Abstract
Sitobion avenae is a major agricultural pest of wheat in China. Using microsatellite markers, we studied the potential gene flow, genetic diversity, genetic differentiation, and genetic structure of seven S. avenae populations from different regions of China (Beijing, Hebei, Henan, Hubei, Jiangsu, Shandong, and Shanxi provinces). The populations from Henan, Shandong, and Jiangsu showed high levels of genic and genotypic diversity. By contrast, the genic diversity in the Beijing and Hebei populations was much lower. Despite this low genic diversity, the genotypic diversity of the Beijing population was higher than that of all of the other populations, except those from Jiangsu and Shandong. Overall, the genetic divergence among the seven S. avenae populations tested was high, though there was almost no differentiation between the Shandong and Henan populations. We observed significant negative correlation between the strength of gene flow and the geographic distances among populations. Based on genetic analysis, the seven S. avenae populations studied can be divided into four distinct clusters; (i) Hubei, (ii) Shanxi, (iii) Beijing and Hebei, and (iv) Shandong, Henan, and Jiangsu. The present results provide a basis for potentially optimizing integrated pest management (IPM) programs in China, through adapting control methods that target biological traits shared by various populations of the same genotype.
Introduction
Several wheat aphid species are major agricultural pests in China, notably Sitobion avenae, Rhopalosiphum padi (L.), Schizaphis graminum (Rondani), and Acyrthosiphon dirhodum Walker [1]. Of these, S. avenae is the most dominant and destructive wheat aphid in China [2], [3]; it affects about 13 million hm2 per year, and yield losses can be up to 40% [4]. Wheat aphids infest cereal crops and cause direct economic losses through sucking sap, and indirect losses by vectoring plant viruses [5].
Wheat aphids have complex life cycles that are known to be highly affected by climate [6]. Some aphid species exhibit great flexibility in the selection of their reproductive mode [7]–[9]. All lifecycle types reproduce parthenogenetically for most of the year; some species have an annual sexual phase [10]. The lifecycle of cyclic parthenogenic species have a sexual reproduction phase once a year, and overwinter as asexually produced eggs; this is a more reliable strategy when winters are harsh [11]–[16]. Cyclic parthenogenesis is the dominant mode of reproduction, especially in the regions with harsh winters [15], [17]. However, if the climatic conditions allow, some clones are obligate parthenogenic, they do not respond to autumnal cues and have no sexual phase. There are also intermediate types which employ both sexual and parthenogenetic reproduction [6], [9], [15], [18]. A study showed that the asexual genotypes of Rhopalosiphum padi had higher genetic variation in fitness compared to the sexual genotypes [19], indicating that the reproductive mode greatly impacts the fitness of wheat aphids.
S. avenae can survive on numerous plant species, including all of the cereals, many other monocots, and certain dicots [20]. Divergent selection on different host plants greatly influences the diversification of the aphids, and imposes considerable selective pressure on aphids' evolution [21]. Host-transfer experiments for clones of S. avenae collected from oat and barley showed that their fitness traits differed significantly, indicating a genetic basis for their differentiation [22].
Aphids have complex lifecycles and adaption to diverse hosts; this means that any given geographical population of aphids will not be homogeneous [23]. If the migration scale is large enough, the population genetic structure and evolutionary trajectory will be influenced [24]. As different kinds of aphids have different abilities to fly and face different pressures leading to migration, they act out different migration behaviors [25]. The flight ability of winged aphids is very weak [26], [27], and they migrate for long distances by largely depending on wind forces [28].
Clonal selection is very important in the study of population genetic structures of wheat aphids. Ecological adaption may occur quickly because of the rapid propagation of asexual offspring [29]. There are many factors influencing clonal selection. These include climatic, host plant, microclimates, crop density, natural enemy pressure, and resistance to pesticides [15], [30], [31].
It is difficult to understand the ecology of wheat aphids, as they tend to migrate with the wind [32]. Additionally, wheat aphids are small and have short life spans and large population sizes. Populations can be diluted rapidly in the air and migrate frequently [26]. Therefore, it is difficult to monitor the migration of wheat aphids using ecological approaches. Currently, polymorphic genetic markers have been widely used in the studies of population ecology [25].
In the past 20 years, many studies addressing the flying behavior and genetic structure of aphids populations have been conducted using microsatellite markers [15], [26], [27], [33], [34]. As noted, wheat aphids have different lifecycles based on the climate conditions of various geographic regions; the most suitable clone lineages for a given area are selected. Due to lifecycle strategy and clonal selection, studies of the aphids in the same region, but in different years, may also present different population genetic structures [15], [35]. However, the gene flow among different geographical populations can also be reflected in population genetic differentiation and population structure [34]–[36]. The management methods used to control wheat aphids can lead to new phenotypes emerging to overcome strong pressures such as pesticide applications, so quantification of the genetic diversity of populations is very important for the management of wheat aphids [37].
In the present study, we used five microsatellite loci (Sm10, Sm11, Sm12, Sm17, and Sa∑4), which had been used previously [15], [25], [38], [39], [40], to characterize the potential gene flow, genetic diversity, genetic differentiation, and genetic structure of seven S. avenae populations from seven different geographic regions in China. To interpret the population structure and dynamics of S. avenae, we need to better understand the importance of migration and clonal selection, which may have relevance in forecasting aphid outbreaks. Our results also provide a foundation for optimizing integrated pest management (IPM) programs through adapting control methods according to biological traits shared by various populations of the same genotype.
Materials and Methods
Aphid Sample Collection
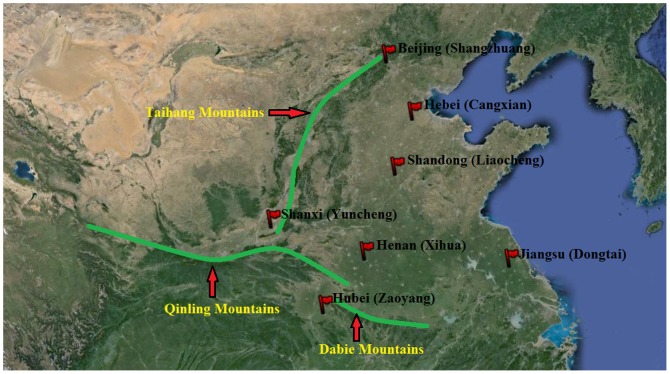
Wingless adults of S. avenae were collected from seven sites across six provinces (Hebei, Shandong, Henan, Shanxi, Jiangsu, and Hubei province) and the Beijing municipality of China. These are the main wheat producing provinces of China. The site locations are presented in Figure 1. The fields, where the S. avenae collected from, are agricultural experiment fields belong to plant protection research stations at Cangxian (Hebei province, China), Liaocheng (Shandong province, China), Xihua (Henan province, China), the Yanhu distriction of Yuncheng (Shanxi province, China), Dongtai (Jiangsu province, China), Zaoyang (Hubei province, China) and the Agricultural Experiment Station of China Agricultural University (Beijing, China), respectively. 30∼40 individuals were collected from each site, and each aphid was taken from a different field in order to minimize resampling of the same clone. Samples that were collected from the same region were considered as one ‘population’. The collected specimens were taken back to the laboratory in 100% ethanol.
Figure 1. S. avenae sampling sites in China.
DNA Extraction and PCR of Microsatellite Loci
Genomic DNA was extracted from wingless adult aphid individuals using DNAVzol (Vigorous Biotechnology Beijing Co., Ltd.), according to the manufacturer's instructions.
Two hundred forty six aphids were examined at five microsatellite loci (Sm10, Sm11, Sm12, Sm17, and Sa∑4). The microsatellite loci of Sm10, and Sm11, Sm12, Sm17 were isolated from S. miscanthi [38], and Sa∑4 was isolated from S. avenae. Sm11 is x-linked, and the other three loci are autosomal [40]. The SaΣ4 locus is also autosomal [15]. The primer sequences of 4 microsatellite loci (Sm10: 5′-TCTGCTGCATTACTGTTGGC-3′, 5′ -TCGTCTACTTCGCCGTCA-3′; Sm11: 5′-TCTGCTGCATTACTGTTGGC-3′, 5′ -TCGTCTACTTCGCCGTCA-3′; Sm17: 5′-TCTGCTGCATTACTGTTGGC-3′, 5′ -TCGTCTACTTCGCCGTCA-3′; Sa∑4: 5′-TCTGCTGCATTACTGTTGGC-3′, 5′ -TCGTCTACTTCGCCGTCA-3′) were reported by Simon et al. (1999) [15]. The primer sequences of locus Sm12 (5′-CACCATCGCGTTTCATCTTA-3′; 5′-ACTCCCAACCTCTGATGAGC-3′) were reported by Llewellyn et al. (2003) [25]. They have verified before application.
The PCR conditions were the same as those of Simon et al. (1999) [15]. PCR reactions were performed in a 20 µL reaction volume. Each reaction mixture contained 0.2 µL of 5 U/µL rTaq polymerase, 2.0 µL of each 2.5 umol dNTP and 2.0 µL of ×10 buffer (TaKaRa Biotechnology (Dalian) Co.,Ltd.), 1 µL of 10 µM of each primer (SANGON), and 1 µL of DNA template (approximately 10 ng). The PCR products were examined using an ABI3730*1 instrument and the allele sizes were analyzed using Genemapper 3.0 software (Applied Biosystems).
Data Analysis
The genotype data is presented in File S1. Departure from Hardy-Weinberg equilibrium (HWE) was tested under the hypothesis of heterogeneity deficit and excess using Genepop [41], [42]. The score test (U test) was used. Linkage disequilibrium among the microsatellite loci were also tested using Genepop [41], [42]. The null hypothesis was “genotypes at one loci are independent from genotypes at the other loci”, and the default test statistic was the log likelihood ratio statistic (G-test). During the processes of identification and isolation of microsatellite sequences using primers and amplification by PCR, the following errors can occur: 1. One or more alleles fail to amplify during PCR (null alleles); 2. Slight changes occur in the allele sizes during PCR (stuttering); 3. Large alleles do not amplify as efficiently as small alleles. We detected these errors using Micro-Checker v2.2.3 [43]. This application calculates the frequency of any null alleles detected, using the methods described by Chakraborty et al. (1992) [44] and Brookfield (1996) [45]. The indices of genetic diversity including the number of alleles (Na), the richness of alleles (Ar), the observed heterozygosity (HO), the expected heterozygosity (HE), gene diversity (HS), and the inbreeding index (FIS) for each population were calculated using FSTAT v2.9.3 [46]. Na is the mean number of alleles in each sample. It can be calculated according to the following equation:
Nj is the number of alleles at microsatellite loci j in one sample. r is the number of all loci. The richness of alleles (Ar) is the mean of Rs (allelic richness per locus and population) across all microsatellite loci. Rs is a measure of the number of alleles and is independent of sample size, hence allowing one to compare Rs between different sample sizes. However, the observed number of alleles in a sample is highly dependent on sample size. To bypass this problem, El Mousadik and Petit (1996) [47] suggested the adaption of the rarefaction index of Hurlbert (1971) [48] to population genetics. The principle is to estimate the expected number of alleles in a sub-sample of 2n genes, given that 2N genes have been sampled (N≥n). In FSTAT, n is fixed as the smallest number of individuals typed for a locus in a sample. Allelic Richness is then calculated according to the following equation:
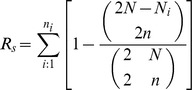 |
Where Ni is the number of alleles of type i among the 2N genes. Note that each term under the sum corresponds to the probability of sampling allele i at least once in a sample of size 2n. In FSTAT, estimates of gene diversity per locus and sample use an unbiased estimator [49]:
Where n is the size of a sample. pi is the frequency of allele Ai in a sample. Ho is the observed proportion of heterozygotes in a sample. The multilocus genotypes (MLG) were classified based on the length of alleles, using software that we developed in house. The genotype diversity (K) of each population was calculated as follows: K = G/N, where G is equal to the number of MLGs, and N is the number of samples [25].
F-statistics (FIT, FIS, and FST) and pairwise FST
[50], were estimated using FASTAT v2.9.3 [46]. Nei's standard genetic distance (DS) [51] and Nei's genetic distance (DA) [52] were calculated using the Dispan program [53]. An indirect estimate of gene flow was calculated as 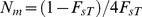 [54]. To analyze the relationship between FST and geographic distance, a Mantel test was carried out. The matrices of pairwise FST and the geographic distance between two populations was transformed by Genepop (the geographic distances were transformed to the natural logarithm (Ln) of geographic distances in kilometers), and a regression was performed using SPSS 17.0.
[54]. To analyze the relationship between FST and geographic distance, a Mantel test was carried out. The matrices of pairwise FST and the geographic distance between two populations was transformed by Genepop (the geographic distances were transformed to the natural logarithm (Ln) of geographic distances in kilometers), and a regression was performed using SPSS 17.0.
To identify the population structure, two clustering methods were used. First, we constructed phylogenetic trees (dendrograms) using the unweighted pair group-method with arithmetic mean (UPGMA) [55], based on Nei's genetic distance (DA) using the Dispan program. Second, factorial correspondence analysis (FCA) was implemented in Genetix v4.04 [56], to examine the three-dimensional spatial distribution of genetic variation for each individual.
An analysis of molecular variance (AMOVA) was implemented in Arlequin v3.0 [57], to confirm population clusters and to examine the component variance of genetic differentiation among populations.
Results
Genetic Diversity
We screened 246 aphids collected from six provinces and the Bejing municipality of China for five microsatellite loci. There was no large allele dropout or stuttering observed in the result. Of the 35 tests, there were 6 tests in which null alleles were found to exist; they appeared at the Sm11 and Sm17 microsatellite loci. However, no particular microsatellite loci had null alleles in all of the populations. The frequencies of the null alleles were not more than 0.08 (Table 1). In the 70 linkage disequilibrium tests, there were 12 tests with significant linkage (Table 1). However, no fixed combination of any two microsatellite loci showed significant linkage in all of the populations, and the 5 microsatellite loci selected here can estimate genetic mutation independently. Five populations deviated from HWE significantly because of heterozygote deficit; both Beijing and Shanxi populations were the exception to this. The Shanxi population deviated from HWE because of heterozygote excess (Table 1). The FIS values of populations were positive, except for the Beijing and Shanxi populations. The FIS of the Shanxi population was the most negative (Table 2).
Table 1. Outcomes of null allele frequency, linkage disequilibrium test, and HWE test.
| Populations | Beijing | Hubei | Hebei | Henan | Jiangsu | Shandong | Shanxi |
| Null allele frequency (r) | |||||||
| Sm10 | — | — | — | — | — | — | — |
| Sm11 | — | — | — | 0.06 | — | 0.07 | — |
| Sm12 | — | — | — | — | — | — | — |
| Sm17 | 0.08 | — | 0.03 | 0.06 | 0.08 | — | — |
| Sa∑4 | — | — | — | — | — | — | — |
| Linkage disequilibrium test (p-value) | |||||||
| Sm10-Sm11 | 0.51 | 0.01* | 0.03* | 0.75 | 0.34 | 0.64 | 0.04* |
| Sm10-Sm12 | 0.09 | 0.07 | 0.00* | 1.00 | 0.74 | 1.00 | 0.12 |
| Sm11-Sm12 | 1.00 | 0.00* | 0.01* | 0.19 | NA | 1.00 | 1.00 |
| Sm11-Sm17 | NA | 0.01* | 0.25 | 0.00* | 0.94 | 0.00* | 1.00 |
| Sm12-Sa∑4 | 0.46 | 0.06 | NA | NA | 0.03* | NA | 1.00 |
| Sm10-Sm17 | 0.28 | 0.02* | 0.21 | 0.04 | 0.11 | 0.77 | 0.06 |
| Sm10-Sa∑4 | 0.24 | 0.76 | NA | NA | 0.66 | 0.61 | 0.97 |
| Sm11-Sa∑4 | 1.00 | 0.26 | NA | NA | 1.00 | 0.46 | 1.00 |
| Sm12-Sm17 | 0.21 | 0.02* | 0.22 | 0.13 | 0.05 | 1.00 | 0.62 |
| Sm17-Sa∑4 | NA | 0.39 | NA | NA | 0.60 | 0.74 | 0.17 |
| HWE test (p-value) | |||||||
| heterozygote deficit | 0.58 | 0.00a | 0.01a | 0.00a | 0.00a | 0.00a | 1.00 |
| heterozygote excess | 0.42 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.00a |
“—”: There were no null alleles detected.
“*”: p<0.05, significant linkage; “NA”: no contingency table.
: P<0.05, significant deviation from Hardy-Weinberg equilibrium.
Table 2. Indices of genetic diversity for the seven S. avenae populations of China sampled in 2012.
| Beijing | Hubei | Hebei | Henan | Jiangsu | Shandong | Shanxi | |
| N | 40 | 35 | 36 | 32 | 36 | 31 | 36 |
| HS | 0.390 | 0.530 | 0.487 | 0.741 | 0.637 | 0.728 | 0.546 |
| Na | 6.0 | 7.6 | 6.8 | 15.2 | 13.0 | 13.0 | 7.2 |
| Ar | 3.65 | 4.35 | 3.69 | 8.36 | 6.11 | 7.15 | 4.10 |
| FIS | −0.075 | 0.027 | 0.036 | 0.079 | 0.003 | 0.122 | −0.413 |
| HO | 0.42 | 0.52 | 0.59 | 0.85 | 0.64 | 0.65 | 0.77 |
| HE | 0.39 | 0.52 | 0.60 | 0.92 | 0.62 | 0.72 | 0.54 |
| MLG | 37 | 31 | 25 | 29 | 36 | 31 | 26 |
| K | 0.925 | 0.886 | 0.694 | 0.906 | 1.000 | 1.000 | 0.722 |
| within | 3 | 4 | 4 | 3 | 0 | 0 | 6 |
| among | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
HO, observed heterozygosity; HE, expected heterozygosity; HS, gene diversity; Na, mean numbers of alleles; Ar, allelic richness based on eight samples per population; MLGs, number of multilocus genotypes; #within, number of MLGs shared within a population; #among, number of MLGs shared among populations; K, index of global genotypic diversity (MLGs /N); FIS, the inbreeding index. The seven populations were named by their sampling locations, Beijing, Hubei, Hebei, Henan, Jiangsu, Shandong, and Shanxi, respectively.
Our results suggested that S. avenae in China have a high level of genetic diversity. The indices of genetic diversity of the seven populations are listed in Table 2. The results show that the populations of Henan, Shandong, and Jiangsu have relatively high levels of genic and genotypic diversity. The values of Na, Ar, and HS present in the Henan population (15.2, 8.36, and 0.74, respectively) were higher than the values for the other populations. The genic diversity level of the Beijing population was the lowest; the values of Na, Ar, and HS of the Beijing population were 6, 3.65, and 0.39, respectively. However, the genotypic diversity of Beijing population was higher (K = 0.925) than all of the other populations, except for the Jiangsu and Shandong populations.
We observed 215 multilocus genotypes from the 246 aphids tested. The same genotypes were observed within the Hubei, Henan, Shanxi, Hebei, and Beijing populations, but the same genotype was not found within the Jiangsu and Shandong populations. However, one individual in 31 aphids from Shandong was found to share the same genotype with one individual from Jiangsu province. There were not any genotypes shared among the other five populations.
Genetic Differentiation
Population differentiation was analyzed using pairwise FST values (Table 3). The pairwise FST values between the Beijing and the other populations, except for Hubei, were generally high (0.2589∼0.4186); high levels of differentiation were also detected between Hubei and the other populations (0.2137∼0.3626). The pairwise FST value between the Shandong and Jiangsu populations (FST = 0.1012) was close to the value between the Beijing and Hebei populations (FST = 0.1055), which were lower than the values for the other populations, with the exception of the value between the Henan and Shandong populations (F ST = 0.0383).
Table 3. Pairwise FST value among the seven S. avenae populations of China.
| Beijing | Hubei | Hebei | Henan | Jiangsu | Shandong | |
| Hubei | 0.4186 | |||||
| Hebei | 0.1055 | 0.3626 | ||||
| Henan | 0.2604 | 0.2137 | 0.2015 | |||
| Jiangsu | 0.3547 | 0.3182 | 0.2852 | 0.1611 | ||
| Shandong | 0.2589 | 0.2253 | 0.1911 | 0.0383 | 0.1012 | |
| Shanxi | 0.3836 | 0.3320 | 0.2648 | 0.1654 | 0.2724 | 0.1858 |
FST≤0.05, indicates little differentiation among populations; 0.05<FST≤0.15, middling differentiation; (iii). 0.15<FST≤0.25, high differentiation; FST>0.25, significant divergence. The seven S. avenae populations were named by their sampled locations, Beijing, Hubei, Hebei, Henan, Jiangsu, Shandong, and Shanxi, respectively.
According to an analysis of molecular variance (AMOVA), the S. avenae populations sampled had high levels of genetic differentiation (FST = 0.2547) (Table 4). Genetic variation between populations accounted for 25.47% of the total genetic variation.
Table 4. The composition of variation (average over 5 loci).
| Source of variation | Sum of squares | Variance components (Vi) | Percentage variation | Fixation Indices |
| Among population | 176.279 | 0.49323Va | 25.47 | FST = 0.2547 |
| Among individuals within populations | 272.715 | −0.09396Vb | −4.85 | FIS = −0.0651 |
| Within individuals | 319.500 | 1.53724Vc | 79.38 | FIT = 0.2062 |
| Total | 768.494 | 1.93651 |
Va, the variance among populations; Vb, the variance whin population; Vc: the variance within individuals. FST, the average of genetic differentiation among populations; FIS, the average of inbreeding coefficient among individuals within populations; FIT, the average of total inbreeding coefficient.
The migration number among different geographical populations has been proposed to be represented as gene flow (Nm) [54]. There is little divergence if Nm>1. However, if Nm<1, it means that gene flow could not counteract the divergence caused by genetic drift [58]. In the present study, the level of genetic differentiation among the seven S. avenae populations was Nm = 0.899 (Table 5), and the gene flow was not large enough to offset the genetic drift. The biggest gene flow (Nm = 6.277) was observed between the Henan and Shandong populations (Table 6). The values of Nm between Beijing and the other populations were less than 1, except for the Hebei population. The gene flow between the Hubei population and the other populations was also low (Nm<1).
Table 5. Summary of the F-statistics at five microsatellite loci.
| FIT | FIS | FST | Nm | |
| Sm10 | 0.012 | −0.086 | 0.090 | 2.528 |
| Sm11 | 0.502 | 0.082 | 0.457 | 0.297 |
| Sm12 | 0.033 | −0.183 | 0.182 | 1.124 |
| Sm17 | 0.282 | 0.021 | 0.267 | 0.686 |
| Sa∑4 | 0.126 | 0.038 | 0.092 | 2.467 |
| Mean | 0.191 | −0.026 | 0.218 | 0.899 |
FIT: the average of total inbreeding coefficient; FIS: the average of inbreeding coefficient among populations; FST: the average of genetic differentiation; Nm: gene flow, 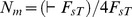 .
.
Table 6. The number of individuals that migrated among differential regions.
| Nm | Beijing | Hubei | Hebei | Henan | Jiangsu | Shandong |
| Hubei | 0.3472 | |||||
| Hebei | 2.1197 | 0.4395 | ||||
| Henan | 0.7101 | 0.9199 | 0.9907 | |||
| Jiangsu | 0.4548 | 0.5357 | 0.6266 | 1.3018 | ||
| Shandong | 0.7156 | 0.8596 | 1.0582 | 6.2774 | 2.2204 | |
| Shanxi | 0.4017 | 0.5030 | 0.6941 | 1.2615 | 0.6678 | 1.0955 |
Nm, gene flow, 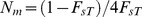 .
.
Overall, the genetic divergence of the populations had significant positive correlation (r2 = 0.401, p = 0.002) with geographic distance, according to the results of the Mantel test.
Population Structure
We used two methods for clustering analysis, and a small divergence appeared between the two sets of results.
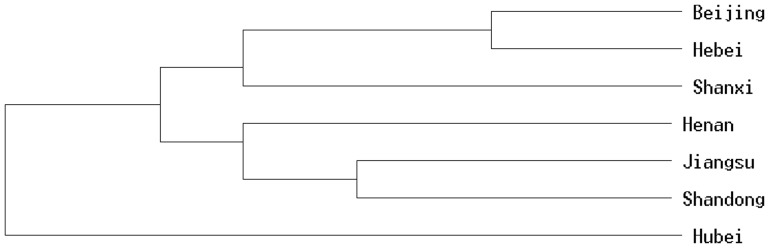
First, we classified the seven geographical populations into three clusters using the UPGMA method (Figure 2): (i) Hubei, (ii) Shanxi, Beijing and Hebei, and (iii) Shandong, Henan, and Jiangsu.
Figure 2. UPMGA Phylogenetic tree of seven different S. avenae geographic populations in China.
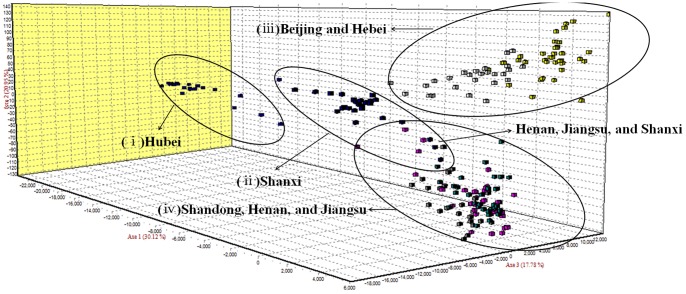
Second, the FCA results divided the seven populations into four groups (Figure 3); (i) Hubei, (ii) Shanxi, (iii) Beijing and Hebei, and (iv) Shandong, Henan, and Jiangsu. The only differentiation between results of the two clustering methods was that the Shanxi population formed its own cluster in the results of the FCA method.
Figure 3. Three-dimensional factorial correspondence analysis (FCA) of S. avenae sampled in 2012.
The circles indicate populations that cluster according to geography.
However, the two kinds of genetic distance (DS and DA) (Table 7) between the Shanxi and Beijing populations were larger than between Shanxi and the other populations, except for Hubei. Therefore, the Shanxi and Beijing populations cannot be classified into a single group.
Table 7. The standard genetic distance (DS) and Nei's unbiased genetic distance (DA) of pairwise comparisons among the seven populations.
| Beijing | Hubei | Hebei | Henan | Jiangsu | Shandong | Shanxi | |
| Beijing | — | 1.0144 | 0.0890 | 0.5421 | 0.9047 | 0.5294 | 0.8219 |
| Hubei | 0.7344 | — | 0.8859 | 0.5784 | 1.0331 | 0.6196 | 0.8590 |
| Hebei | 0.1914 | 0.6757 | — | 0.4638 | 0.7195 | 0.4123 | 0.4870 |
| Henan | 0.5674 | 0.7286 | 0.5835 | — | 0.5557 | 0.1233 | 0.3740 |
| Jiangsu | 0.5864 | 0.7111 | 0.5447 | 0.5470 | — | 0.2811 | 0.7536 |
| Shandong | 0.5050 | 0.7147 | 0.4976 | 0.3532 | 0.3294 | — | 0.4354 |
| Shanxi | 0.5382 | 0.6810 | 0.3746 | 0.5354 | 0.5196 | 0.5318 | — |
“upper triangle”, Nei's standard genetic distance (DS); “lower triangle”: Nei's genetic distance (DA). The seven populations were named by their sampled locations, Beijing, Hubei, Hebei, Henan, Jiangsu, Shandong, and Shanxi, respectively.
We classified the seven populations into 4 groups (i. Hubei; ii. Shanxi; iii. Beijing and Hebei; iv. Shandong, Henan, and Jiangsu.), and conducted AMOVA to detect the variance components. The results showed that the variances among groups and among populations within groups accounted for 18.34% and 9.53% of the total variance, respectively (Table 8), and that the variance between groups was significantly larger than the variance among populations within a particular group (FCT = 0.18399). This result indicates that gene flow among the four groups was strictly restricted. The AMOVA results thus confirmed the conclusions of the clustering analysis.
Table 8. Analysis of molecular variances (AMOVA) of microsatellites in the seven populations.
| Source of variation | Sum of squares | Variance components | Percentage variation | Fixation Indices |
| Among groups | 142.425 | 0.36698 | 18.33888 | FCT = 0.18399 |
| Among populations within groups | 33.855 | 0.19082 | 9.53602 | FSC = 0.11678 |
| Among individuals within population | 272.715 | −0.09396 | −4.69544 | FIS = −0.06510 |
| Within individuals | 319.5 | 1.53724 | 76.82053 | FIT = 0.23179 |
| Total | 768.494 | 2.00108 |
Va, variance components among groups; Vb, variance components among populations but within groups; Vc, variance components among individuals within population; Vd, variance components within individuals; FCT, F SC, FIS, and FIT are inbreeding coefficients.
Discussion
S. avenae is a migratory pest insect that is widely distributed throughout the wheat growing regions of China [59]. In the present study, we analyzed the population genetic diversity, population genetic differentiation, and genetic structure of seven different S. avenae geographical populations in China using microsatellite marker technology. These research results provide a basis for efforts to optimize integrated pest management (IPM) programs through adapting control methods which target the biological traits shared by various populations from the same genotype.
High Level of Genetic Diversity
The genetic diversity of aphids is influenced by many factors such as climate, host plants, topography, and physiognomy [60]. Wheat aphid populations frequently exhibit high levels of genetic variation [61]. In the present study, a high level of genetic diversity was detected at all of the five microsatellite loci. There were 32, 41, 35, 60, and 13 alleles at the Sm10, Sm11, Sm12, Sm17, and Sa∑4 loci, respectively. The genetic diversity level in our study (average = 36.2 alleles/locus) was much higher than that detected by Guo et al. (2005) (average = 9.6 alleles/locus) [61]. In particular, the genic diversity of the Henan and Shandong populations was higher than the diversity of the other populations. This may be due to the fact that their geographic location had more gene flow with the surrounding wheat production regions. In our results, genotypic diversity was also quite high. Two hundred fifteen multilocus genotypes were distinguished in the 246 individual samples. A given genotype was rarely present in the different geographic populations, which may result from a prevalence of sexual reproduction.
Many factors can affect the heterozygosity of populations. The expected heterozygosity (HE) of the Hebei, Henan, and Shandong populations was lower than their observed heterozygosity (HO). According to previous aphid genetic studies, factors like inbreeding, the Wahlund effect, and the presence of null alleles and/or clonal selection have been implicated in the generation of heterozygote deficits [39], [62]. As S. avenae has the ability for long-range dispersal, and their host plants are common and widespread, strong inbreeding and the Wahlund effect might not occur in this aphid [39], [63]. There are also many reasons for heterozygote excess, including rarely migrating between locations, asexual lineage expansion, and/or the accumulation of heterozygosity by mutation in longer-term parthenogenesis [64]. In our results, the greatest heterozygosity excess was present in the Shanxi population, and the FIS value of the Shanxi population was extremely negative. This might be associated with the geography of Yuncheng of Shanxi. Shanxi province is mountainous, which might restrict aphid migration.
Genetic Differentiation and Genetic Structure
The genetic structure of populations reflects the interaction of genetic drift, mutation, gene flow, migration, and natural selection [25]. There is absolutely no genetic differentiation among populations when FST = 0, and when the FST = 1, the populations can be recognized to have differentiated completely. The threshold of FST value can be divided into 4 levels: (i). FST≤0.05, indicates little differentiation among populations; (ii). 0.05<FST≤0.15, middling differentiation; (iii). 0.15<FST≤0.25, high differentiation; (iv). FST>0.25, significant divergence [65].
In the present study, there was a high level of genetic differentiation (FST = 0.2547) present in the S. avenae populations of China. Various factors might affect the divergence of S. avenae populations in different sites, including reproductive mode, clonal selection, migration, and so on. Additionally, genetic differentiation might be accelerated by the high level of variation in S. avenae population size [66].
A study showed that the aphid clones on the winter host plant did not significantly contribute to the spring/summer population build-up in the cereal fields over short distances, but that the density of sources for early migrants, on a regional scale, is important for the establishment of the population [67]. Therefore, the migration of aphids plays an important role in genetic structure. Aphids can become suspended in the air, and thus fly for long distances with the wind [10], [68]. This flight phenomenon can overcome the combination of the forces of selection, genetic drift, and mutation, if the scale of the migration of S. avenae is large enough [25]. The time we collected the wheat aphids samples was the peak time of their breeding. It is known that when the population density of S. avenae is high enough and if the host plant cannot supply adequate nutrition, winged aphids will be produced, and a large scale migration can occur. In our results, there was little differentiation between the Henan and Shandong populations (FST = 0.0383), which might be attributed to large scale migration and/or the local pest management strategies. The Mantel test showed that the genetic divergence of the populations had a significant positive correlation with the geographic distances between the populations. This means that the gene flow between populations was significantly associated with the distance among the locations, and that the gene exchange among the different geographic populations could be affected by geographical isolation. For example, the genetic divergence between Beijing (the most northern site in our study) and Hubei (the most southern site in our study) populations was the highest as compared to the others.
However, geographical distance didn't explain the study results completely. According to the results published by Huang et al. (2013) [69], some vital life-history traits (the developmental time of the instars of nymphs, postreproductive timing, and total lifespan of adults, and so on) were different for aphid populations living North or South of the Qinling Mountains. The divergence and gene flow among S. avenae populations might be impacted by the barrier of the Qinling Mountains. In the present study, the Qinling mountains and Dabie Mountains separate Zaoyang city of Hubei from the other 6 sites. There was great genetic differentiation between Hubei and the other populations, which might result from a limited dispersal of aphids. Similar phenomena of genetic isolation resulting from by geographical barriers were also found in Eriosoma lanigerum [70], two tansy-feeding aphids (Macrosiphoniella tanacetaria and Metopeurum fuscoviride) [33], and Diuraphis noxia [34].
Populations of S. avenae in China might be classified into 4 clusters based on our research results. The population from Shanxi had less communication with other populations because of Taihang mountain, which may have obstructed the migration of the wheat aphids. However, the geographic location of Shanxi is adjacent to Henan, Hebei, and Shandong, and the location of sample collection for Shanxi was Yuncheng, which is at the southern limit of the Taihang mountain range, so gene flow was affected relatively little and there was overlap between Shanxi, Henan, and Shandong in the FCA analysis (Figure 3).
Conclusions
In the present study, we analyzed the population genetics and predicted the gene flow among the seven different S. avenae geographical populations in China using microsatellite marker technology. The genetic diversity of the Henan and Shandong populations was higher than the others populations. The genic diversity of the Hebei and Beijing populations were the lowest. For genetic differentiation, there was high or significant differentiation among the Hubei population and the others. The genetic differentiation between Beijing and Hebei, and Henan and Shandong were both at very low levels, indicating that the gene flow occurs frequently between these populations. Finally, the 7 S. avenae geographic populations can be divided into 4 clusters: (i) Hubei, (ii) Shanxi, (iii) Beijing and Hebei, and (iv) Shandong, Henan, and Jiangsu. The present results provide a basis for efforts to optimize Integrated Pest Management programs through adapting control methods which target the biological traits shared by various populations from the same genotype.
Supporting Information
Microsatellite genotypes of the seven Sitobion avenae populations in China. “individual ID”, the tag of each tested aphid individual. “sampling location”, the corresponding location where each aphid individual was collected. The other ten columns were the corresponding allele sizes of each aphid individual at each microsatellite loci.
(XLSX)
Acknowledgments
This work was supported by the National Basic Research and Development Program of China (2012CB114103).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This research was supported by National Basic Research and Development Program of China (Contract No. 2012CB114103) (http://program.most.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lu YH, Gao XW (2009) Multiple mechanisms responsible for differential susceptibilities of Sitobion avenae (Fabricius) and Rhopalosiphum padi (Linnaeus) to pirimicarb. B Entomol Res 99: 611–617. [DOI] [PubMed] [Google Scholar]
- 2. Lu YH, He YP, Gao XW (2013) Comparative studies on acetylcholinesterase characteristics between the aphids, Sitobion avenae and Rhopalosiphum padi . J Insect Sci 13: 9 Available: http://www.insectscience.org/13.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang YJ, Jiang YY, Feng XD, Xia B, Zeng J (2009) Occurring trends of major crop pests in national significances in 2009. China Plant Prot 29: 33–36. [Google Scholar]
- 4. Xu ZH, Chen JL, Cheng DF, Liu Y, Francis F (2011) Genetic variation among the geographic population of the grain aphid, Sitobion avenae (Hemiptera: Aphididae) in China inferred from mitochondrial COI gene sequence. Agricultural Sciences in China 10: 1041–1048. [Google Scholar]
- 5.Power AG, Gray S (1995) Aphid transmission of barley yellow dwarf viruses: interactions between viruses, in-Barley Yellow Dwarf: 40 years of progress (C.J. D'Arcy and P. Burnett). The American Phytopalogical Society, St. Paul. pp. 259–291.
- 6. Simon JC, Rispe C, Sunnucks P (2002) Ecology and evolution of sex in aphids. Trends Ecol Evol 17: 34–39. [Google Scholar]
- 7. Blackman RL (1971) Variation in the photoperiodic response within natural populations of Myzus persicae (Sulz). B Entomol Res 60: 533–546. [DOI] [PubMed] [Google Scholar]
- 8. Simon JC, Blackman RL, Le Gallic JF (1991) Local variability in the life-cycle of the bird-cherry oat aphid, Rhopalosiphum padi (Homoptera: Aphididae) in western France. B Entomol Res 81: 315–322. [Google Scholar]
- 9. Dedryver CA, Le Gallic JF, Gauthier JP, Simon JC (1998) Lifecycle of the cereal aphid Sitobion avenae F.: polymorphism and comparison of life-history traits associated with sexuality. Ecol Entomol 23: 123–132. [Google Scholar]
- 10.Dixon AFG (1988) Aphid Ecology, 2nd edn. London: Chapman and Hall.
- 11. Hand SC (1980) Overwintering of cereal aphids. SROP/WPRS Bulletin 3: 59–61. [Google Scholar]
- 12. Leather SR (1980) Egg survival in the bird cherry-oat aphid, Rhopalosiphum padi (L.). Entomol Exp Appl 27: 96–97. [Google Scholar]
- 13. James BD, Luff ML (1982) Cold-hardiness and development of eggs of Rhopalosiphum insertum . Ecol Entomol 7: 277–282. [Google Scholar]
- 14. Rispe C, Pierre JS, Simon JC, Gouyon PH (1998) Models of sexual and asexual coexistence in aphids based on constraints. J Evolution Biol 11: 685–701. [Google Scholar]
- 15. Simon JC, Baumann S, Sunnucks P, Hebert PDN, Pierre JS (1999) Reproductive mode and population genetic structure of the cereal aphid Sitobion avenae studied using phenotypic and micro satellite markers. Mol Ecol 8: 531–545. [DOI] [PubMed] [Google Scholar]
- 16. Nespolo RF, Halkett F, Figueroa CC, Plantegenest M, Simon JC (2009) Evolution of trade-offs between sexual and asexual phases and the role of reproductive plasticity in the genetic architecture of aphid life histories. Evolution 63: 2402–2412. [DOI] [PubMed] [Google Scholar]
- 17. Blackman RL (1972) The inheritance of life cycle differences in Myzus persicae (Sulz) (Hem., Aphididae). B Entomol Res 62: 281–294. [Google Scholar]
- 18. Demotte F, Leterme N, Bonhomme J, Rispe C, Simon JC (2001) Multiple routes to asexuality in an aphid. Proc Roy SocSer B. 263: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carter MJ, Simon JC, Nespolo RF (2012) The effects of reproductive specialization on energy costs and fitness genetic variances in cyclical and obligate parthenogenetic aphids. Ecol Evol 2: 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackman R, Eastop VF (2006) Aphids on the world's herbaceous plants and shrubs. Available: http://www.aphidsonworldsplants.info/index.htm.
- 21. Peccoud J, Simon JC, Dohlen CV, Coeur d'acier A, Plantegenest M (2010) Evolutionary history of aphid-plant associations and their role in aphid diversification. CR Bilo 333: 474–487. [DOI] [PubMed] [Google Scholar]
- 22.Gao SX, Liu DG, Chen H, Meng XX (2013) Fitness traits and underlying genetic variation related to host plant specialization in the aphid Sitobion avenae. Insect Sci. doi:10.1111/1744-7917.12085 [DOI] [PubMed]
- 23. Lushai G, Loxdale HD (2002) The biological improbability of a clone (mini-review). Genetical Research, Cambridge. 79: 1–9. [DOI] [PubMed] [Google Scholar]
- 24. Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236: 787–792. [DOI] [PubMed] [Google Scholar]
- 25. Llewellyn KS, Loxdale HD, Harrington R, Brookes CP, Clark SJ (2003) Migration and genetic structure of the grain aphid (Sitobion avenae) in Britain related to climate and clonal fluctuation as revealed using microsatellites. Mol Ecol 12: 21–34. [DOI] [PubMed] [Google Scholar]
- 26. Loxdale HD, Hardie J, Halbert S, Foottit R, Kidd NAC (1993) The relative importance of short- and long-range movement of flying aphids. Bilo Rev 68: 291–311. [Google Scholar]
- 27. Zhang Y, Wang L, Wu KM, Wyckhuys KAG, Heimpel GE (2008) Flight performance of the soybean aphid, Aphis glycines (Hemiptera: Aphididae) under different temperature and humidity regimens. Environ Entomol 37: 301–306. [DOI] [PubMed] [Google Scholar]
- 28. Venette RC, Ragsdale DW (2004) Assessing the invasion by soybean aphid (Homoptera: Aphididae): where will it end? Ann Entomol Soc Am 97: 219–226. [Google Scholar]
- 29. Lushai G, Loxdale HD (2003) Maintenance of aphid clonal lineages: images of immortality. Infect Genet Evol 3: 259–269. [DOI] [PubMed] [Google Scholar]
- 30. Margaritopoulos JT, Kasprowicz L, Malloch GL, Fenton B (2009) Tracking the global dispersal of a cosmopolitan insect pest, the peach potato aphid. BMC Ecology 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vorburger C (2006) Temporal dynamics of genotypic diversity reveal strong clonal selection in the aphid Myzus persicae . J Ecolution Bilo 19: 97–107. [DOI] [PubMed] [Google Scholar]
- 32. Hardie J (1993) Flight behaviour in migrating insects. J Agr Urban Entomol 10: 239–245. [Google Scholar]
- 33. Loxdale HD, Schofl G, Wiesner KR, Nyabuga FN, Heckel DG (2011) Stay at home aphids: comparative spatial and seasonal metapopulation structure and dynamics of two specialist tansy aphid species studied using microsatellite markers. Biol J Linn Soc 104: 838–865. [Google Scholar]
- 34. Zhang B, Edwards OR, Kang L, Fuller SJ (2012) Russian wheat aphids (Diuraphisnoxia) in China: native range expansion or recent introduction? Mol Ecol 21: 2130–2144. [DOI] [PubMed] [Google Scholar]
- 35. Guillemaud T, Mieuzet L, Simon JC (2003) Spatial and temporal genetic variability in French populations of the peach-potato aphid, Myzus persicae . Heredity 91: 143–152. [DOI] [PubMed] [Google Scholar]
- 36. Massonnet B, Simon JC, Weisser WW (2002) Metapopulation structure of the specialized herbivore Macrosiphoniella tanacetaria (Homoptera, Aphididae). Mol Ecol 11: 2511–2521. [DOI] [PubMed] [Google Scholar]
- 37. Marcelo LDS, Vieira LGE (2010) Temporal genotypic diversity of Schizaphis graminum (Rondani 1852) (Hemiptera: Aphididae) in a black oats (Avena strigosa) field. Braz Arch Biol Techn 53: 911–916. [Google Scholar]
- 38. Sunnucks P, England PR, Taylor AC, Hales DF (1996) Microsatellite and chromosome evolution of parthenogenetic Sitobion aphids in Australia. Genetics 144: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sunnucks P, De Barro PJ, Lushai G, Maclean N, Hales D (1997) Genetic structure of an aphid studied using microsatellites: cyclic parthenogenesis, differentiated lineages, and host specialisation. Mol Ecol 6: 1059–1073. [DOI] [PubMed] [Google Scholar]
- 40. Wilson ACC, Sunnucks P, Hales DF (1997) Random loss of X chromosome at male determination in an aphid, Sitobion near Fragariae, detected using an X-linked polymorphic microsatellite marker. Genetical Research,Cambridge 69: 233–236. [Google Scholar]
- 41. Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity 86: 248–249. [Google Scholar]
- 42. Rousset F (2008) Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resources 8: 103–106. [DOI] [PubMed] [Google Scholar]
- 43. Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538. [Google Scholar]
- 44. Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5: 453–455. [DOI] [PubMed] [Google Scholar]
- 45. Chakraborty R, De Andrade M, Daiger SP, Budowle B (1992) Apparent heterozygote deficiencies observed in DNA typing data and their implications in forensic applications. Annals Human Genetics 56: 45–57. [DOI] [PubMed] [Google Scholar]
- 46.Goudet J (2001) FSTAT: a program to estimate and test gene diversities and fixation indices, version 2.9.3. Available: http://www.unil.ch/izea/softwares/Fstat.html.
- 47. El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor. Appl Genet 92: 832–839. [DOI] [PubMed] [Google Scholar]
- 48. Hurlbert SH (1971) The nonconcept of species diversity: a critique and alternative parameters. Ecology 52: 577–586. [DOI] [PubMed] [Google Scholar]
- 49.Nei M (1987) Molecular Evolutionary Genetics Columbia. New York: University Press.
- 50. Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- 51. Nei M (1972) Genetic distance between populations. Am Nat 106: 283–293. [Google Scholar]
- 52. Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19: 153–170. [DOI] [PubMed] [Google Scholar]
- 53.Ota T (1993) DISPAN: Genetic distance and phylogenetic analysis. Institute of Molecular Evolutionary Genetics, Pennsylvania State University, PA, USA.
- 54.Wright S (1969) Evolution and the genetics of populations: the theory of gene frequencies. Illinois, Chicago: The University of Chicago Press.
- 55.Sneath PHA, Sokal RR (1973) Numerical taxonomy. San Francisco, CA: W.H. Freeman.
- 56.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996) GENETIX 4.05, Logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier, France.
- 57. Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 58. Slatkin M (1981) Estimating levels of gene flow in natural populations. Genetics 99: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu ZH, Chen JL, Cheng DF, Sun JR (2011) Discovery of English grain aphid (Hemiptera: Aphididae) biotypes in China. J Econ Entomol 104: 1080–1086. [DOI] [PubMed] [Google Scholar]
- 60. Cai QN, Wang Y, Hu MH, Gu P (2004) Influence factors and molecular basis of the genetic diversity of aphid populations. Entomological Knowledge 41: 285–290. [Google Scholar]
- 61. Wei G, Zuorui S, Zhihong L, Lingwang G (2005) Migration and population genetics of the grain aphid Macrosiphum miscanti (Takahashi) in relation to the geographic distance and gene flow. Prog Nat Sci 15: 1000–1004. [Google Scholar]
- 62. Simon JC, Hebert PDN (1995) Patterns of genetic variation among Canadian populations of the bird cherry-oat aphid, Rhopalosiphum padi L. (Homoptera: Aphididae). Heredity 74: 346–353. [Google Scholar]
- 63. Loxdale HD, Tarr IJ, Weber CP, Brookes CP, Digby PGN (1985) Electrophoretic study of enzymes from cereal aphid populations. III- Spatial and temporal genetic variation of Sitobion avenae F. (Hemiptera: Aphididae). B Entomol Res 75: 121–141. [Google Scholar]
- 64. Lynch M (1984) Destabilizing hybridization, general-purpose genotypes and geographic parthenogenesis. Q Rev Biol 59: 257–290. [Google Scholar]
- 65. Wright S (1978) Evolution and genetic populations. University of Chicago Press, Chicago, London: University of Chicago Press 4: 79–103. [Google Scholar]
- 66. Whitlock MC (1992) Temporal fluctuations in demographic parameters and the genetic variance among populations. Evolution 46: 608–615. [DOI] [PubMed] [Google Scholar]
- 67. Klueken AM, Simon JC, Hondelmann P, Mieuzet L, Gilabert A (2011) Are primary woody hosts ‘island refuges’ for host-alternating aphids and important for colonization of local cereals? J Appl Entomol 136: 347–360. [Google Scholar]
- 68. Delmotte F, Leterme N, Gauthier JP, Rispe C, Simon JC (2002) Genetic architecture of sexual and asexual populations of the aphid Rhopalosiphum padi based on allozyme and microsatellite markers. Mol Ecol 11: 711–723. [DOI] [PubMed] [Google Scholar]
- 69. Huang XL, Liu DG, Gao SX, Chen H (2013) Differential performance of Sitobion avenae populations from both sides of the Qinling Mountains under common garden conditions. Environ Entomol 42: 1174–1183. [DOI] [PubMed] [Google Scholar]
- 70. Lavandero B, Miranda M, Ramirez CC, Fuentes-Contreras E (2009) Landscape composition modulates population genetic structure of Eriosoma lanigerum (Hausmann) on Malus domestica Borkh in central Chile. B Entomol Res 99: 97–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microsatellite genotypes of the seven Sitobion avenae populations in China. “individual ID”, the tag of each tested aphid individual. “sampling location”, the corresponding location where each aphid individual was collected. The other ten columns were the corresponding allele sizes of each aphid individual at each microsatellite loci.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.