Summary
RLIP76 (RalBP1) is a multidomain protein that interacts with multiple small G protein families: Ral via a specific binding domain and Rho and R-Ras via a GTPase activating domain. RLIP76 interacts with endocytosis proteins and has also been shown to behave as a membrane ATPase that transports chemotherapeutic agents from the cell. We have determined the structure of the Ral binding domain of RLIP76 and show that it comprises a coiled-coil motif. The structure of the RLIP76-RalB complex reveals a novel mode of binding compared to the structures of RalA complexed with the exocyst components Sec5 and Exo84. RLIP76 interacts with both nucleotide-sensitive regions of RalB and key residues in the interface have been identified using affinity measurements of RalB mutants. Sec5, Exo84 and RLIP76 bind Ral proteins competitively and with similar affinities in vitro.
Introduction
Cell migration is a normal, essential process but in cancer progression it is the basis for the ability of tumour cells to metastasize to new areas of the body, a process that leads to 90% of cancer deaths (Sporn, 1996). Approximately 50% of metastatic tumours contain mutations in the small G protein Ras and one of the three main effector pathways downstream of Ras is controlled by RalGEFs, which are exchange factors (and therefore activators) for another pair of small G proteins, RalA and RalB. Cells transformed with either an activated Ras variant (12V, 37G) that only interacts with the RalGEFs or with constitutively active RalGEF produced aggressive, infiltrative metastases when injected into mice. These were inhibited by dominant negative RalB (Ward et al., 2001), demonstrating that the RalGEF pathway alone is sufficient to induce a metastatic phenotype. In bladder cancer lines, EGF stimulation activates Ral and elevated levels of activated Ral are confined to metastatic cells (Gildea et al., 2002). It is therefore clear that the Ral pathway represents a potential target for the treatment of human metastatic cancers.
The Ral signalling pathway(s) responsible for conferring metastatic potential on cancer cells is less well defined. The effectors for the Ral GTPases regulate a wide variety of cellular functions and include phospholipase D (Jiang et al., 1995), the actin filament crosslinking protein filamin A (Ohta et al., 1999), the Y-box transcription factor ZONAB (ZO-1 associated nucleic acid binding protein) (Frankel et al., 2005), phospholipase C δ1 (Sidhu et al., 2005), two components of the exocyst complex, Sec5 and Exo84, (Moskalenko et al., 2002), (Sugihara et al., 2002), (Moskalenko et al., 2003) and RLIP76 (RalBP1/RIP1) (Jullien-Flores et al., 1995), (Cantor et al., 1995), (Park and Weinberg, 1995). RLIP76 is a multifunctional protein, containing a variety of domains and motifs (Figure 1A). Its RhoGAP domain acts on Rac1 and Cdc42, linking Ral with Rho family signalling (Jullien-Flores et al., 1995), (Cantor et al., 1995), (Park and Weinberg, 1995) and therefore control of the actin cytoskeleton and cell motility. RLIP76 is also involved in endocytosis and tyrosine kinase receptor signalling via its ability to bind to AP2 and POB1 through its N-terminal and C-terminal regions respectively (Yamaguchi et al., 1997), (Jullien-Flores et al., 2000).
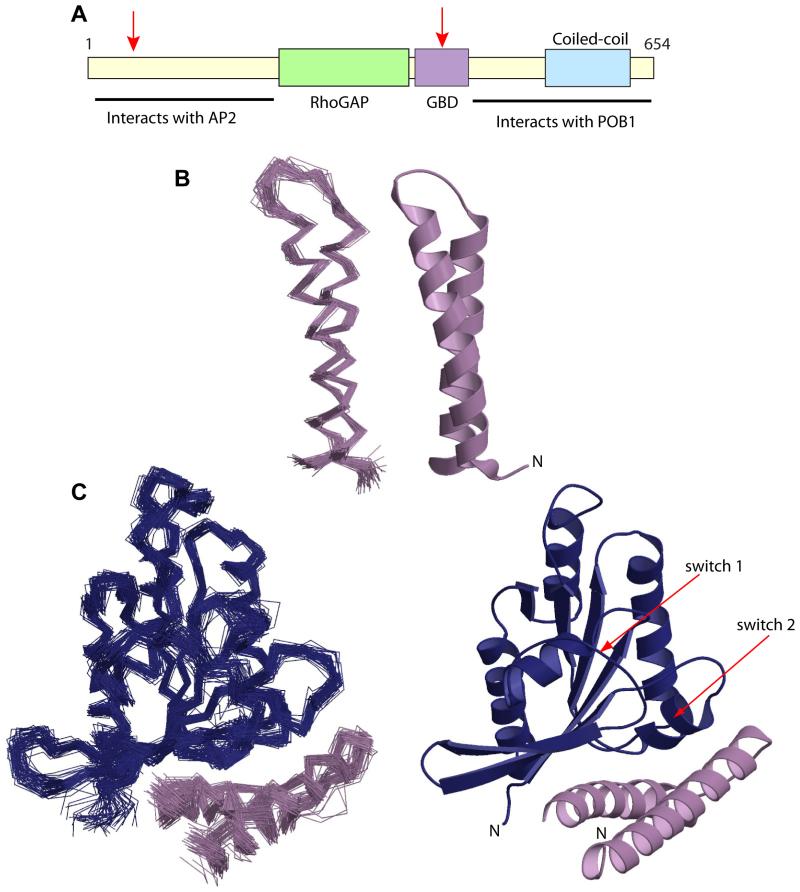
Figure 1. Structures of RLIP76 GBD alone and in complex with RalB.
A. Domain structure of RLIP76. The approximate positions of the RhoGAP domain, the Ral binding domain (GBD) and coiled-coil region are represented as coloured boxes, the regions found to interact with AP2 and POB1 are indicated and the locations of the putative ATP binding sites are marked with red arrows (Awasthi et al., 2001).
B. Structure of free RLIP76 GBD. On the left is the backbone trace of the family of structures consistent with the NMR restraints, on the right is the closest structure to the mean. All structure figures were produced using Molscript (Kraulis, 1991) and rendered with Raster3D (Merritt and Bacon, 1997).
C. Structure of the RLIP76 GBD-RalB complex. Ral is shown in blue and RLIP76 is lilac. On the left is the backbone trace of the lowest energy structures, on the right is the closest structure to the mean.
Although RLIP76 is thought to be primarily cytosolic it translocates to the membrane upon binding by Ral. RLIP76 contains two ATP binding sites (Awasthi et al., 2001), which allow it to function as an ATP-dependent transporter protein and efflux pump for small molecules, including anticancer drugs and endogenous metabolites (Awasthi et al., 2002).
We have solved the structure of the minimal Ral GTPase binding domain (GBD) of RLIP76 by NMR, both alone and in complex with RalB. This represents the first structural data available for RLIP76 and also reveals, for the first time, the conformation of RalB in complex with one of its effectors. The RLIP76 GBD is a coiled-coil, which although identified as a binding motif for other small G protein families (e.g. (Modha et al., 2008), (Panic et al., 2003)), is unique for an effector of the Ras family of small G proteins. Comparison of the free and bound structures of RalB and RLIP76 GBD shows that there are small changes in the orientation of the RLIP GBD α-helices and in the loop between them. The regions of RalB that are involved in the interaction with RLIP76 GBD include both the switch regions that change conformation on nucleotide exchange, however 31P NMR experiments reveal that there is still residual dynamics in switch 1 after complex formation. We have used the structure to design mutants to investigate the thermodynamics of the binding interface between RalB and RLIP76. This combination of structural and thermodynamic information could be used to assist rational drug design of therapeutics directed towards disrupting the RalB-RLIP76 complex.
Results and Discussion
Structure of the RLIP76 GBD
The quality of the NMR data of wild-type RLIP76 GBD protein, comprising residues 393-446, suggested that the domain was partially dimerized in solution and this was confirmed by analytical gel filtration experiments. Electrospray ionization mass spectrometric analysis in the presence and absence of the reducing agent tris(2-carboxyethyl)phosphine (TCEP) revealed that dimerization was the result of disulphide bond formation (data not shown). The single Cys, residue 411, in the RLIP76 GBD was mutated to Ser and the resulting protein was judged to be fully monomeric in solution by analytical gel filtration, mass spectrometry and analytical ultracentrifugation. Analytical gel filtration analyses were also used to confirm that the mutation of Cys-411 to Ala did not affect the ability of the RLIP GBD to complex with RalB (data not shown).
The RLIP76 GBD backbone resonances were assigned as previously described (Fenwick et al., 2008b). Distance restraints were generated from a 3D 15N-separated NOESY and from a 2D 1H NOESY recorded on unlabelled RLIP76. A total of 998 distance restraints were used in the first round of structure calculation, of which 575 were unambiguous and 423 were ambiguous. After 8 rounds of structure calculation, where at each round the ambiguity of the restraints was reduced, there were 737 unambiguous and 560 ambiguous restraints. 39 hydrogen bond restraints were included for residues whose backbone amides were undergoing negligible exchange with the solvent in a CLEANEX series of experiments (Hwang et al., 1998) and whose NOE patterns indicated that they were within an α-helix. The final family of 50 structures (Figure 1B) shows that the RLIP76 GBD forms an anti-parallel coiled-coil, comprising two α-helices (α1RLIP76 and α2RLIP76) that are each of ~20 amino acid residues in length (395-415 and 423-442). The coiled-coil is held together by hydrophobic interactions between aliphatic residues: Leu, Ile and a single Val sidechain comprise the core of the coil.
A coiled-coil motif is used by several effector proteins for binding members of various families of the small G protein superfamily, such as the Rho family: HR1 domains that bind to RhoA and Rac1 (Maesaki et al., 1999), (Modha et al., 2008); the Arf family: the GRIP domains that bind to Arl1 (Panic et al., 2003), (Wu et al., 2004) and the Rab family: Rab-binding proteins such as Rabenosyn-5 (Eathiraj et al., 2005). The RLIP76 GBD structure, however, represents the first structural information obtained for a coiled-coil motif that binds to a Ral protein and indeed to any member of the Ras family. Although it is perhaps not surprising however, given that coiled-coil domains have been found to bind to all of the other small G protein families, with the exception of Ran, that a coiled-coil would also be found in a Ras family effector.
Structure of the RLIP76 GBD-RalB complex
The backbone resonances of the RLIP76 GBD and RalB in the complex were assigned as previously described (Fenwick et al., 2008a; Fenwick et al., 2008b). All RLIP76 backbone atoms were assigned in the complex, with the exception of the 3 residues at the extreme N-terminus (393-395), and the sidechain assignments were essentially complete. RalB backbone atoms were fully assigned with the exception of Glu-44 and Thr-46 in switch 1: the resonances for these atoms were also missing or weak in the spectra of free RalB-GMPPNP (Prasannan et al., 2007). All sidechains were assigned in RalB, although Lys-27, Glu-44, Asp-49, Ser-50 and Tyr-51 were not complete. Hydrogen bond restraints were included for RLIP76, as above, and pairs of dihedral angle restraints, which were calculated using the TALOS program (Cornilescu et al., 1999) based on the experimentally determined backbone chemical shifts for both proteins (Table 1). Distance restraints were obtained from 15N-separated and 13C-separated NOESY experiments, recorded on appropriately labelled samples. These were translated using ARIA into 3,547 unique unambiguous restraints and 2,200 ambiguous restraints. Of these there were 45 unambiguously assigned distance restraints between the two components of the complex. After 8 rounds of structure calculation, where the ambiguity of the restraints was reduced at each round, there were 4,461 unambiguous restraints and 1,408 ambiguous distance restraints.
The RLIP76 GBD binds to one face of RalB in such a way that it contacts the two main regions that change conformation when the G protein is activated by GDP exchanging for GTP (Figure 1C). These regions, known as switch 1 (Glu-41RalB – Tyr-51RalB) and switch 2 (Thr-69RalB – Asn-81RalB), are generally involved in interactions with G protein effectors since their molecular topography is sensitive to the nucleotide status of the G protein.
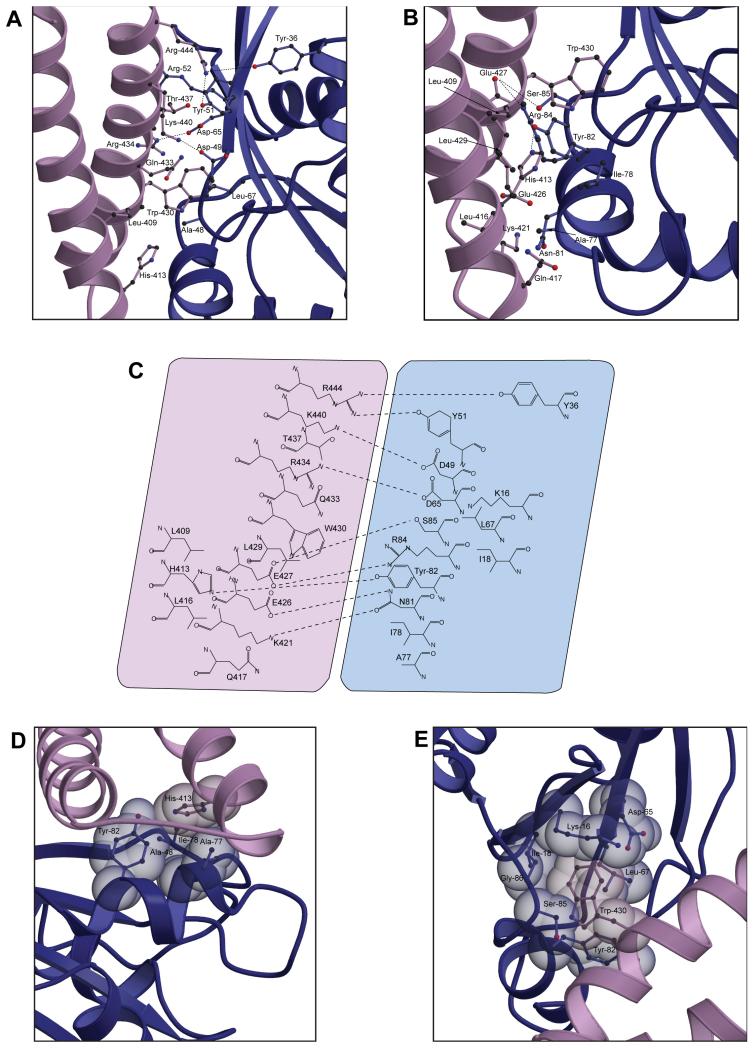
The N-terminal region of switch 1 (Glu-41RalB to Lys-47RalB) forms a flexible loop that includes the conserved residue Thr-46 (equivalent to Thr-35 in Ras). Thr-46 forms a hydrogen bond with the γ-phosphate of GTP, contributing to the conformational change that ensues when the nucleotide is exchanged. The C-terminal amino acids of switch 1, residues 48-52RalB, make significant interactions with RLIP76, whereas the N-terminus of switch 1, including Thr-46RalB, does not interact with RLIP76 at all. Ser-50RalB to Arg-52RalB comprises the beginning of the second β-strand in RalB (β2RalB) and contacts residues in α2RLIP76 (Figure 2A, C). In particular, intermolecular NOEs were observed between the amide of Ser-50RalB and the sidechains of Thr-437RLIP76 and Gln-433RLIP76 and also between the sidechains of Thr-437RLIP76 and Tyr-51RalB. Leu-67RalB, which is in the region between the switches, also contacts α2RLIP76 (Figure 2A), predominantly via interactions with Trp-430RLIP76 (Figure 2E).
Figure 2. Details of the interactions between RalB and the RLIP76 GBD.
A. Interactions involving the RalB switch 1 and interswitch regions. RalB is shown in blue and the RLIP76 GBD is in lilac. The sidechains are shown in a ball-and-stick representation with sticks in the same colours as the ribbon for each molecule and the atoms coloured as follows: carbon, dark grey; oxygen, red; nitrogen blue.
B. Interactions involving the RalB switch 2. The colour scheme is the same as in part (a).
C. Summary of all interactions between the proteins. Putative hydrogen bonds and salt bridges are shown as dotted lines between the participating atoms. RalB is shown in blue and RLIP76 is shown in lilac.
D. Interactions involving His-413RLIP76. Sidechains are shown in a spacefilling representation superimposed with a ball-and-stick. The colour scheme is the same as in part (a).
E. Trp-430 of RLIP76 is surrounded by a cage of RalB sidechains. The colour scheme is the same as in part (a).
Interactions between RLIP76 and switch 2 of RalB are more extensive than those of switch 1 and involve most of switch 2 interacting with both α-helices of RLIP76. The N-terminus of switch 2, comprising residues Asp-74RalB, Tyr-75RalB, Ala-76RalB, Ala-77RalB and Ile-78RalB, interacts with α1RLIP76 (Figure 2B, C) and multiple intermolecular contacts were observed between these residues and His-413RLIP76, Leu-416RLIP76 and Gln-417RLIP76. Asn-81RalB and Tyr-82RalB at the C-terminal end of switch 2 contact residues in both α-helices of RLIP76, while residues just C-terminal to switch 2, Arg-84RalB and Ser-85RalB, interact almost exclusively with α2RLIP76 (Figure 2B, C, D).
Comparison of free and bound structures
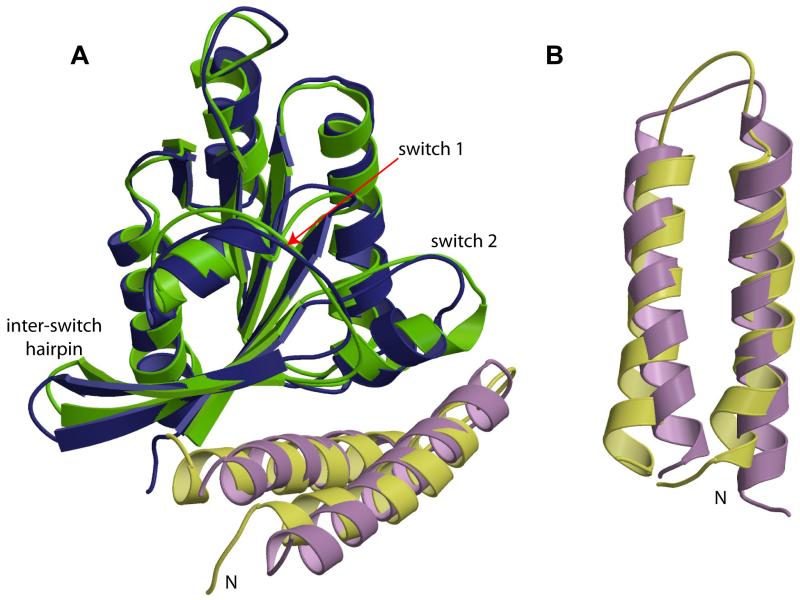
A comparison of the structure of the RLIP76 GBD in its free and RalB-bound form (Figure 3A) reveals that the overall topology of the GBD does not change. There are however, minor changes in the two α-helices that comprise the GBD (Figure 3B). The helices are slightly different lengths: 395-414 and 424-444 in the free GBD, compared to 393-418 and 423-444 in the complex (averaged over the ensemble of structures, using Procheck-NMR (Laskowski et al., 1993)). The slight differences at the extreme N- and C-termini of the GBD are unlikely to be significant, since there are few restraints tying down these regions of the structure. The N-terminal α-helix is however one turn longer at its C-terminal end in the complex than in the free GBD, finishing at Arg-414 in the free GBD and Gly-418 in the complex. The differences in the end of this helix and in the inter-helix loop are due to the interaction with RalB: there are unambiguous intermolecular NOEs locking His-413 and Gln-417 against switch 2 of RalB. The consequence of these interactions is that the sidechains 413 and 417 must face the same way and the logical solution to this is to extend the α-helix. The shorter α-helix in the free GBD results in a somewhat longer inter-helix loop, which is less well-defined in the family of structures and thus presumably more mobile. The overall inter-helical angle in the GBD is approximately the same, around −155 to −160°, although some reorientation of the helices is evident (Figure 3). This slight reorientation does not change the overall sidechain packing between the helices.
Figure 3. Comparison of free RalB·GMPPNP (pdb code 2KE5) and RLIP76 GBD with their structures in the complex. The closest structure to the mean is shown in each case.
A. The RalB-RLIP76 complex with the contributing free proteins overlaid. RalB is shown in blue (complex) and green (free), RLIP76 GBD is shown in lilac (complex) and yellow (free). The regions of greatest divergence in RalB, the switch regions and the interswitch hairpin, are labelled.
B. The RLIP76 GBD alone, in the structures adopted in the free and complex forms, in an orientation that shows the small changes in the lengths and orientations of the α-helices. The structure of free RLIP76 GBD is yellow and that in the complex is lilac.
Comparison of the RalB structure with that of the published free RalB structure (Fenwick et al., 2009) shows that the overall fold is, as expected, very similar. The changes are confined to the two switch regions and the β-hairpin between the switches (Figure 3A), which are the main points of contact with RLIP76.
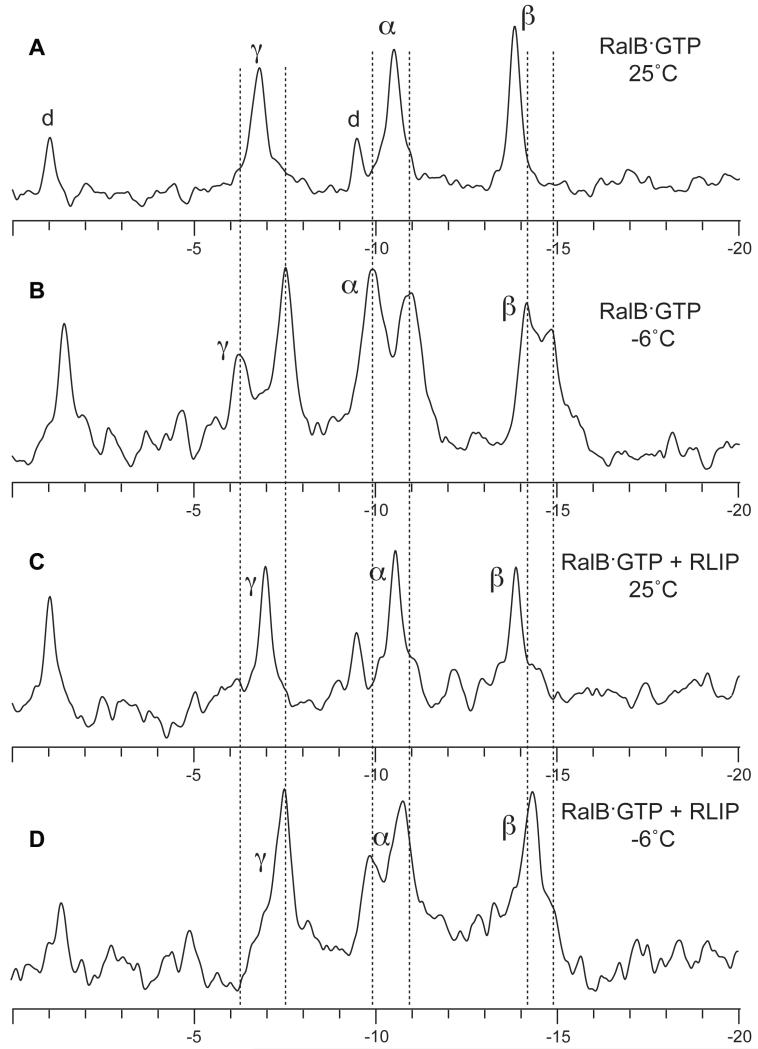
As mentioned above, some resonances for residues within switch I, including those for Thr-46, were missing in both the free and the complex RalB·GMPPNP NMR spectra, presumably due to conformational exchange. It has been observed previously that 31P NMR spectra of small G proteins, including Ha-Ras {Geyer, 1996 #1364}, at low temperatures, reveal that there are two sets of NMR signals for each phosphate group in the GTP. These have been suggested to be due to the existence of two states (known as state 1 and state 2) whose interconversion is slow enough at low temperature to be visible by NMR. The first of these states is unable to bind to effectors, whereas state 2 is competent for effector binding. The addition of an excess of the effector Raf led to a single set of peaks for Ha-Ras, presumably due to the stabilization of state 2 (Geyer et al., 1996). When similar experiments were performed on RalB·GTP, we found that there were also two states apparent at low temperatures, but that in contrast to the situation with Ha-Ras, the addition of an excess of Sec5 only led to partial stabilization of state 2 (Fenwick et al., 2009) and that state 1 was still visible. We have investigated the effect of binding of RLIP76 GBD to RalB·GTP on the 31P NMR spectra. As expected from previous work, the free RalB·GTP spectrum exhibited a splitting of phosphate signals into state 1 (−9.94 ppm) and state 2 (−10.93 ppm) when the temperature was lowered from 25°C to −6°C (compare Figure 4A and Figure 4B). When RLIP76 was added in a 1.5-fold excess, we found that, as for Sec5, the splitting of 31P resonances persisted at low temperatures (Figure 4D). The α resonance is split into two discrete components and the β resonance has a distinct shoulder at ~15 ppm.
Figure 4. 31P NMR spectra recorded on RalB·GTP.
A. RalB·GTP recorded at 25°C. The phosphorus resonances for the GTP attached to RalB are labelled and were assigned as described previously (Fenwick et al., 2009). The resonances labeled ‘d’ are likely to be due to small amounts of contaminating GDP in the sample {Geyer, 1996 #1364}. B. The same sample recorded at −6°C. Each of the three phosphorus resonances is split into two at low temperatures and their positions are marked by dotted lines. Note that the GDP peak that is close to the α resonance contributes to the state 1 component of the α resonance at −9.94 ppm at low temperatures. This has the effect of making the state 1 component larger than the state 2 component (−10.93 ppm), whereas previously the state 2 component was larger {Fenwick, 2009 #1345}. C. The spectrum of RalB·GTP recorded in the presence of excess RLIP76 GBD at 25°C. D. The same sample recorded at −6°C. The splitting is still visible for the α and β resonances. Although the GDP peak is again contributing to the state 1 component of the α resonance, it cannot account for all of the intensity of the state 1 peak, since the GDP peak at −9.5 ppm is of a lower intensity than the peak at −1.0 ppm in this sample.
It has been proposed that the two states observed in Ha-Ras are due to the effects of the Tyr-32 ring (equivalent to Tyr-43 in RalB), which points towards the β-phosphate in the more active state 2 and away from the β-phosphate in the less active state 1. In the RalB-RLIP76 complex, Tyr-43 is not well-defined, although its sidechain points away from the nucleotide. The dynamic nature of this sidechain in the complex is consistent with the splitting in the 31P NMR spectrum even in the presence of RLIP76. It is noteworthy that the splittings observed for the α resonance are the most pronounced, even though it is the β-phosphate that would be most affected by the orientation of the Tyr-43 sidechain. This implies that the position of Tyr-43 may not be the only factor that causes the splitting observed for the phosphate groups in RalB·GTP.
It has been suggested that the preference for state 1 in M-Ras results in rather low effector affinities (Ye et al., 2005). In the case of RalB·GTP, the interaction with Sec5 and with RLIP76 are still of a high affinity: 150 nM (Fenwick et al., 2009) and 184 nM (see below) respectively even though the 31P spectra show that both states are present in solution in the presence of excess Sec5 or RLIP76. This suggests that for RalB·GTP at least, both states are competent for binding to effectors.
Interactions conserved in evolution are important for affinity
The RLIP76 GBD, in common with most coiled-coil domains, is held together mainly through hydrophobic interactions between the two α-helices. The residues involved in these interactions tend to have aliphatic sidechains, such as leucine and isoleucine. It is notable that this domain has just three aromatic residues, Phe-407, His-413 and Trp-430, and that all of them are solvent-exposed in the free GBD structure. Furthermore, His-413RLIP76 and Trp-430RLIP76 are involved in multiple interactions with RalB in the complex, mainly with residues in switch 2. His-413RLIP76 forms a hydrogen bond with Tyr-82RalB as well as interacting with Ala-48RalB, Ala-77RalB and Ile-78RalB although it is not completely buried in the complex (Figure 2C, D). Trp-430RLIP76 on the other hand is completely buried when RLIP76 forms a complex with RalB and is caged by the sidechains of multiple residues in switch 2 and in β1RalB (Figure 2E). The hydrophobic sidechains of Leu-67RalB and Ile-18RalB lie on either side of the Trp-430RLIP76 ring, while the top and bottom of the ring is capped by the hydrophobic portions of Lys-16RalB and Tyr-82RalB.
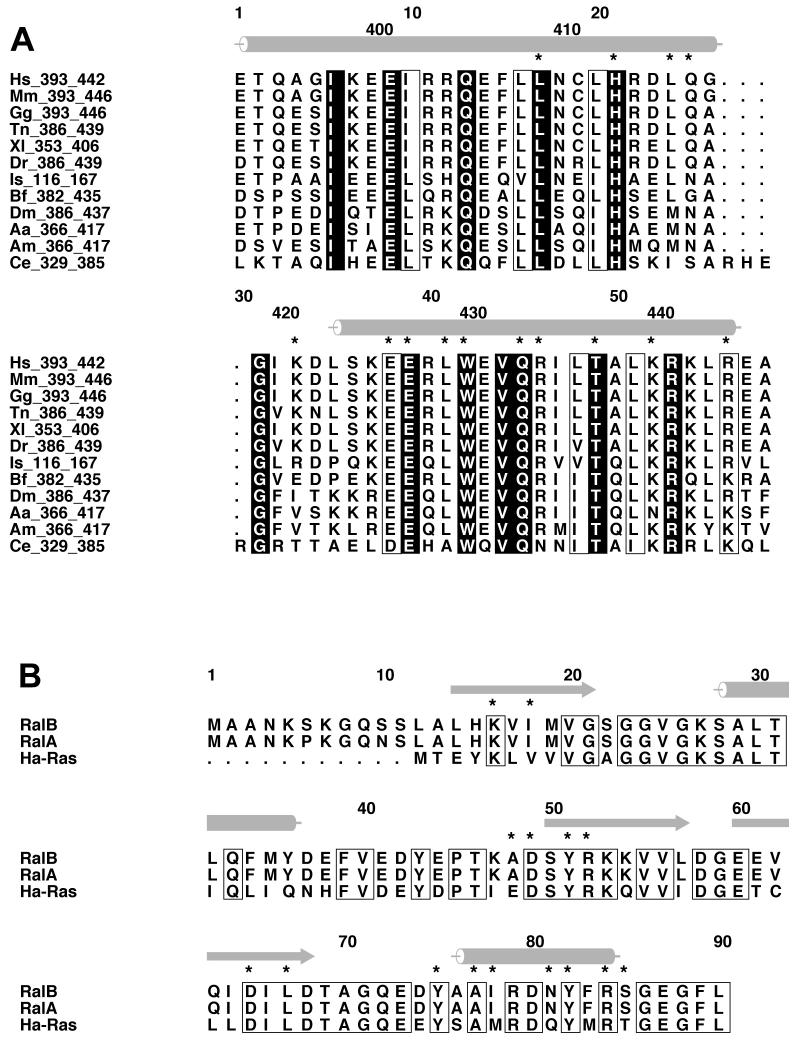
Both His-413 and Trp-430 are completely conserved within RLIP76 orthologues in a range of organisms from C. elegans to man (Figure 5A). The Ral proteins are also highly conserved between these organisms and within the RLIP76-interacting regions are 100% identical (Figure 5B). Overall this suggests that the interactions involving these sidechains will also be conserved.
Figure 5. Sequence alignments.
A. Alignment of RLIP76 GBD from different organisms. The positions of α-helices in the structure are shown as grey cylinders on the top of the alignment. Residues that are conserved in all sequences are coloured white on a black background. Residues whose properties are conserved are boxed. The stars above the sequences mark the position of residues that interact with RalB.
Hs – Homo sapiens; Mm – Mus musculus; Gg – Gallus gallus; Tn –Tetraodon nigroviridis; Xl – Xenopus laevis; Dr – Danio rerio; Is – Ixodes scapularis; Bf – Branchiostoma floridae; Dm – Drosophila melanogaster; Aa – Aedes aegypti; Am – Apis mellifera; Ce – Caenorhabditis elegans.
B. Alignment of the N-terminal regions of RalA, RalB and Ha-Ras. The positions of α-helices in the Ral B structure are shown as grey cylinders and the β-strands as grey arrows, on the top of the alignment. Residues that are conserved between all three sequences are boxed. The stars above the sequences indicated the residues that interact with RLIP76 GBD.
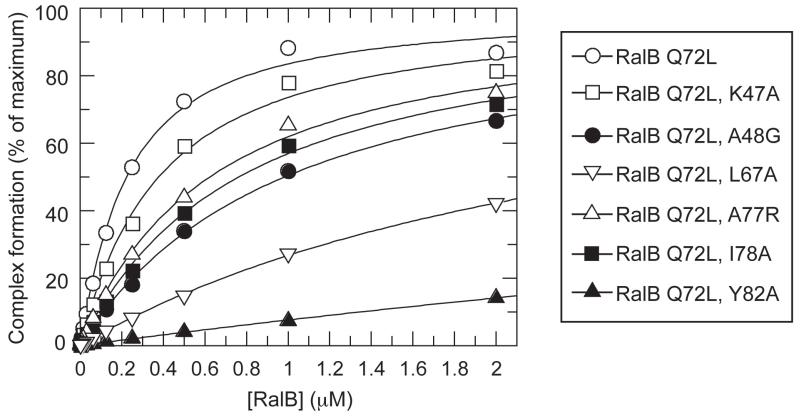
We set out to test the importance of some of these interactions by making a series of point mutations in RalB where the residue of interest was changed to alanine. All mutants were constructed in a background of Q72L RalB. This mutation decreases the GTP hydrolysis rate, allowing the GTP-bound form to be stable during the course of the experiment. Using scintillation proximity assays (SPA) with Q72L RalB-GTP and C-terminally His-tagged RLIP76 GBD, a Kd of 184 nM was obtained (Figure 6). Some of the residues that contact His-413 can be mutated with relatively modest effects on the affinity of the interaction e.g. I78A and A77R bind with a Kd of 724nM and 585 nM respectively, equivalent to only a 4.0-fold and 3.2-fold decrease in affinity respectively. Mutation of other residues however has a more dramatic effect. Tyr-82RalB, which forms a hydrogen bond with His-413RLIP76, is also involved in hydrophobic contacts with Trp-430RLIP76 (Figure 2C, D, E). Mutation of Tyr-82RalB to Ala reduced the binding to RLIP76 significantly, increasing the Kd to >1μM thus increasing ΔG by at least 0.9 kcal/mol. This interaction therefore contributes significantly to the binding energy of the complex (ΔG Q72L RalB-RLIP76 is −9 kcal/mol). Leu-67RalB also makes hydrophobic interactions with Trp-430 (Figure 2E) and the L67A mutant also increases the Kd to >1 μM. Overall, these mutational data suggest that contacts involving the Trp-430RLIP76 sidechain are important for the affinity of the interaction.
Figure 6. Measurement of the affinities of Q72L RalB and selected mutants for the RLIP76 GBD.
The indicated concentration of [3H] GTP-labelled RalB were incubated with 80nM RLIP76 GBD-His in SPAs. The SPA signal was corrected by subtraction of a blank from which RLIP76 GBD-His was omitted. The effect of RalB on this corrected SPA counts/min signal was fitted to a binding isotherm to give an apparent Kd value and the signal at saturating concentrations of RalB. The data are expressed as a percentage of this maximum signal. The calculated Kds were: RalB Q72L, 183.9 ± 19.9nM; RalB Q72L K47A, 339.5 ± 34.7nM; RalB Q72L A48G, 945.5 ± 60.7; RalB Q72L L67A, >1μM; RalB Q72L A77R, 585.0 ± 50.2 nM; RalB Q72L I78A, 724.2 ± 55.1nM; RalB Q72L Y82A, >1μM.
Discrimination between Ral and Ras effectors
The structure presented here is the first example of a complex involving RalB, one of the two human variants of Ral. Comparison of the structures of RalA-GMPPNP (Nicely et al., 2004) and RalB-GMPPNP (Fenwick et al., 2009) shows that they are broadly similar. The residues that interact with RLIP76 are identical in RalA and RalB (Figure 5B) and this conservation is reflected in a comparable in vitro affinity of both G proteins for this effector (data not shown and (Bauer et al., 1999)). We observed similar parity in vitro with RalA and RalB for the effector Sec5 (Fenwick et al., 2009). Most of the Ral residues that interact with RLIP76 are also identical in Ha-Ras and the switch regions in particular are well conserved between the G proteins (Figure 5B). Ha-Ras does not, however, bind to RLIP76 and Ral does not bind to Ras effector proteins (Jullien-Flores et al., 1995), (Cantor et al., 1995), (Park and Weinberg, 1995). A binding analysis of RalA and Ha-Ras combined with mutagenesis of both proteins pinpointed two residues in switch 1, Lys-47 and Ala-48 (Ral numbering), as important for discrimination between Ras and Ral effectors (Bauer et al., 1999). These residues are replaced by Ile-36 and Glu-37 respectively in Ras and represent the most drastic changes in sequence within the switches. Essentially this region is positively charged in Ral proteins and negatively charged in Ras. The RalB K47I mutant showed little change in its ability to bind to RLIP76, A48E binding was compromised and the double K47I/A48E mutant showed no binding at all to RLIP76 by yeast two-hybrid and GST-pulldown assays (Bauer et al., 1999). Our SPA data are in broad agreement with these results, with the A48G mutant showing a 5-fold decrease in affinity and the K47A mutant having little effect (Figure 6). Ala-48RalB is in the interface and packs against Leu-409RLIP76 and His-413RLIP76 (Figure 2A,D): its replacement in Ras with the negatively charged Glu would cause a rearrangement of this packing and the loss of hydrophobic interactions. Lys-47RalB does not interact directly with RLIP76: it points away from the interface towards Glu-44RalB, which could pin Lys-47RalB away from RLIP76 by forming a salt bridge. Lys-47RalB replacement with an Ile residue would remove any potential interaction with Glu-44RalB and allow rearrangement of this loop in RalB in the double K47I/A48E mutant. The importance of these residues to the affinity for RLIP76 is underlined by the observation that the Ras double mutant I36K/E37A (i.e. to the equivalent residues) is able to bind RLIP76 (Bauer et al., 1999). Lys-47 is also important in the interaction between RalA and the exocyst component Exo84 and is proposed to be a specificity determinant for binding to this effector (Jin et al., 2005).
Comparison with RalA-Exocyst Component Complexes
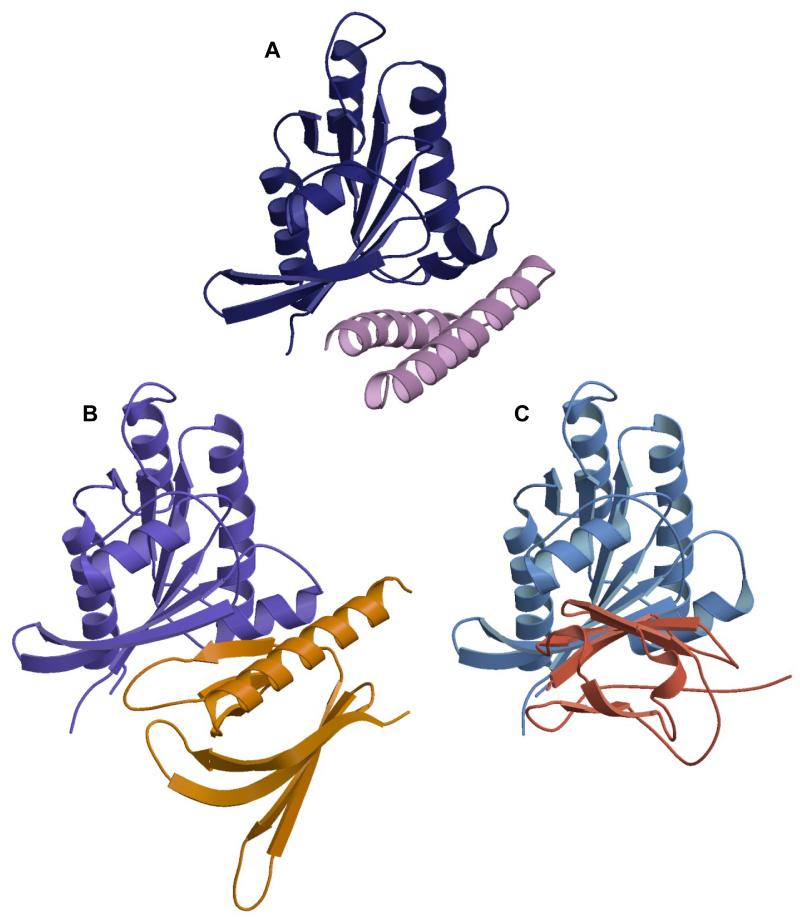
RalA structures have been solved in complex with two different components of the exocyst complex, Sec5 and Exo84. A comparison of the three Ral complex structures reveals that the effectors themselves are all strikingly different (Figure 7). Sec5 GBD has an all β-sheet, Ig-like fold, which forms an intermolecular antiparallel β-sheet with β2RalA (Figure 7C) interacting exclusively with residues in and around switch 1 and burying ~1000 Å2 in the interface (Fukai et al., 2003). In contrast, the Exo84 GBD is a PH domain (Figure 7B) that interacts with both switch 1 and switch 2 of RalA (Jin et al., 2005). Exo84 does not form an intermolecular β-sheet with RalA and buries ~1700 Å2 when it binds. The RLIP76 GBD-RalB interaction also interacts with both switch 1 and switch 2 and buries ~1700 Å2. In this way, it would appear to be more similar to the Exo84 interaction. The Exo84 PH domain contains a single α-helix, which, although it makes some contacts with RalA, is not the sole determinant in binding since Exo84 also utilises a β-strand in the binding interface. The 5th β-strand of Exo84 interacts with β2RalA, in a manner that resembles an intermolecular parallel β-sheet, although it is only held together by three hydrogen bonds, between Ala-48/Ser-50RalA and Asn-231/Lys-233Exo84 (Jin et al., 2005). The long α-helix at the C-terminus of the PH domain is in approximately the same orientation as the N-terminal α-helix of the RLIP76 GBD (Figure 7A, B). The two helices are however in slightly different positions and this is reflected in the interactions that they make: the Exo84 PH domain helix contacts residues further towards the N-terminus of Ral compared to those contacted by the RLIP76 GBD.
Figure 7. Comparison of structures of Ral-effector complexes shows that they bind to overlapping interfaces of the G protein.
A. RalB-RLIP76: RLIP76 uses a coiled-coil and binds to both switch 1 and switch 2. RalB is blue and RLIP76 is lilac
B. RalA-Exo84: Exo84 uses a PH domain and binds to both switch 1 and switch 2. RalA is purple and Exo84 is orange.
C. RalA-Sec5: Sec5 uses an immunoglobulin-like domain and binds exclusively to switch 1 and the interswitch region. RalA is blue and Sec5 is red.
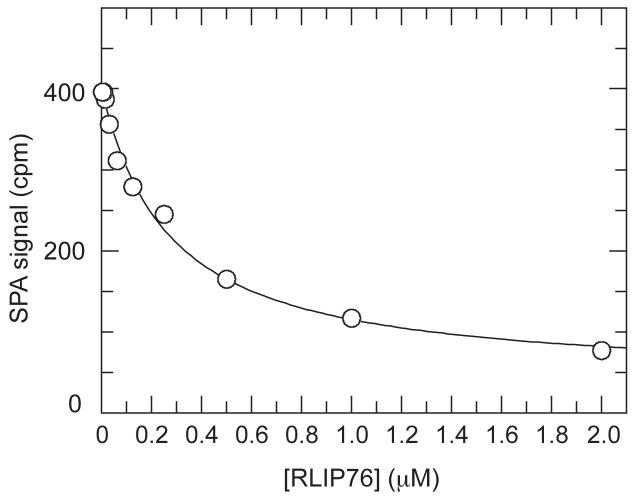
The binding sites for Exo84 and Sec5 were considered to be partially overlapping because three residues in RalA, Ala-58, Ser-50 and Arg-52 are shared in both binding interfaces. These three residues are also in the binding interface with RLIP76 and it is not surprising therefore that the Sec5 GBD and RLIP76 GBD bind competitively (Figure 8). In addition to these three interactions, the RLIP76 interface shares several residues with the Exo84 interface: Lys-16, Asp-65, Ile-78, Asn-81 and Tyr-82 and it is thus likely that the binding of these two effectors will also be mutually exclusive.
Figure 8. Displacement of [3H]GTP·RalB from GST-Sec5 by RLIP76.
Increasing concentrations of RLIP76 GBD were titrated into fixed concentrations of [3H]GTP·RalB (20nM) and GST-Sec5 GBD (20nM) in competition SPAs. The fit of the inhibition of the [3H]GTP·RalB/GST-Sec5 GBD interaction is shown and yields a Kd of 199.4 ± 33.3 nM. This is the same as the Kd measured by direct binding (Figure 6).
Functional Consequences of the RLIP76-Ral Interaction
RLIP76 has several domains apart from the GBD, which have been ascribed different functions. The best-characterized sequence is the GAP domain, which is just N-terminal to the GBD in the RLIP76 sequence. RLIP76 appears to behave as a GAP towards both Cdc42 and Rac1, albeit with low activity (Cantor et al., 1995), (Jullien-Flores et al., 1995), (Park and Weinberg, 1995), but its activity towards other Rho family proteins has not been systematically investigated. As the GAP domain and the GBD are juxtaposed, it was an attractive possibility that Ral binding would modulate the GAP activity in vivo but it seems that Ral has little effect on RLIP76 RhoGAP activity in vitro (Park and Weinberg, 1995).
It has however recently been shown that phosphorylation of RalA Ser194 by Aurora-A leads to its translocation from the plasma membrane to endosomes and increased interaction with RLIP76. This localizes RLIP76 to the same internal compartments and results in a concomitant loss of filopodia and lamellipodia, presumably because Cdc42 and Rac1 activity is decreased (Lim et al., 2010). These results have two possible explanations: either localizing RLIP76 to the correct compartment brings it into contact with its substrates Rac1/Cdc42 or RalA does have a direct affect on modulating the GAP activity of RLIP76. RalA·GTP phosphorylated at Ser194 co-immunoprecipitates RLIP76 better than GTP-loaded RalA that is not phosphorylated, suggesting that the RalA C-terminus may be involved in the RLIP76 interaction in vivo. However analytical gel filtration analyses we performed on full-length and C-terminally truncated RalB show that both readily form complexes with RLIP76 (data not shown), implying that the C-terminus of RalB does not affect the interaction with RLIP76, at least in vitro. Furthermore, Ser194 is not conserved in RalB, suggesting that the two Ral isoforms may differ in the nature of their interaction with RLIP76 in this region. The C-terminal polybasic region of Rac1 has been shown to contribute to interaction with the coiled-coil HR1b domain of the effector PRK1 (Modha et al., 2008) so there is a precedent for the C-terminus of small G proteins to be involved in effector binding.
RLIP76 has also been shown to bind to ATP, which is necessary for its function as a xenobiotic transporter (Awasthi et al., 2001). One ATP motif is within the RLIP76 GBD (418GGIKDLSK425) and encompasses the loop between the two helices. It is likely however that more than the simple coiled-coil that comprises the minimal GBD would be required to form an ATP binding site. It is therefore not surprising that we have been unable to detect ATP binding to the RLIP76 GBD either alone or in the presence of RalB (data not shown).
Given the importance of Ral and RLIP76 in diseases such as cancer, it is crucial to determine the precise effects of Ral binding on these two major functions of RLIP76. This knowledge will depend on further structural and biochemical investigations on the remainder of the RLIP76 protein. This first insight into the Ral/RLIP76 interaction at a molecular level is a starting point to elucidate the functions of these two proteins in disease progression.
Experimental Procedures
Protein Expression and Purification
Human RalB residues 1-185, containing the activating mutation Q72L (henceforth referred to as RalB) was cloned into pET16b (Novagen) and expressed in E. coli BL21(DE3) (Invitrogen). Unlabelled and uniformly labelled RalB proteins incorporating 15N or 15N and 13C were expressed and purified as previously described (Prasannan et al., 2007). The protein was concentrated to ~0.6 mM and the bound nucleotide exchanged for GTP or its non-hydrolysable analogue GMPPNP (Sigma) as described previously (Fenwick et al., 2009), (Thompson et al., 1998). Presence of the bound nucleotide was confirmed by HPLC analysis.
The GBD of human RLIP76 (393-446) was amplified by PCR and cloned into pGEX-4T3 (Invitrogen) using BamHI and XhoI restriction sites that had been incorporated into the PCR primers. The C411S mutation was introduced into the RLIP76 GBD expression construct by site-directed mutagenesis using the QuikChange Multi Site Directed Mutagenesis Kit (Stratagene) following the manufacturer’s instructions. The construct was expressed in E. coli BL21 (Invitrogen). The unlabelled and uniformly labelled RLIP76 GBD proteins were produced as previously described (Fenwick et al., 2008b).
The GBD of human RLIP76 (393-446) was also cloned into a modified version of pGEX-His-2 (Strugnell et al., 1997). A thrombin cleavage site was engineered into pGEX-His-2, 5′ to the BamHI cloning site. RLIP76 (393-446) was amplified by PCR and cloned into modified pGEX-His-2 using BamHI and XhoI restriction sites that had been incorporated into the PCR primers. The resulting construct expressed GST-RLIP76 GBD with a C-terminal His tag. The C411S mutation was introduced as previously described. The construct was expressed in E. coli BL21 (Invitrogen). A 50mL overnight culture of the construct was diluted into 500mL 2TY, grown to an A600 of ~0.8, induced with 0.1mM IPTG and grown for a further 5hr. Cells were lysed and the fusion protein purified using glutathione agarose (Sigma-Aldrich) following manufacturer’s instructions. The fusion protein was cleaved with thrombin to remove the GST tag and further purified by gel filtration (S30 16/60, GE Healthcare). This protein was then used directly in SPAs.
Mutations were introduced, as specified, into the coding region of RalB using the QuikChange Multi Site Directed Mutagenesis Kit (Stratagene) following manufacturer’s instructions. The sequences of the coding regions of all mutants were verified using an automated DNA sequencer (Applied Biosystems Inc.) by the DNA Sequencing Facility, Department of Biochemistry, University of Cambridge. Proteins were expressed and purified as previously described (Prasannan et al., 2007).
NMR Spectroscopy
The sample used to determine the structure of the free RLIP76 GBD C411S contained 0.8 mM 15N labelled protein in 50 mM sodium phosphate, pH 6.3, 100 mM NaCl, 1 mM MgCl2, 10 mM 2-mercaptoethanol, 0.05% NaN3 and 10% (v/v) D2O. Experiments on free RLIP76 were recorded on a Bruker DRX500 at 25 °C and included; 2D 15N HSQC, 3D 15N-separated NOESY (mixing time 130 ms), 3D 15N-separated TOCSY, 2D NOESY (mixing time 130 ms), 2D TOCSY, and 2D DQF-COSY.
Mixed samples of the RalB-RLIP76 GBD C411S complex were prepared containing one labelled and one unlabelled protein. The samples contained a 10% excess of the unlabelled component, which was sufficient to ensure saturation of the labelled component. NMR samples contained 50 mM sodium phosphate, pH 7.6 or pH 6.8, 100 mM NaCl, 1 mM MgCl2, 0.05 % NaN3 and 10% (v/v) D2O. Two pH conditions were used to optimize the NMR spectra for the labelled component in the complex. The affinities were measured at the different conditions and they were not affected by the pH change (data not shown). All experiments were recorded on Bruker DRX spectrometers at 25 °C. The following experiments were recorded on the labelled RalB (reviewed in (Cavanagh et al., 2007)): 2D 15N HSQC, 3D HNCA, HNCO, HN(CO)CA, HNCACB, CBCA(CO)NH, (H)C(CCO)NH, 2D 13C HSQC, 2D methyl-selective CT HSQC, HC(C)H TOCSY, H(C)CMeHMe TOCSY HMeCMeCγβα, (H)CCMeHMe TOCSY (D.N. unpublished). The experiments recorded on the labelled RLIP76 GBD C411S were; 2D 15N HSQC, 3D HNCA, HNCO, HN(CO)CA, HNCACB, 13C HSQC, 3D (H)C(CCO)NH, HBHA(CBCACO)NH and HC(C)H TOCSY. For NOE restraint generation 15N-separated NOESY, 13C-separated NOESY and 13C-filtered, 13C-separated NOESY experiments were recorded on each sample on a Bruker DRX800. The samples for 31P NMR experiments contained 0.3 mM RalB·GTP in 10 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM MgCl2, 0.05 % NaN3 and 10% (v/v) D2O or 0.2 mM RalB·GTP and 0.3 mM RLIP-76 GBD in the same buffer. The experiments were recorded on a Bruker DRX500 at 25°C or −6°C.
Assignment and Structure Calculation
The assignment of spectra and generation of NOE distance restraints was achieved using the CCPN Analysis program (Vranken et al., 2005) and the assignments are reported elsewhere (Fenwick et al., 2008a; Fenwick et al., 2008b). Dihedral restraints were generated from the chemical shifts using TALOS (Cornilescu et al., 1999) and the structure calculations were run using CNS 1.2 with the Aria 1.2 protocols (Brunger et al., 1998), (Linge et al., 2001). Hydrogen bonds used in the structure calculations were inferred for the RLIP76 GBD from amide exchange rates (Hwang et al., 1998). Restraints were added to model a magnesium ion with octahedral geometry, coordinated to two molecules of water, the β and γ phosphates and residues Ser-28RalB and Thr-46RalB. The structure calculations of the unbound RLIP76 GBD started from a structure of an extended chain, while the calculations of the complex started from the coordinates of free RalB (Fenwick et al., 2009) and the RLIP76 GBD structures, with randomized orientations with respect to one another. 100 structures were calculated in the final iteration and the 50 lowest energy structures selected for further analysis.
Scintillation Proximity Assays
Direct Binding SPAs
Affinities of RalB proteins for RLIP76 GBD-His were measured using scintillation proximity assays (SPAs), in which His-tagged fusion protein was attached to a fluoromicrosphere via an anti-His antibody (Sigma) in the presence of Q72L RalB·[3H]GTP or mutant variants. Binding of the G protein to the RLIP76 GBD-His brings the labelled nucleotide close enough to the scintillant to obtain a signal. Apparent Kd s for Q72L RalB·[3H]GTP and proteins incorporating further mutations were measured as described previously (Thompson et al., 1998) by varying the concentration of RalB·[3H]GTP at a constant concentration of RLIP76 GBD-His. These assays were performed with 80 nM RLIP76 GBD-His. Using this method, the upper and lower limits of the Kd that can accurately be measured are 1000 and 1nM, respectively. For each affinity determination, data points were obtained for at least ten different RalB concentrations. Binding curves were fitted using the appropriate binding isotherms to obtain Kd values and their standard errors (Thompson et al., 1998), (Graham et al., 1999).
Competition SPAs
For competition assays, free RLIP76 GBD was titrated into a mixture of 20nM [3H]GTP-RalB and 20nM GST-Sec5 RBD immobilised on fluoromicrospheres as above. The added RLIP76 GBD competes with the GST-Sec5 GBD/[3H]GTP-RalB interaction, abolishing the scintillation signal. The highest sample concentrations of competitor used were 2 μM. In each case, a blank was performed in the absence of GST-Sec5 GBD. For the RalB-RLIP76 GBD affinity determination, data points were obtained for at least 10 different competitor concentrations. The Kd value and its standard errors were obtained by fitting the dose-response curve to binding isotherms that describe competition between two proteins binding to one site on another protein and account for mutual depletion of the interacting components. The value of Kd for the GST-Sec5/RalB interaction was also required and this was obtained from direct binding SPAs (Fenwick et al., 2009). The equations used were adapted for SPA from the previously published derivations (Wang, 1995) and have been fully described elsewhere (Elliot-Smith et al., 2007).
Table I. Structural statistics for RLIP76 GBD and RalB·GMPPNP–RLIP76 GBD.
|
Experimental restraints used in structure calculation
| ||||
|---|---|---|---|---|
| RLIP76 | RalB·GMPPNP–RLIP76 | |||
| Unambiguous NOEs | 737 | 4470 | ||
| Ambiguous NOEs | 560 | 1393 | ||
| Dihedral angle restraints (φ + (ψ) | - | 292 | ||
| Hydrogen bonds (RLIP76) | 39 | 39 | ||
| Structural statistics | ||||
|---|---|---|---|---|
| RLIP76 | RalB·GMPPNP–RLIP76 | |||
| Coordinate precision (Ǎ) | <SA>a | <SA>cb | <SA>a | <SA>cb |
| RMSD of backbone atoms (12-180, 394-444) | 0.54±0.15 Ǎ | 0.35 Ǎ | 0.79 ± 0.11 Ǎ | 0.59 Ǎ |
| RMSD of heavy atoms (12-180, 393-444) | 1.11±0.13 Ǎ | 0.87 Ǎ | 1.03 ± 0.12 Ǎ | 0.85 Ǎ |
| RMS deviations | ||||
| From experimental restraints: | ||||
| NOE distances (Ǎ) | 2.00×10−2±1.4×10−3 | 1.94×10−2 | 1.57×10−2±8.0×10−4 | 1.51×10−2 |
| Dihedral angles (°) | - | 0.58 ± 9.2 × 10−2 | 0.40 | |
| From idealised geometry: | ||||
| Bonds (Ǎ) | 3.89×10−3±1.2×10−4 | 3.95×10−3 | 3.58×10−3±6.4×10−5 | 3.57×10−3 |
| Angles (°) | 0.51±1.2×10−2 | 0.48 | 0.50±1.2×10−2 | 0.49 |
| Impropers (°) | 1.21±0.1 | 1.22 | 1.32±7.5×10−2 | 1.37 |
| Ramachandran analysis d | ||||
| Most favoured regions | 92.2% | 94.1% | 84.6% | 85.0% |
| Allowed regions | 7.5% | 5.9% | 13.1% | 13.1% |
| Generously allowed regions | 0.1% | 0.0% | 1.0% | 0.9% |
| Disallowed regions | 0.3% | 0.0% | 1.3% | 0.9% |
<SA> represents the average RMS deviations for the ensemble.
<SA>c represents values for the structure that is closest to the mean.
PROCHECK (Laskowski et al., 1993)
Acknowledgements
This work was supported by Cancer Research UK, grant C9467/A4658 and the Medical Research Council, grant G0700057 (to DO and HRM) and Association de Recherche sur la Cancer (ARC; grant 4845), Agence Nationale de la Recherche (ANR; grant 08-BLAN-0290-01) and Association Christelle Bouillot (to JC). RBF was supported by the Cambridge Commonwealth Trust and the Overseas Research Students Awards Scheme. All structure calculations were performed on CamGRID.
Footnotes
The authors declare that they have no conflict of interest
Accession Numbers: NMR assignments have been deposited in BioMagResBand accession numbers 15524 (RLIP76) and 15525 (RLIP76-RalB complex). The coordinates have been deposited in the RCSB protein data bank, accession numbers 2KWH (RLIP76) and 2KWI (RLIP76-RalB complex).
References
- Awasthi S, Cheng JZ, Singhal SS, Pandya U, Sharma R, Singh SV, Zimniak P, Awasthi YC. Functional reassembly of ATP-dependent xenobiotic transport by the N- and C-terminal domains of RLIP76 and identification of ATP binding sequences. Biochemistry. 2001;40:4159–4168. doi: 10.1021/bi002182f. [DOI] [PubMed] [Google Scholar]
- Awasthi S, Sharma R, Yang YS, Singhal SS, Pikula S, Bandorowicz-Pikula J, Singh SV, Zimniak P, Awasthi YC. Transport functions and physiological significance of 76 kDa Ral-binding GTPase activating protein (RLIP76) Acta Biochim Pol. 2002;49:855–867. [PubMed] [Google Scholar]
- Bauer B, Mirey G, Vetter IR, Garcia-Ranea JA, Valencia A, Wittinghofer A, Camonis JH, Cool RH. Effector recognition by the small GTP-binding proteins Ras and Ral. J. Biol. Chem. 1999;274:17763–17770. doi: 10.1074/jbc.274.25.17763. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cantor SB, Urano T, Feig LA. Identification and Characterization of Ral-Binding Protein-1, a Potential Downstream Target of Ral Gtpases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. 2nd edn Academic Press; San Diego: 2007. [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Eathiraj S, Pan XJ, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot-Smith AE, Owen D, Mott HR, Lowe PN. Double mutant cycle thermodynamic analysis of the hydrophobic Cdc42-ACK protein-protein interaction. Biochemistry. 2007;46:14087–14099. doi: 10.1021/bi701539x. [DOI] [PubMed] [Google Scholar]
- Fenwick R, Prasannan S, Campbell LJ, Evetts KA, Nietlispach D, Owen D, Mott HR. H-1, C-13 and N-15 resonance assignments for the active conformation of the small G protein RalB in complex with its effector RLIP76. Biomol. NMR Assign. 2008a;2:179–182. doi: 10.1007/s12104-008-9115-7. [DOI] [PubMed] [Google Scholar]
- Fenwick RB, Prasannan S, Campbell LJ, Evetts KA, Nietlispach D, Owen D, Mott HR. Resonance assignments for the RLIP76 Ral binding domain in its free form and in complex with the small G protein RalB. Biomol. NMR Assign. 2008b;2:191–194. doi: 10.1007/s12104-008-9118-4. [DOI] [PubMed] [Google Scholar]
- Fenwick RB, Prasannan S, Campbell LJ, Nietlispach D, Evetts KA, Camonis J, Mott HR, Owen D. Solution Structure and Dynamics of the Small GTPase RalB in Its Active Conformation: Significance for Effector Protein Binding. Biochemistry. 2009;48:2192–2206. doi: 10.1021/bi802129d. [DOI] [PubMed] [Google Scholar]
- Frankel P, Aronheim A, Kavanagh E, Balda MS, Matter K, Bunney TD, Marshall CJ. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 2005;24:54–62. doi: 10.1038/sj.emboj.7600497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai S, Matern HT, Jagath JR, Scheller RH, Brunger AT. Structural basis of the interaction between RalA and Sec5, a subunit of the sec6/8 complex. Embo J. 2003;22:3267–3278. doi: 10.1093/emboj/cdg329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer M, Schweins T, Herrmann C, Prisner T, Wittinghofer A, Kalbitzer HR. Conformational transitions in p21(ras) and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry. 1996;35:10308–10320. doi: 10.1021/bi952858k. [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Harding MA, Seraj MJ, Gulding KM, Theodorescu D. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002;62:982–985. [PubMed] [Google Scholar]
- Graham DL, Eccleston JF, Lowe PN. The conserved arginine in Rho-GTPase-activating protein is essential for efficient catalysis but not for complex formation with rho CDP and aluminum fluoride. Biochemistry. 1999;38:985–991. doi: 10.1021/bi9821770. [DOI] [PubMed] [Google Scholar]
- Hwang TL, van Zijl PCM, Mori S. Accurate quantitation of water-amide proton exchange rates using the Phase-Modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- Jiang H, Luo JQ, Urano T, Frankel P, Lu ZM, Foster DA, Feig LA. Involvement of Ral Gtpase in V-Src-Induced Phospholipase-D Activation. Nature. 1995;378:409–412. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- Jin RS, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. Embo J. 2005;24:2064–2074. doi: 10.1038/sj.emboj.7600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis JH. Bridging Ral Gtpase to Rho-Pathways - Rlip76, a Ral Effector With Cdc42/Rac Gtpase-Activating Protein Activity. J. Biol. Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- Jullien-Flores V, Mahe Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, Camonis JH. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 2000;113:2837–2844. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. Molscript - a Program to Produce Both Detailed and Schematic Plots of Protein Structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- Lim KH, Brady DC, Kashatus DF, Ancrile BB, Der CJ, Cox AD, Counter CM. Aurora-A Phosphorylates, Activates, and Relocalizes the Small GTPase RalA. Mol. Cell Biol. 2010;30:508–523. doi: 10.1128/MCB.00916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linge JP, O’Donoghue SI, Nilges M. Automated assignment of ambiguous NOEs with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- Maesaki R, Ihara K, Shimizu T, Kuroda S, Kaibuchi K, Hakoshima T. The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1. Mol. Cell. 1999;4:793–803. doi: 10.1016/s1097-2765(00)80389-5. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Modha R, Campbell LJ, Nietlispach D, Buhecha HR, Owen D, Mott HR. The Rac1 Polybasic Region Is Required for Interaction with Its Effector PRK1. J. Biol. Chem. 2008;283:1492–1500. doi: 10.1074/jbc.M706760200. [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat. Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- Nicely NI, Kosak J, de Serrano V, Mattos C. Crystal structures of Ral-GppNHp and Ral-GDP reveal two binding sites that are also present in Ras and Rap. Structure. 2004;12:2025–2036. doi: 10.1016/j.str.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic B, Perisic O, Veprintsev DB, Williams RL, Munro S. Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol. Cell. 2003;12:863–874. doi: 10.1016/s1097-2765(03)00356-3. [DOI] [PubMed] [Google Scholar]
- Park SH, Weinberg RA. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene. 1995;11:2349–2355. [PubMed] [Google Scholar]
- Prasannan S, Fenwick RB, Campbell LJ, Evetts KA, Nietlispach D, Owen D, Mott HR. H-1, C-13, and N-15 resonance assignments for the small G protein RalB in its active conformation. Biomol. NMR Assign. 2007;1:147–149. doi: 10.1007/s12104-007-9040-1. [DOI] [PubMed] [Google Scholar]
- Sidhu RS, Clough RR, Bhullar RP. Regulation of phospholipase C-delta 1 through direct interactions with the small GTPase Ral and calmodulin. J Biol Chem. 2005;280:21933–21941. doi: 10.1074/jbc.M412966200. [DOI] [PubMed] [Google Scholar]
- Sporn MB. The war on cancer. Lancet. 1996;347:1377–1381. doi: 10.1016/s0140-6736(96)91015-6. [DOI] [PubMed] [Google Scholar]
- Strugnell SA, Wiefling BA, DeLuca HF. A modified pGEX vector with a C-terminal histidine tag: Recombinant double-tagged protein obtained in greater yield and purity. Anal. Biochem. 1997;254:147–149. doi: 10.1006/abio.1997.2425. [DOI] [PubMed] [Google Scholar]
- Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y. The exocyst complex binds the small GTPase RaIA to mediate filopodia formation. Nat. Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- Thompson G, Owen D, Chalk PA, Lowe PN. Delineation of the Cdc42/Rac-binding domain of p21-activated kinase. Biochemistry. 1998;37:7885–7891. doi: 10.1021/bi980140+. [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas P, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wang ZX. An Exact Mathematical Expression for Describing Competitive-Binding of 2 Different Ligands to a Protein Molecule. FEBS Lett. 1995;360:111–114. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- Ward Y, Wang W, Woodhouse E, Linnoila I, Liotta L, Kelly K. Signal pathways which promote invasion and metastasis: Critical and distinct contributions of extracellular signal-regulated kinase and Ral-specific guanine exchange factor pathways. Mol Cell Biol. 2001;21:5958–5969. doi: 10.1128/MCB.21.17.5958-5969.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MS, Lu L, Hong WJ, Song HW. Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat. Struct. Mol. Biol. 2004;11:86–94. doi: 10.1038/nsmb714. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Urano T, Goi T, Feig LA. An eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J Biol Chem. 1997;272:31230–31234. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- Ye M, Shima F, Muraoka S, Liao JL, Okamoto H, Yamamoto M, Tamura A, Yagi N, Ueki T, Kataoka T. Crystal structure of M-Ras reveals a GTP-bound “off” state conformation of Ras family small GTPases. J. Biol. Chem. 2005;280:31267–31275. doi: 10.1074/jbc.M505503200. [DOI] [PubMed] [Google Scholar]