HES6 | HNF4G | LEF1 | LEF1 | SIX4 |
| HOXD11 | LEF1 | LITAF | LITAF | UXT |
| LEF1 | LITAF | LTF | LTF | ZNF227 |
| LITAF | LTF | NMI | NMI | ZNF85 |
| LTF | NMI | PRIC285 | RUNX1 | |
| PRIC285 | PCGF1 | RUNX1 | SHOX2 | |
| RUNX1 | RUNX1 | SHOX2 | SNAI2 | |
| SNAI2 | SNAI2 | SNAI2 | SPI1 | |
| SP100 | SP100 | SP100 | TEAD3 | |
| SPI1 | UXT | SPI1 | UXT |
Table 2.
List of downregulated master regulators for each GBM subtype.
| CLASSICAL | NEURAL | MESENCHYMAL | PRONEURAL− | PRONEURAL+ |
|---|---|---|---|---|
| CHD3 | ARID4A | CHD3 | DACH2 | ARNTL2 |
| DACH2 | CHD3 | DACH2 | EZH1 | KLF16 |
| EZH1 | DACH2 | EZH1 | IKZF5 | LZTFL1 |
| IKZF5 | EZH1 | IKZF5 | KLF16 | MEF2A |
| KLF13 | IKZF5 | JMY | MMS19 | NPAS2 |
| LCOR | JMY | KLF13 | MTA3 | NR3C2 |
| LDB1 | KLF13 | LDB1 | NCOA1 | PCGF5 |
| MMS19 | LCOR | MTA3 | NPAS2 | PHF15 |
| MTA3 | LDB1 | MYST4 | NR3C2 | PLAGL1 |
| NR3C2 | MTA3 | NCOA1 | PHF15 | RNF14 |
| TCEAL7 | MYST4 | NR3C2 | TCEAL7 | SATB2 |
| TIAM1 | SUDS3 | TIAM1 | TIAM1 | STAT6 |
| ZMYND11 | ZMYND11 | ZMYND11 | ZMYND11 | TCEAL7 |
| ZNF208 | ZNF208 | ZNF248 | ZNF208 | TFPT |
| ZNF248 | ZNF248 | ZNF248 | TLE4 |
Figure 2.
Kaplan–Meier survival plots based on the expression status of four master regulators of GBM subtypes computed by FastMEDUSA. Each regulator name is below the survival plot. Log-rank P < 0.05 for each plot.
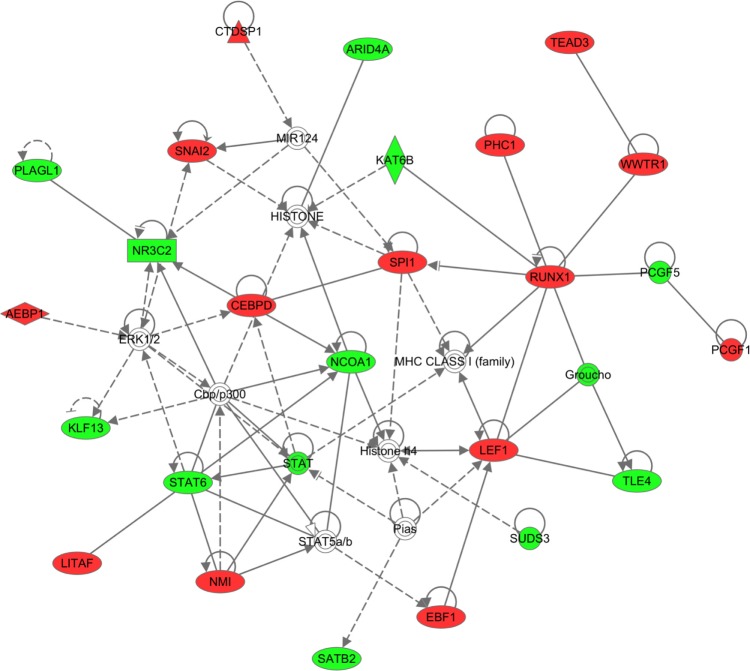
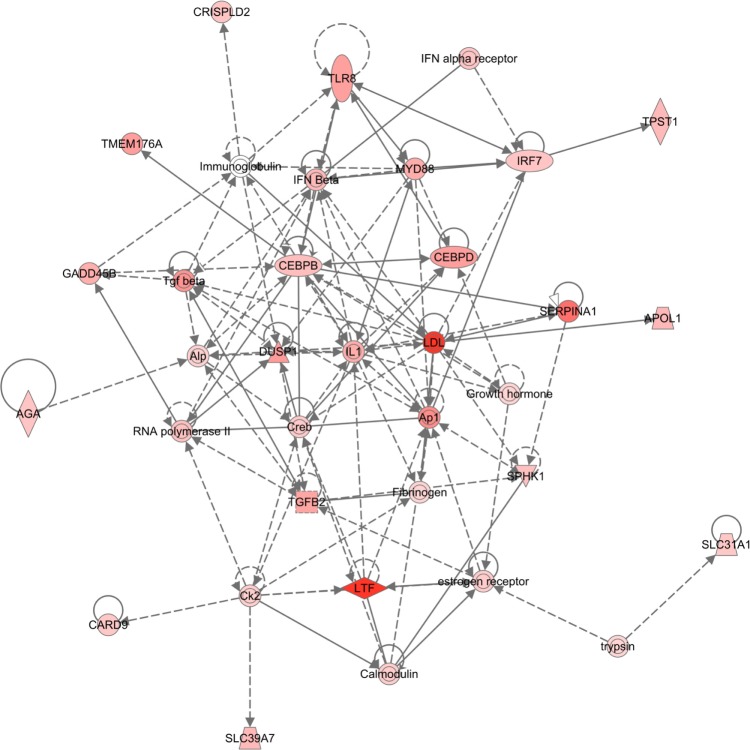
To examine biological relationship among these TFs based on literature, we uploaded the master regulators into IPA and ran core analysis to build networks from these regulators. The top-scoring network was associated with functions of gene expression, cellular function and maintenance, and cellular growth and maintenance (Fig. 3).
Figure 3.
IPA network built from master regulators of GBM subtypes computed by FastMEDUSA. Colored nodes are the master regulators (Red: upregulated, Green: downregulated).
Subtype-specific gene regulatory networks
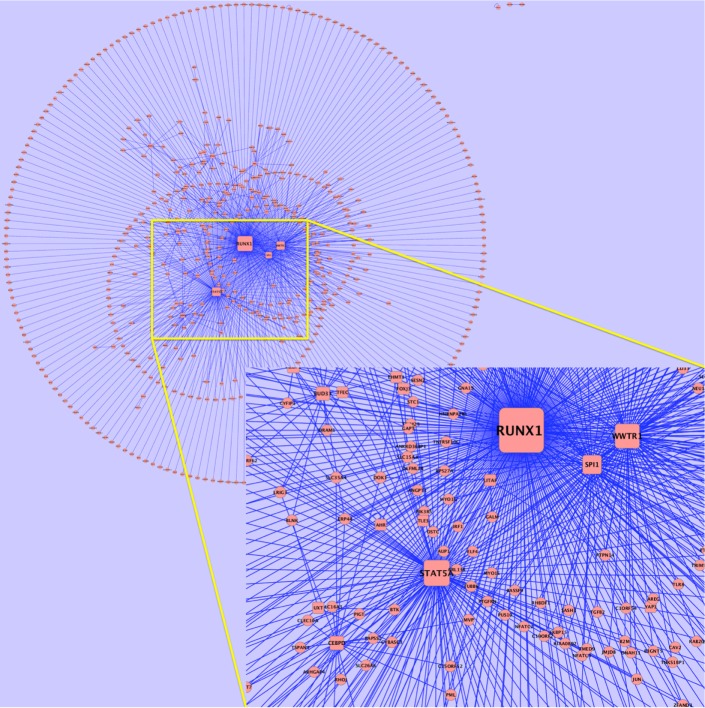
We also computed TF–gene significance score and computed gene interaction networks (see Fig. 4 for the mesenchymal subtype and Supplementary Figs. 5–8 for the other subtypes). We observed that some of the hub TFs (ie, TFs with high connectivity) in these networks were also master regulators (eg, RUNX1, SP1, and HES6). There were also some hub TFs that were not in the master regulator list, but occurred in all networks, particularly STAT5A and WWTR1.
Figure 4.
Gene regulatory network of mesenchymal subtype computed by FastMEDUSA. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score. The inset figure shows the zoomed view of central area.
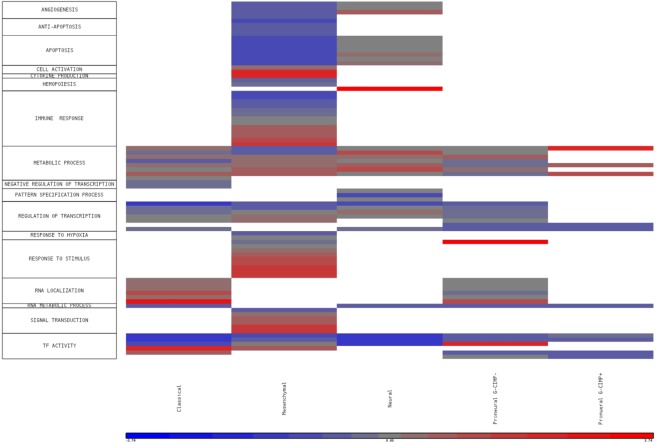
We checked the functional enrichment of genes in each network and plotted the enrichment P values in a heatmap (Fig. 5). GO terms related to TF activity, regulation of transcription, and metabolic processes were common in all subtypes. Mesenchymal group was uniquely enriched in GO terms related to immune response, response to stimulus, response to hypoxia, signal transduction, and anti-apoptosis. Apoptosis and angiogenesis terms were enriched in both mesenchymal and neural networks. GO terms related to RNA localization were enriched in both classical and proneural−. The classical group network was also uniquely enriched with GO terms related to negative regulation of transcription.
Figure 5.
Heatmap of GO term enrichment of genes in FastMEDUSA gene regulatory networks. Only the significant P values are colored via log transformation (White: no significance, Blue: low enrichment, Red: high enrichment). GO terms are categorized into more general terms in rows.
To further annotate these genes, we also uploaded them into IPA and examined networks with similar functions as found by GO terms. The network for the mesenchymal subtype is shown in Figure 6 and other networks are shown in Supplementary Figure 9.
Figure 6.
IPA network built from genes in gene regulatory network of the mesenchymal subtype. Colored nodes are the genes in the mesenchymal gene regulatory network (Red: upregulated, Green: downregulated).
Identify abnormal pathways in GBM subtypes by IPA and GSEA analysis
We ran IPA core analysis on DEGs of TCGA gene expression dataset with respect to normal samples and found 246 canonical pathways that were statistically significant in at least one subtype (FDR ≤0.05). There were 157, 177, 225, 154, and 105 canonical pathways enriched in classical, neural, mesenchymal, proneural− and proneural+ samples, respectively (Supplementary Table 1). We also computed IPA networks that have enrichment of DEGs. Some of these networks contained significant TFs found by Fast-MEDUSA (Supplementary Fig. 10).
We also ran GSEA on the gene expression dataset. Figure 7 shows the number of shared upregulated and downregulated pathways found in each subtype. We observed that mesenchymal and proneural− groups had about 100 unique significant upregulated pathways. The majority of downregulated pathways were shared by all subtypes, whereas there were only nine upregulated pathways shared by all subtypes.
Figure 7.
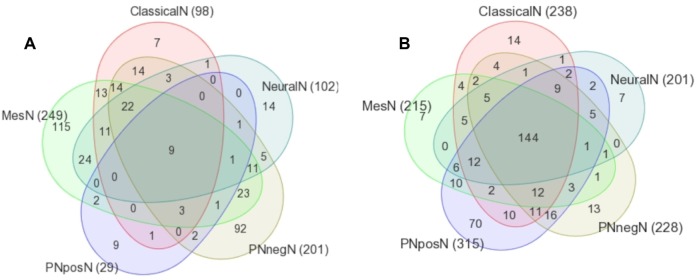
Venn diagram of (A) upregulated and (B) downregulated GSEA pathways enriched in GBM subtypes (P < 0.05). PNpos: proneural+, PNneg: proneural−.
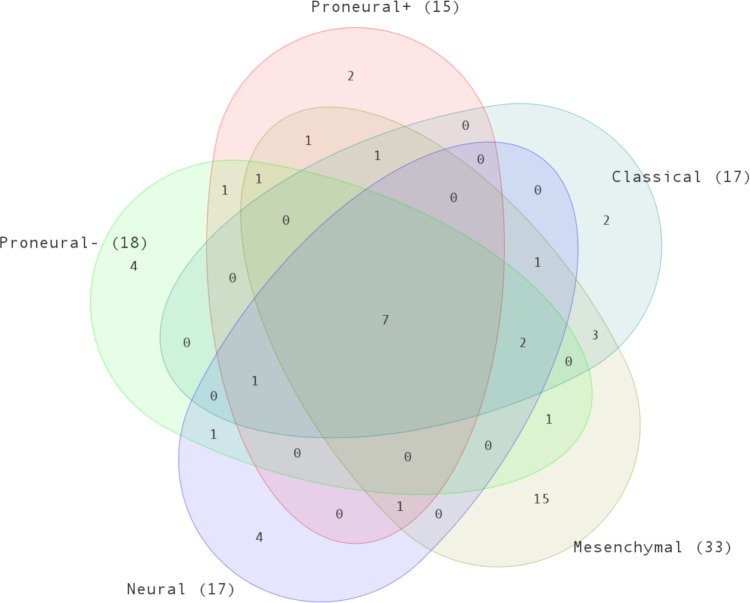
We compared IPA and GSEA pathway results to find common pathways. In this comparison, we tended to use a less-stringent threshold for each subtype (P ≤ 0.15) to increase sensitivity. The number of common pathways found for each subtype is listed in Table 3. The list of pathways that were significant in both IPA and GSEA is listed in Supplementary Table 2. Figure 8 shows the Venn diagram of these pathways. Here, we focused on the intersection list and subtype-specific pathways reported as significantly enriched by both GSEA and IPA. The common pathways identified by both IPA and GSEA and enriched in all GBM subtypes are listed in Table 4. As expected, glioma pathway and notch signaling pathway were in this list.
Table 3.
Number of subtype-specific significant pathways in IPA and GSEA, and number of common significant pathways. The last row shows the number of pathways common to both IPA and GSEA.
| CLASSICAL | MESENCHYMAL | NEURAL | PRONEURAL+ | PRONEURAL− | |
|---|---|---|---|---|---|
| IPA significant (FDR <0.15) | 34 | 46 | 36 | 27 | 34 |
| GSEA significant (P < 0.15) | 23 | 38 | 25 | 21 | 26 |
| Common to IPA and GSEA | 17 | 33 | 17 | 15 | 18 |
Figure 8.
Venn diagram of number of common pathways of each subtype. Common pathways are the significant pathways found by both GSEA and IPA.
Table 4.
The list of significant common pathways found by GSEA and IPA and that were significantly enriched in all subtypes. The pathway names in GSEA database are listed.
| PATHWAY NAME (IN GSEA DATABASE) | ANNOTATION |
|---|---|
| BIOCARTA_G1_PATHWAY | Cell cycle regulation |
| KEGG_GLIOMA | Cancer |
| KEGG_LONG_TERM_DEPRESSION | Neurotransmitters and Other Nervous System Signaling |
| KEGG_LONG_TERM_POTENTIATION | Neurotransmitters and Other Nervous System Signaling |
| KEGG_NOTCH_SIGNALING_PATHWAY | Cancer, Organismal Growth and Development |
| KEGG_O_GLYCAN_BIOSYNTHESIS | Glycan biosynthesis and metabolism |
| KEGG_RENAL_CELL_CARCINOMA | Cancer |
There were 15 significant pathways uniquely enriched in the mesenchymal subtype (Table 5). All these pathways except for KEGG_VEGF_SIGNALING_PATHWAY were upregulated. We observed that immune response-related pathways comprised the vast majority of the list as in the mesenchymal gene-regulated network computed by FastMEDUSA. The higher rate of immune response activity in mesenchymal GBMs has been reported recently.29 The VEGF signaling pathway was enriched in the mesenchymal subtype uniquely. VEGF activity is associated with angiogenesis,30 which was one of the GO terms associated with mesenchymal gene networks in FastMEDUSA results (Fig. 5). There were also apoptosis-related pathways.
Table 5.
List of significant common pathways found by IPA and GSEA that were enriched in mesenchymal subtype only.
| PATHWAY NAME | ANNOTATION |
|---|---|
| KEGG_REGULATION_OF_ACTIN_CYTOSKELETON | Organismal growth and development |
| REACTOME_SEMAPHORIN_INTERACTIONS | Cellular Growth, Proliferation and Development |
| KEGG_VEGF_SIGNALING_PATHWAY | Cellular growth, Proliferation and development, Growth factor signaling |
| BIOCARTA_DEATH_PATHWAY | Apoptosis |
| BIOCARTA_HIVNEF_PATHWAY | Apoptosis |
| BIOCARTA_TOLL_PATHWAY | Apoptosis, Cellular Immune response |
| REACTOME_TOLL_LIKE_RECEPTOR_4_CASCADE | Apoptosis, Cellular Immune response |
| KEGG_ACUTE_MYELOID_LEUKEMIA | Cancer |
| KEGG_GALACTOSE_METABOLISM | Carbohydrate metabolism |
| KEGG_STARCH_AND_SUCROSE_METABOLISM | Carbohydrate metabolism |
| KEGG_GRAFT_VERSUS_HOST_DISEASE | Immune response |
| KEGG_AUTOIMMUNE_THYROID_DISEASE | Immune response |
| BIOCARTA_COMP_PATHWAY | Immune response |
| BIOCARTA_IL2RB_PATHWAY | Immune response |
| KEGG_ANTIGEN_PROCESSING_AND_PRESENTATION | Immune response |
Two pathways, namely BIOCARTA_STRESS_PATHWAY and ST_INTEGRIN_SIGNALING_PATHWAY were enriched uniquely in the classical subtype. The BIOCARTA_STRESS_PATHWAY, which was upregulated in the classical subtype, involves two cell surface receptors, TNFR1 and TNFR2, to regulate apoptotic pathways31 and activate stress-activated protein kinases.32 The ST_INTEGRIN_SIGNALING_PATHWAY, which was downregulated in the classical subtype, involves integrins as primary sensors of the extracellular matrix (ECM) environment for cell migration, growth, and survival.33
In the neural subtype, BIOCARTA_SHH_PATHWAY, BIOCARTA_TCR_PATHWAY, and KEGG_FC_EPSILON_RI_SIGNALING_PATHWAY were uniquely and significantly downregulated, and KEGG_ONE_CARBON_POOL_BY_FOLATE pathway was upregulated. In the BIOCARTA_SSH_PATHWAY, Sonic HedgeHog (SSH) plays a distinct and crucial role in development such as proliferation of neuronal precursor cells in the developing cerebellum and other tissues.34,35 The BIOCARTA_TCR_PATHWAY (T-cell receptor pathway) plays a key role in the immune system. The KEGG_FC_EPSILON_RI_SIGNALING_PATHWAY (Fc epsilon RI-mediated signaling pathway) in mast cells are initiated by the interaction of antigen (Ag) with IgE bound to the extracellular domain of the alpha chain of Fc epsilon RI. The activated mast cells release especially histamines and heparin. Genes in the neural gene regulator network were enriched in GO terms related to hemopoiesis, which is modulated by histamine receptor signaling.36 The KEGG_ONE_CARBON_POOL_BY_FOLATE pathway is in the category of metabolic pathways. Metabolic process-related GO terms were enriched in genes in neural gene regulatory network.
In the proneural− subtype, KEGG_MISMATCH_ REPAIR, KEGG_PYRIMIDINE_METABOLISM, REACTOME_ACTIVATION_OF_THE_PRE_REP-LICATIVE_COMPLEX, and REACTOME_G2_M_ CHECKPOINTS pathways were uniquely and significantly upregulated. The KEGG_MISMATCH_REPAIR and the REACTOME_G2_M_CHECKPOINTS pathways play a key role in correcting DNA mismatches during DNA replication, thus maintains genomic stability. The REACTOME_ACTI-VATION_OF_THE_PRE_REPLICATIVE_COMPLEX is associated with initiation of DNA replication, and the KEGG_ PYRIMIDINE_METABOLISM pathway has been studied in human gliomas for its relation to chromosomal aberrations.37
In proneural+ subtype, REACTOME_DOUBLE_ STRAND_BREAK_REPAIR and BIOCARTA_KERATINOCYTE_PATHWAY were uniquely enriched. REACTOME_DOUBLE_STRAND_BREAK_REPAIR pathway, which has a key role in DNA repair, was upregulated, and BIOCARTA_KERATINOCYTE_PATHWAY, which has a role in inducing differentiation and inhibiting apoptosis, was downregulated.
Discussion
In this work, we analyzed the gene expression dataset of TCGA GBM samples to compute master regulators and gene regulatory networks for each subtype, namely classical, mesenchymal, neural, proneural+, and proneural−. We also performed pathway analysis by using GSEA and IPA. We found some master regulators that are known previously to play a role in GBM biology, as well as some other potentially important regulators. In pathway analysis, we focused on subtype-specific and GBM-specific pathways by taking the intersection of IPA and GSEA results.
We ran FastMEDUSA five times on the expression and promoter sequence dataset to compute master regulators and gene regulatory networks for each subtype. Some of the master regulators were reported to play a role in GBM biology, such as CEBPD,21 RUNX1,21 LEF1,22 HES6,23 ASCL1,24 EBF1,25,26 SP100,27 and AEBP1.28 Some of the master regulators were common to several subtypes; for instance, BUD31, EN2, LTF, and RUNX1 were computed as a master regulator for each subtype except for proneural+, UXT was a master regulator for proneural−, proneural+, and neural subtypes only, and PRIC285 was a master regulator for classical and mesenchymal subtype only. There were also subtype-specific master regulators. For instance, HES6 was a master regulator for proneural+ subtype (Table 1) and it was also a hub TF in the gene regulatory network of proneural+ subtype (Supplementary Fig. 8). HES6 expression is known to be associated with proneural group (Supplementary Fig. 11) and could play a role in cell proliferation and migration.23
Gene regulatory networks computed by FastMEDUSA do not necessarily demonstrate direct interactions between TFs and genes. Due to the complexity of GBM biology, and high dimensionality and noise of the data, FastMEDUSA gene regulatory networks are not completely accurate. However, these networks would give an overall representation of the subtype biology. For instance, we observed that some of the hub TFs in these networks were also identified as master regulators (Table 1, Fig. 4, Supplementary Figs. 5–8). We used IPA to retrieve existing literature data to build networks from these genes in these regulatory networks (Fig. 6 and Supplementary Fig. 9). We also observed that the GO term enrichment of these networks recapitulate the overall subtype biology. Thus, we believe that FastMEDUSA gene regulatory networks encode a significant number of useful interactions that merit further experimental validation.
There were two TFs, STAT5A and WWTR1, that were not reported as master regulators, but were hub TFs in all gene regulatory networks. WWTR1 (TAZ) has a role in regulating the mesenchymal differentiation in GBMs11 and has been reported as master regulator of glioblastoma in a previous computational study,38 and STAT5 is known to regulate glioma cell invasion.38 Another interesting TF was TLE3, which was a hub TF in proneural+ gene regulatory network (Supplementary Fig. 8). TLE3 (transducin-like enhancer of split 3) encodes a transcriptional co-repressor protein that belongs to the transducin-like enhancer family of proteins. Its expression is known to be associated with sensitivity to taxane treatment in ovarian carcinoma.39 There is no current study to discuss the association of TLE3 with proneural+, so it merits the further exploration of the role of TLE3 in proneural+ biology.
Performing pathway analysis in GSEA and IPA and taking the intersection of the findings, we found some GBM-and GBM subtype-specific pathways. As expected, we found glioma and notch signaling pathways as GBM specific. We also observed that the methylation subtype was enriched in immune response-related pathways as reported recently.29 The proneural+ group was enriched in DNA repair-related pathways, which could explain the better survival of proneural+ samples.8 The pathway analysis did not reveal a clear unique pathway for classical samples. This could be because classical samples could be split into the remaining subtypes found by Verhaak et al7 when the classification scheme by Phillips et al.40 is followed.41
Supplementary Materials
Supplementary Figure 1. Clustering of GBM samples based on discretized gene expression. (A) PCA after discretizing genes with t = 2, TFs with t = 1.5; (B) PCA after discretizing genes with t = 1.5, TFs with t = 1.2; (C) PCA after discretizing genes with t = 1.2, TFs with t = 1.1; (D) hierarchical clustering after discretizing genes with t = 1.2, TFs with t = 1.1. Sample coloring: Red, classical; blue, mesenchymal; green, neural; purple, proneural−; orange, proneural+.
Supplementary Figure 2. Percentage of test loss for different number of boosting iterations.
Supplementary Figure 3. The source of variation of batch ID and GBM subtype in TCGA exon data based on a two-way ANOVA model.
Supplementary Figure 4. Kaplan–Meier survival plots for nine master regulators of GBM subtypes. Each regulator name is below the survival plot. Log-rank P < 0.05.
Supplementary Figure 5. Gene regulatory network of the classical subtype. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score.
Supplementary Figure 6. Gene regulatory network of the neural subtype. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score.
Supplementary Figure 7. Gene regulatory network of the proneural− subtype. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score.
Supplementary Figure 8. Gene regulatory network of the proneural+ subtype. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score.
Supplementary Figure 9. IPA networks built from genes in gene regulatory network of the (A) classical (B) proneural−, (C) neural, (D) proneural+ subtypes. Colored nodes are the genes in the corresponding gene regulatory network (Red: upregulated, Green: downregulated).
Supplementary Figure 10. IPA networks derived from DEGs of (A) classical, (B) mesenchymal, (C) neural, (D) proneural− samples. Nodes annotated with a star are significant master regulators.
Supplementary Figure 11. Expression dot plot of HES6 in GBM subtypes.
Supplementary Table 1. Significant IPA canonical pathway for each GBM subtype. All P values are <0.05. Blank cells refer to P value > 0.05.
Supplementary Table 2. List of pathways that was significant in both IPA and GSEA. For each pathway, the subtypes that have that pathway have a “1” in that row.
Footnotes
SUPPLEMENT: Computational Advances in Cancer Informatics (B)
ACADEMIC EDITOR: JT Efird, Editor in Chief
FUNDING: Funding for this paper was provided by the Intramural Research Program of the National Institutes of Health and the National Cancer Institute. Startup funds for SB were provided by the Department of Mathematics, Statistics and Computer Science at Marquette University. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants. Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived and designed the experiments: SB, AL, HAF. Analyzed the data: SB, AL, MB, HAF. Wrote the first draft of the manuscript: SB. Contributed to the writing of the manuscript: AL, MB, HAF. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109(1):93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 2.Adamson C, Kanu OO, Mehta AI, et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18(8):1061–83. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- 3.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–7. [PubMed] [Google Scholar]
- 4.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102(16):5814–9. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mclendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li A, Walling J, Ahn S, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69(5):2091–9. doi: 10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a cpg island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–25. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhat KPL, Salazar KL, Balasubramaniyan V, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25(24):2594–609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57:289–300. [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozdag S, Li A, Wuchty S, Fine HA. FastMEDUSA: a parallelized tool to infer gene regulatory networks. Bioinformatics. 2010;26(14):1792–3. doi: 10.1093/bioinformatics/btq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karolchik D, Baertsch R, Diekhans M, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31(1):51–4. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guberman JM, Ai J, Arnaiz O, et al. BioMart central portal: an open database network for the biological community. Database (Oxford) 2011;2011:bar041. doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 18.Kundaje A, Xin X, Lan C, et al. A predictive model of the oxygen and heme regulatory network in yeast. PLoS Comput Biol. 2008;4(11):e1000224. doi: 10.1371/journal.pcbi.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmulevich I, Schwikowski B, Warner G, Ideker T. Integration of biological networks and gene expression data using cytoscape. Nat Protoc. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Cooper LAD, Gutman DA, Chisolm C, et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol. 2012;180(5):2108–19. doi: 10.1016/j.ajpath.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Yan W, Zhang W, et al. MiR-218 reverses high invasiveness of glioblastoma cells by targeting the oncogenic transcription factor LEF1. Oncol Rep. 2012;28(3):1013–21. doi: 10.3892/or.2012.1902. [DOI] [PubMed] [Google Scholar]
- 23.Haapa-Paananen S, Kiviluoto S, Waltari M, et al. HES6 gene is selectively overexpressed in glioma and represents an important transcriptional regulator of glioma proliferation. Oncogene. 2012;31(10):1299–310. doi: 10.1038/onc.2011.316. [DOI] [PubMed] [Google Scholar]
- 24.Somasundaram K, Reddy SP, Vinnakota K, et al. Upregulation of ASCL1 and inhibition of notch signaling pathway characterize progressive astrocytoma. Oncogene. 2005;24(47):7073–83. doi: 10.1038/sj.onc.1208865. [DOI] [PubMed] [Google Scholar]
- 25.Guilhamon P, Eskandarpour M, Halai D, et al. Meta-analysis of idh-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol Cancer Res. 2009;7(12):1893–901. doi: 10.1158/1541-7786.MCR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Held-Feindt J, Hattermann K, Knerlich-Lukoschus F, Mehdorn HM, Mentlein R. SP100 reduces malignancy of human glioma cells. Int J Oncol. 2011;38(4):1023–30. doi: 10.3892/ijo.2011.927. [DOI] [PubMed] [Google Scholar]
- 28.Ladha J, Sinha S, Bhat V, Donakonda S, Rao SMR. Identification of genomic targets of transcription factor AEBP1 and its role in survival of glioma cells. Mol Cancer Res. 2012;10(8):1039–51. doi: 10.1158/1541-7786.MCR-11-0488. [DOI] [PubMed] [Google Scholar]
- 29.Doucette T, Rao G, Rao A, et al. Immune heterogeneity of glioblastoma subtypes: extrapolation from the cancer genome atlas. Cancer Immunol Res. 2013;1:112. doi: 10.1158/2326-6066.CIR-13-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noy P, Williams H, Sawasdichai A, Gaston K, Jayaraman PS. PRH/hhex controls cell survival through coordinate transcriptional regulation of vascular endothelial growth factor signaling. Mol Cell Biol. 2010;30(9):2120–34. doi: 10.1128/MCB.01511-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh WC, Shahinian A, Speiser D, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7(5):715–25. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 32.Natoli G, Costanzo A, Ianni A, et al. Activation of SAPK/JNK by TNF receptor 1 through a noncytotoxic traf2-dependent pathway. Science. 1997;275(5297):200–3. doi: 10.1126/science.275.5297.200. [DOI] [PubMed] [Google Scholar]
- 33.Martin KH, Slack JK, Boerner SA, Martin CC, Parsons JT. Integrin connections map: to infinity and beyond. Science. 2002;296(5573):1652–3. doi: 10.1126/science.296.5573.1652. [DOI] [PubMed] [Google Scholar]
- 34.Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130(1):15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 35.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron. 1999;22(1):103–14. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 36.Schneider E, Bertron A-F, Dy M. Modulation of hematopoiesis through histamine receptor signaling. Front Biosci (Schol Ed) 2011;3:467–73. doi: 10.2741/s165. [DOI] [PubMed] [Google Scholar]
- 37.Bardot V, Dutrillaux AM, Delattre JY, et al. Purine and pyrimidine metabolism in human gliomas: relation to chromosomal aberrations. Br J Cancer. 1994;70(2):212–18. doi: 10.1038/bjc.1994.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao S, Wang C, Zheng Q, et al. STAT5 regulates glioma cell invasion by pathways dependent and independent of STAT5 DNA binding. Neurosci Lett. 2011;487(2):228–33. doi: 10.1016/j.neulet.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 39.Samimi G, Ring BZ, Ross DT, et al. TLE3 expression is associated with sensitivity to taxane treatment in ovarian carcinoma. Cancer Epidemiol Biomarkers Prev. 2012;21(2):273–9. doi: 10.1158/1055-9965.EPI-11-0917. [DOI] [PubMed] [Google Scholar]
- 40.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 41.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59(8):1190–9. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Clustering of GBM samples based on discretized gene expression. (A) PCA after discretizing genes with t = 2, TFs with t = 1.5; (B) PCA after discretizing genes with t = 1.5, TFs with t = 1.2; (C) PCA after discretizing genes with t = 1.2, TFs with t = 1.1; (D) hierarchical clustering after discretizing genes with t = 1.2, TFs with t = 1.1. Sample coloring: Red, classical; blue, mesenchymal; green, neural; purple, proneural−; orange, proneural+.
Supplementary Figure 2. Percentage of test loss for different number of boosting iterations.
Supplementary Figure 3. The source of variation of batch ID and GBM subtype in TCGA exon data based on a two-way ANOVA model.
Supplementary Figure 4. Kaplan–Meier survival plots for nine master regulators of GBM subtypes. Each regulator name is below the survival plot. Log-rank P < 0.05.
Supplementary Figure 5. Gene regulatory network of the classical subtype. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score.
Supplementary Figure 6. Gene regulatory network of the neural subtype. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score.
Supplementary Figure 7. Gene regulatory network of the proneural− subtype. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score.
Supplementary Figure 8. Gene regulatory network of the proneural+ subtype. The network shows the association of upregulated TFs with upregulated gene. Each rectangle node is a TF and each oval node is a gene. The width of the edge is proportional to the TF–gene significance score.
Supplementary Figure 9. IPA networks built from genes in gene regulatory network of the (A) classical (B) proneural−, (C) neural, (D) proneural+ subtypes. Colored nodes are the genes in the corresponding gene regulatory network (Red: upregulated, Green: downregulated).
Supplementary Figure 10. IPA networks derived from DEGs of (A) classical, (B) mesenchymal, (C) neural, (D) proneural− samples. Nodes annotated with a star are significant master regulators.
Supplementary Figure 11. Expression dot plot of HES6 in GBM subtypes.
Supplementary Table 1. Significant IPA canonical pathway for each GBM subtype. All P values are <0.05. Blank cells refer to P value > 0.05.
Supplementary Table 2. List of pathways that was significant in both IPA and GSEA. For each pathway, the subtypes that have that pathway have a “1” in that row.