Abstract
Genetics is believed to have an important role in intellectual disability (ID). Recent studies have emphasized the involvement of de novo mutations (DNMs) in ID but the extent to which they contribute to its pathogenesis and the identity of the corresponding genes remain largely unknown. Here, we report a screen for DNMs in subjects with moderate or severe ID. We sequenced the exomes of 41 probands and their parents, and confirmed 81 DNMs affecting the coding sequence or consensus splice sites (1.98 DNMs/proband). We observed a significant excess of de novo single nucleotide substitutions and loss-of-function mutations in these cases compared to control subjects, suggesting that at least a subset of these variations are pathogenic. A total of 12 likely pathogenic DNMs were identified in genes previously associated with ID (ARID1B, CHD2, FOXG1, GABRB3, GATAD2B, GRIN2B, MBD5, MED13L, SETBP1, TBR1, TCF4, WDR45), resulting in a diagnostic yield of ∼29%. We also identified 12 possibly pathogenic DNMs in genes (HNRNPU, WAC, RYR2, SET, EGR1, MYH10, EIF2C1, COL4A3BP, CHMP2A, PPP1CB, VPS4A, PPP2R2B) that have not previously been causally linked to ID. Interestingly, no case was explained by inherited mutations. Protein network analysis indicated that the products of many of these known and candidate genes interact with each other or with products of other ID-associated genes further supporting their involvement in ID. We conclude that DNMs represent a major cause of moderate or severe ID.
Author Summary
Intellectual disability (ID) is the most frequent severe handicap of childhood. Several observations indicate that genetic factors explain a large fraction of cases with ID. We and others have recently found that de novo mutations (DNMs; genetic changes not transmitted from the parents) represent a common cause of ID. To further assess the contribution of DNMs to the development of ID, we interrogated virtually all the genes of the genome in 41 affected children with moderate or severe ID and in their healthy parents. In 12 of the cases, we identified disease-causing DNMs in genes known to be associated with ID, resulting in a molecular diagnostic yield of 29%. We also found 12 possibly disease-causing DNMs in genes that were not previously causally linked to ID. Interestingly, many of the genes with deleterious DNMs uncovered by this study encode proteins that interact with each other and affect specific processes in brain cells. In contrast, we did not identify any inherited mutations that could explain our cases. We conclude that DNMs play a predominant role in moderate or severe ID.
Introduction
Intellectual disability (ID) is defined by significant impairment of cognitive and adaptive functions with onset before 18 years of age. It has an estimated worldwide prevalence of 1–3%, with moderate or severe forms of ID (IQ<50) affecting up to 0.5 % of the population in Western countries [1]. We and others have reported that de novo point mutations (including single nucleotide substitutions (SNVs) and small insertions/deletions, referred herein collectively as DNMs) play a significant role in the genetics of ID [2]–[5]. Similarly, DNMs were found to be implicated in the etiology of other neurodevelopmental disorders overlapping with ID, such as autism spectrum disorders (ASD), epileptic encephalopathy and schizophrenia [6]–[10]. DNMs represent the most extreme form of rare genetic variations; they are more deleterious, on average, than inherited variations because they have been subjected to less stringent evolutionary selection. Importantly, they provide a mechanism by which early-onset reproductively lethal diseases remain frequent in the population. This makes these mutations prime candidates for causing diseases that occur sporadically, and that decrease the reproductive fitness and incur a large degree of selection against phenotypes such as ID. Based on these considerations, we hypothesized that the contribution of DNMs is greater in more severe forms of ID. In order to explore this hypothesis, we performed high-depth exome sequencing in 41 trios consisting of individuals with moderate or severe ID and their healthy parents and assessed the contribution of DNMs to this condition.
Results/Discussion
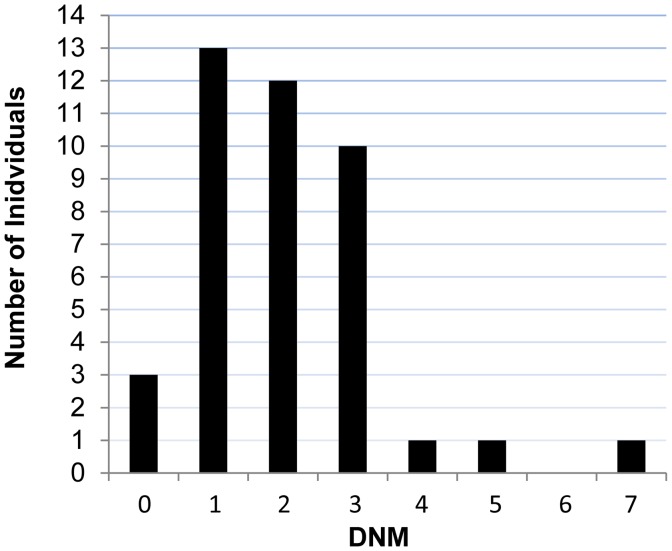
We performed exome sequencing in 41 individuals with ID and their unaffected parents. We identified a total of 83 putative DNMs in as many genes within both coding and consensus splice site sequences. Sanger sequencing confirmed 81 of these as de novo and 2 as inherited from one of the parents (Table S1). All of these DNMs were represented by ≥25% of reads, suggesting that they are unlikely to be associated with somatic mosaicism. The fact that the mutant and wild-type peaks on Sanger chromatograms were comparable in size is consistent with this conclusion. The average DNM rate per trio was 1.98, with only 3 trios containing no detectable DNMs (Figure 1). The observed de novo SNV rate in the consensus coding sequences (CCDS) was 1.56 events per trio or 2.58×10−8 per base per generation (64 SNVs in 2,477,702,175 CCDS bases sequenced at ≥10× in the 41 affected individuals), which is significantly higher than the expected population rate of 1.65×10−8 (R binomial test, p = 0.0007), or than the ones experimentally determined from exome sequencing studies in control trios (1.28×10−8 and 1.51×10−8) [2], [4]. Considering only de novo SNVs affecting the coding and the canonical splice sites (AG, GT at intronic positions −1/−2 and +1/+2 of the acceptor and donor splice sites, respectively), 73% were missense and 11% were nonsense and canonical splice site mutations. We found a significant excess of these de novo nonsense and splice site mutations in the probands of our cohort when compared to data from exome sequencing of 54 control trios with no family history of ID [4], [11] or of 593 quartets, including unaffected siblings of individuals with ASD (R binomial test, p = 0.0015 and p = 0.02, respectively) (Table 1) [7], [9], [10]. Such an excess of deleterious DNMs suggest that at least a subset of them are pathogenic.
Figure 1. Number of DNMs per affected individual in each trio.
Table 1. Distribution of the DNMs identified in this study and in controls.
| DNM | This study (n = 41) | Trio controls (n = 54) refs [4], [11] | ASD unaffected siblings controls (n = 593) refs [7], [9], [10] |
| Missense | 48 | 26 | 305 |
| Synonymous | 11 | 12 | 117 |
| Nonsense | 5 | 0 | 16 |
| Canonical splice site | 2 | 1 | 3 |
| Consensus splice site* | 1 | 0 | NA |
| Frameshift | 12 | 1 | 0 |
| Inframe insertion/deletion | 2 | 0 | 0 |
| Total DNMs | 81 | 40 | 441 |
| Average DNM/trio | 1.98 | 0.74 | 0.74 |
| Average coding SNVs/trio | 1.56 | 0.86 | 0.74 |
| Total SNVs** | 66 | 39 | 441 |
| LoF SNVs | 7 | 1 | 19 |
| p-value (LoF SNVs) vs controls | - | 0.0015 | 0.02 |
*canonical splice site variants not included.
**Consensus splice site variant not included.
NA, not applicable. LoF SNVs, nonsense and canonical splice site. Nominally significant P values (<0.05) calculated using an R exact binomial test.
Twelve DNMs were found in as many probands in genes previously associated with ID based on the documentation of deleterious DNMs in at least 4 unrelated individuals with similar phenotypes. Nine of these DNMs are Loss-of-Function (LoF) variants (nonsense, frameshift and canonical splice variants) and affect the following genes: ARID1B [OMIM 614556] [12], CHD2 [OMIM 602119] [4], [13], FOXG1 [OMIM 164874] [14], GATAD2B [OMIM 614998] [2], [15], MBD5 [OMIM 611472] [9], [13], [16]–[18], MED13L [OMIM 608771] [7], [19]–[21], SETBP1 [OMIM 611060] [4], [22], TCF4 [OMIM 602272] [4], [23]–[25], and WDR45 [OMIM 300526] [26], [27] (Tables 2 and 3). None of these 9 DNMs were found in public SNP databases. The phenotype of each of the probands is consistent with that of subjects previously described with mutations in these respective genes, with two exceptions (Text S1). Although truncating mutations in CHD2 have been reported in individuals with epileptic encephalopathy [4], [6], [13], the individual described herein with a CHD2 frameshift mutation has no history of epilepsy, suggesting that LoF mutations in CHD2 are associated with greater clinical heterogeneity than initially expected. Another example of a gene associated with clinical heterogeneity in our dataset is SETBP1. Missense mutations clustering in a conserved 11-bp coding region of SETBP1 have been reported to cause Schinzel-Giedon syndrome (OMIM 269150), a condition characterized by severe ID and specific craniofacial features [22]. In contrast, our case carried a de novo truncating mutation in SETBP1 and showed moderate non-syndromic ID without the typical craniofacial features of Schinzel-Giedon syndrome. Recent studies reported a similar phenotype in individuals with a truncating mutation in SETBP1 or microdeletions encompassing SETBP1 [4], [28]. Collectively, these observations suggest that SETBP1 haploinsufficiency results in a different phenotype than that induced by the missense mutations reported in Schinzel-Giedon syndrome, which presumably lead to a gain-of-function or a dominant negative effect [22]. We conclude that all of these 9 DNMs are likely to be pathogenic.
Table 2. Top risk DNMs identified in this study.
| Individual | sex | Gene | Genomic change (hg19) | NCBI RefSeq. | AA | MutationType | Change; prediction (score) |
| 289.143 | F | ARID1B | chr6:157511198delC | NM_020732.3 | 2236 | frameshift del | c.3716delC (Pro1239Hisfs*5) |
| 1396.504 | F | CHD2 | chr2:93470514C>G | NM_001271.3 | 1828 | nonsense | c.335C>G (p.Ser112*) |
| 893.339 | F | FOXG1 | chr14:29236991delG | NM_005249.4 | 489 | frameshift del | c.506delG (p.Gly169Alafs*23) |
| 1907.666 | F | GATAD2B | chr1:153785930T>C | NM_020699.2 | 593 | CSS | c.1217-2A>G |
| 79.65 | M | MBD5 | chr2:149221431_149221438delAAAAGCAT | NM_018328.4 | 1494 | frameshift del | c.340_347del (p.Lys114Glyfs*35) |
| 820.316 | F | MED13L | chr12:116446509_116446510delCT | NM_015335.4 | 2210 | frameshift del | c.1708_1709delCT (p.Ser570Phefs*27) |
| 1861.653 | M | SETBP1 | chr18:42531126delC | NM_015559.2 | 1596 | frameshift del | c.1821delC (p.Ser608Alafs*22) |
| 1045.400 | M | TCF4 | chr18:52921925G>A | NM_001083962.1 | 667 | nonsense | c.1153C>T (p.Arg385*) |
| 1883.659 | F | WDR45 | chrX:48935736G>A | NM_007075.3 | 361 | nonsense | c.C19T (p.Arg7*) |
| 1843.647 | M | GABRB3 | chr15:26866506_26866507InsACC | NM_021912.4 | 473 | insertion | c.413_415dupACC(p.Asn138_Arg139insHis); PVN (−12.3) |
| 121.83 | M | TBR1 | chr2:162274305T>C | NM_006593.2 | 682 | missense | c.811T>C (p.Trp271Arg); SIFT (0.00); PFF2 (1.0), PVN (−11.5) |
| 838.321 | M | GRIN2B | chr12:13720098C>T | NM_000834.3 | 1484 | missense | c.2459G>A (p.Gly820Glu); SIFT (0.00). PFF2 (1.0), PVN (−7.5) |
| 1464.524 | M | HNRNPU | chr1:245027099G>A | NM_031844.2 | 825 | nonsense | c.511C>T (p.Gln171*) |
| 762.297 | F | WAC | chr10:28824675_28824678delAGAG | NM_016628.4 | 647 | frameshift del | c.263_266delAGAG (p.Glu88Glyfs*103) |
| 341.162 | M | RYR2 | chr1:237995907G>A | NM_001035.2 | 4967 | missense | c.14864G>A (p.Gly4955Glu); SIFT (0.00), PFF2 (1.00); PVN(−6.0) |
| 1871.656 | F | MYH10 | chr17:8455445G>A | NM_001256012.1 | 2007 | missense | c.838C>T (p.Arg280Cys); SIFT (0.00), PFF2 (1.00), PVN (−7.5) |
| 702.278 | F | EIF2C1 | chr1:36359357G>A | NM_012199.2 | 857 | missense | c.595G>A (p.Gly199Ser); SIFT (0.00), PFF2 (1.00), PVN (−5.2) |
| 1312.477 | M | COL4A3BP | chr5:74712811C>T | NM_001130105.1 | 752 | missense | c.1111G>A (p.Gly371Arg); SIFT (0.002), PFF2 (0.97), PVN (−7.3) |
| 115.81 | M | SET | chr9:131456084_131456086delCTT | NM_001122821.1 | 290 | frameshift del | c.699_701delCTT (p.Tyr233*) |
| 670.267 | F | EGR1 | chr5:137803485_137803485insA | NM_001964.2 | 543 | frameshift ins | c.1347_1348insA (p.Tyr450Ilefs*92) |
| 1439.518 | F | PPP1CB | chr2:29022094dupA | NM_206876.1 | 327 | frameshift | c.909dupA (p.Tyr304Ilefs*19) |
| 580.240 | M | CHMP2A | chr19:59063688_59063688insG | NM_198426.2 | 222 | frameshift | c.286_287insC (p.Asn96Thrfs*35) |
| 1841.646 | M | PPP2R2B | chr5:146070692C>G | NM_181678.2 | 501 | missense | c.413G>C (p.Arg138Pro); SIFT (0.01); PFF2 (0.48); PVN (−4.9) |
| 985.382 | M | VPS4A | chr16:69353403_69353405delTCC | NM_013245.2 | 437 | deletion | c.577_579delTCC (p.Ser193del); PVN (−12.3) |
AA, total amino acids. All predictions by SIFT (http://sift.jcvi.org/), PFF2 (PolyPhen-2; http://genetics.bwh.harvard.edu/pph2/) and PROVEAN (PVN; (http://provean.jcvi.org/genome_submit.php) were damaging (scores indicated in parenthesis). CSS, Canonical splice site.
Table 3. Genes affected by predicted-damaging DNMs identified herein and their implication in ID.
| Mutation type | ID-associated Genes with likely pathogenic DNMs | Candidate Genes with possibly pathogenic DNMs | Genes of unknown significance to IDψ |
| Missense | GRIN2B, TBR1 | MYH10, RYR2, EIF2C1, COL4A3BP, PPP2R2B | BCORL1, VIPR1, MTUS1, WDR33, R3HDM1, FBXO28, MAPKBP1, KCNH1 |
| Nonsense | TCF4, CHD2, WDR45 | HNRNPU | |
| Canonical splice site* | GATAD2B | ||
| Consensus splice site* | GIT1 | ||
| Frameshift deletion | MBD5, ARID1B, MED13L, FOXG1, SETBP1 | WAC, SET | |
| Inframe insertion | GABRB3 | ||
| Frameshift insertion | EGR1, CHMP2A, PPP1CB | ||
| Inframe deletion | VPS4A | ||
| Synonymous – splicing* | NANS ** |
All missense mutations were predicted damaging by SIFT and Polyphen-2. All in-frame deletions and insertions here were predicted damaging by PROVEAN (http://provean.jcvi.org/genome_submit.php).
*Predicted to affect splicing by both Human Splicing Finder (http://www.umd.be/HSF/) and Mutation Taster (http://www.mutationtaster.org/).
Predicted-damaging DNMs present in cases with no likely or possibly pathogenic DNMs.
**Splicing defect verified by RT-PCR (Figure S1).
The three other DNMs in genes previously associated with ID include an in-frame insertion in GABRB3 [OMIM 137192], a missense in TBR1 [OMIM 604616] and a missense in GRIN2B [OMIM 138252] (Tables 2 and 3). All of these DNMs affect conserved residues and are predicted to be damaging. Moreover, none of them were found in public SNP databases. Damaging missense mutations in GABRB3 have been previously documented in cases with ID and intractable epilepsy with various types of seizures [6]. Individual 1843.647 also showed ID and intractable epilepsy with a similar pattern of seizures as these cases (Text S1). DNMs in TBR1 have been found in patients with ID and the variable presence of ASD or growth retardation [8], [9], [21], [29], [30]. Individual 121.83 displayed a phenotype similar to previously described cases, including ID, ASD and growth retardation (Text S1). Finally, DNMs in GRIN2B have been associated with ID of variable severity with or without ASD and epilepsy [2], [6], [31], [32]. Individual 838.321 showed severe ID, not walking and saying only one word at 16 years of age (Text S1). He has never had any seizures though his EEG revealed multifocal epileptic activity. Similar patterns of cognitive impairment were also reported in other patients with DNMs in GRIN2B [2], [32]. Interestingly, the mutation identified in our case affects a residue located in the ligand-binding domain of the protein, like previously reported de novo missenses in GRIN2B [32]. We conclude that these three DNMs are also likely to be pathogenic.
Among the remaining cases, 22 have predicted-damaging DNMs, including 7 LoF mutations (5 frameshifts, 1 nonsense, 1 consensus splice site), 13 missenses, 1 deletion, and 1 synonymous mutation whose predicted effect on splicing was confirmed by RT-PCR (Figure S1). Interestingly, deleterious DNMs in 6 of these genes (HNRNPU [OMIM 602869], WAC [OMIM 615049], RYR2 [OMIM 180902], MYH10 [OMIM 160776], EIF2C1 [OMIM 606228], COL4A3BP [OMIM 604677]) have previously been reported in at least one individual with ID. We discuss hereafter the DNMs that we identified in these genes (Tables 2 and 3).
HNRNPU [OMIM 602869] codes for a highly conserved protein that binds RNAs and mediates different aspects of their metabolism and transport. Chromosome 1q44 microdeletions have defined a critical region associated with ID and seizures that encompasses HNRNPU as well as two other genes [33], [34]. Two truncating and one splice mutations in HNRNPU were subsequently identified in individuals with ID and seizures [6], [13], [24]. Two of these mutations occurred de novo whereas the origin of the other one was not elucidated. One of these individuals also showed ASD whereas the case with the splice mutation displayed syndromic features, including panhypopituitarism, bifid great toe and vertebral segmental defects. We identified an individual (1464.524) who carries a de novo truncating mutation (c.511C>T, p.Gln171*) in HNRNPU. This mutation is located in an upstream coding exon present in all isoforms, thus having the potential to induce nonsense mRNA mediated decay [35]. Moreover, inspection of the Exome Variant Server (EVS) database (6500 exomes) revealed no LoF variants in HNRNPU, indicating that haploinsufficiency of this gene is not tolerated. Our case displayed ID, epilepsy and ASD (Text S1), a phenotype that is similar to that of the other non-syndromic cases with DNMs in this gene, further supporting its involvement in ID.
WAC encodes a nuclear protein that interacts with RNF20/40 to regulate histone H2B ubiquinitation, chromatin organization, and gene transcription [36]. De novo microdeletions encompassing WAC and a nonsense DNM in WAC in individuals with severe ID were recently reported [2], [37]. Our subject (762.297) carries a truncating mutation in WAC (c.263_266del, p.Glu88Glyfs*103). This mutation is located in an upstream coding exon present in all isoforms. Inspection of the EVS database revealed no LoF variants in WAC. Individual 762.297 showed moderate ID without any distinguishing features on clinical examination and brain imaging, a phenotype that is consistent with that observed in the previously reported patient with a truncating mutation in this gene (Text S1) [2]. Our finding, thus, further supports the involvement of WAC in ID.
RYR2 encodes the cardiac and brain-expressed calcium release channel ryanodine receptor 2. Mutations in RYR2 are typically associated with exercise-induced ventricular and atrial arrhythmias. Virtually all reported mutations in RYR2 are missenses or in-frame deletions that are believed to confer a gain of function, resulting in an increase of Ca+ release [38], [39]. We identified an individual (341.162) with ID, seizures, short stature and severe atrial arrhythmias (Text S1) who carries a predicted-damaging de novo missense mutation in RYR2 (c.14864G>A, p.Gly4955Glu). Interestingly, 3 patients with seizures have previously been reported with DNMs in RYR2: 1) an individual with epileptic encephalopathy but presumably without a history of arrhythmia was recently found to carry a nonsense mutation (c.9568C>T, p.Arg3190*) in RYR2 [6]; this DNM might not be disease-causing considering that the pathogenic impact of truncating mutations in RYR2 remains unclear and that inspection of EVS revealed 5 different heterozygous LoF mutations in RYR2; 2) an individual with cognitive impairment, intractable seizures, short stature and subclinical ventricular tachycardia was found to carry a missense mutation (c.12563T>C, p.Leu4188Pro) [40]; and 3) an individual with intractable seizures but without cognitive impairment and arrhythmia was described with a missense mutation (c.14803G>A, p.Gly4935Arg) [41]. It is noteworthy that the mutation found in this latter individual is in close proximity to that of our subject, affecting a highly conserved C-terminal region of the protein. Interestingly, mice heterozygous for the missense mutation p.R2474S in Ryr2 display generalized seizures and arrhythmias [42]. More recently, two brothers with ID, seizures and atrial arrhythmias were found to carry a missense mutation in CLIC2 (OMIM 300138), which maps to the X chromosome [43]. CLIC2 is a negative regulator of RYR2. The mutation was shown to stimulate the release of Ca2+ by keeping the RYR2 channel in an open state, possibly due to a higher binding affinity for the RYR2 protein. The specificity of the phenotype observed in our subject and its similarity with that of other individuals with DNMs in RYR2 or with the mutation in CLIC2 suggest that the mutation identified herein may be causal.
MYH10 encodes the non-muscle myosin heavy chain IIB that is critical for heart and brain development [44], [45]. Loss of Myh10 function in mice results in embryonic lethality, hydrocephalus and neuronal migration defects but the cognitive and behavioural phenotype of heterozygous mice has not yet been reported. We identified a predicted-damaging de novo missense mutation (c.838C>T, p.Arg280Cys; individual 1871.656) in MYH10, affecting its conserved motor domain, whereas another group recently reported a de novo truncating mutation (c.2722G>T, p.Glu908*) in the same gene [46]. Both individuals displayed severe ID, microcephaly, and feeding difficulties as well as cerebral atrophy with increased intensities in bilateral basal ganglia and thalami on brain MRI (Text S1). The similarities between the phenotypes of these individuals raise the possibility that these mutations in MYH10 are pathogenic. O'Roak et al. (2012) also reported a predicted-damaging de novo missense mutation (c.794A>G, p.Y265C; NM_001256012.1) in the motor domain of MYH10, in close proximity to the mutation identified herein, in a patient with ASD and moderate to severe ID. However no additional phenotypic data was available. Interestingly our patient with the MYH10 mutation also displayed autistic features. Inspection of EVS for potential LoF mutations in MYH10 showed the presence of a heterozygous frameshift deletion and a heterozygous splice site mutation. It is important to note, however, that these EVS variants were seen in single individuals and were not validated.
DNMs in EIF2C1 and COL4A3BP have also been previously reported in single individuals with severe ID [2], [4]. For each of these genes, the phenotype of the affected individuals appears similar to that of our subjects (Text S1). However, because of the lack of specific clinical features in these individuals, the occurrence of DNMs in unrelated subjects does not readily indicate pathogenicity, especially in the case of missense mutations whose functional consequences are not validated.
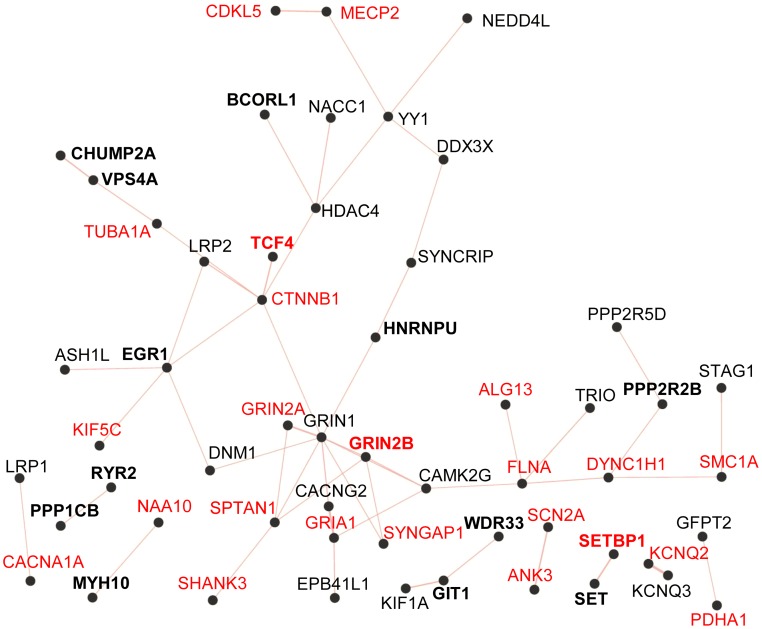
Among the remaining cases, we also identified 6 predicted-damaging DNMs in genes (SET [OMIM 600960], EGR1 [OMIM 128990], PPP1CB [OMIM 600590], CHMP2A [OMIM 610893], PPP2R2B [OMIM 604325], and VPS4A [OMIM 609982]) that play biological functions relevant to ID (Table 2). Inspection of the EVS database revealed no LoF variants in these genes, with the exception of a single heterozygous variant in PPP1C1B (MAF = 1/12518) with a potential effect on splicing. In addition, some of these genes were found in proteomic studies to physically interact with the product of at least one ID-associated gene, further increasing the probability of their involvement in this disorder (see below and Figure 2). Each of these DNMs is discussed hereafter.
Figure 2. Physical protein-protein interaction network generated by GeneMANIA (http://www.GeneMANIA.org/; Gene Ontology molecular function based weighting).
The Query genes included those listed in Table 3 from this study (in bold) and known and candidate ID genes reported with predicted-damaging DNMs from other studies (Table S2). Known ID genes are in red. The resulting network of 38 interconnected proteins was found to be enriched for proteins whose Gene Ontology molecular functions are implicated in the glutamate receptor signalling pathway (GRIN1, GRIN2A, GRIN2B, GRIA1, CACNG2, SHANK3; FDR q-value = 7.04e-6).
SET encodes a widely expressed multifunctional nuclear protein that affects pathways involved in ID, such as chromatin remodelling and gene transcription [47]. SET physically binds SETBP1 [48], whose disruption is known to cause severe ID (see above). In addition, recent studies indicate that SET directly interacts with MCPH1 (OMIM 607117) to ensure the proper temporal activation of chromosome condensation during mitosis [49]. Cells with SET knockdown exhibited abnormal condensed chromosomes similar to those observed in MCPH1-deficient fibroblasts. In addition, mutations that impair binding of MCPH1 to SET affect the ability of the former to rescue the abnormal chromosome condensation phenotype in fibroblasts from Mcph1 mutant mice. Recessive mutations in MCPH1 cause primary microcephaly, which is characterized by reduced brain size, without major structural abnormalities, and mild-to-moderate ID [50]. We identified a de novo deletion resulting in the creation of a premature stop codon in SET (c.699_701del, pTyr233*) in an individual (115.81) with congenital microcephaly, normal brain MRI, and moderate ID without any other distinguishing feature (Text S1). The functional relationship between MCPH1 and SET and the phenotypical similarities between cases with mutations in MCPH1 and our subject suggest that the truncating DNM in SET may be pathogenic.
EGR1 encodes a transcription factor that plays a key role in learning and memory [51]. We identified a de novo truncating mutation (c.1347_1348insA, p.Tyr450Ilefs*92) in EGR1 in an individual (670.267) with severe non-syndromic ID and acquired microcephaly (Text S1). Mice harbouring a heterozygous deletion of Egr1 showed synaptic plasticity, learning and memory impairments [52], [53]. Due to the prominent role of EGR1 in learning and memory and the impact of its haploinsufficiency on cognition in mice, we postulate that the truncating DNM identified herein in EGR1 may be pathogenic.
PPP1CB, which encodes a brain-enriched beta catalytic subunit of protein phosphatase 1 (PP1), and PPP2R2B, which encodes a neuron-specific B regulatory subunit of protein phosphatase 2 (PP2A), have been shown to regulate synaptic plasticity pathways [54], [55]. Individual 1439.518 carries a truncating mutation (c.909dupA, p.Tyr304Ilefs*19) in PPP1CB. This individual displayed severe ID, growth retardation and some dysmorphic features (Text S1). Individual 1841.646 carries a predicted-damaging missense mutation (c.413G>C, p.Arg138Pro) in PPP2R2B. This individual showed ID, intractable seizures and autistic features (Text S1). The pathogenic impact of these mutations remains uncertain at this point.
Among these candidate genes, CHMP2A and VPS4A are of special interest, as the proteins encoded by each are interacting partners. VPS4 ATPases play a critical role in the ESCRT pathway by recognizing membrane-associated ESCRT-III complexes and catalyzing their disassembly, a process that involves a direct interaction between CHMP2A and VPS4A [56]. The ESCRT-III pathway is involved in key cellular processes, including formation of endocytic multivesicular bodies, the abscission stage of cytokinesis, as well as centrosome and spindle maintenance [57]. Specific depletion of either CHMP2A or VPS4A proteins in cultured cells disrupts mitosis by inhibiting abscission and altering centrosome and spindle pole numbers [58]. We identified an individual (580.240) with a de novo frameshift insertion (c.286_287insC, p.Asn96Thrfs*35) in CHMP2A and another individual (985.382) with a predicted-damaging in-frame deletion (c.577_579delTCC, p.Ser193del) in VPS4A (Table 2). Both subjects showed severe ID as they were non-ambulatory and non-verbal at 4 years of age (Text S1). Our findings, thus, raise the possibility that components of the ESCRT-III complex maybe involved in ID.
To determine whether the genes identified here with predicted-damaging DNMs (likely/possibly pathogenic or of yet unknown significance to ID) (Table 3) encode proteins that are physically interconnected, we performed protein-protein interaction network analysis using GeneMANIA (http://www.GeneMANIA.org/) [59]. We also included in this analysis the known and candidate ID genes identified with predicted-damaging DNMs in other ID trio studies (Table S2) [2]–[6], [21]. This analysis showed that 11 out of the 24 proteins encoded by genes found herein with likely/possibly pathogenic DNMs interacted with either known or candidate ID genes, or with each other, further supporting their link to ID. Interestingly, we observed an enrichment for proteins implicated in glutamate receptor signaling pathways (FDR q-value = 7.04e-6) in the generated network (38 interconnected proteins) (Figure 2). Previous studies have shown an excess of functional DNMs over neutral ones in genes associated with glutamatergic systems in cases with non-syndromic ID, further supporting the critical involvement of this pathway in ID [3].
We also searched for the presence of rare inherited deleterious mutations (truncating, splicing, predicted-damaging missense and insertions or deletions) in genes associated with autosomal recessive or X-linked forms of ID, epilepsy or ASD (see Table S3 for the complete list of inherited rare variants in each proband). We identified only one case (692.274) that could potentially be explained by such mutations. This individual is hemizygous for a predicted-damaging missense (c.7949G>A [p.Arg2650His]; NM_031407.6) in the E3 ubiquitin ligase gene HUWE1, which is inherited from his healthy mother. Missense mutations in HUWE1 have been associated with moderate to severe X-linked ID with normocephaly or macrocephaly [60]. Our case showed severe ID (non-verbal, non-ambulatory at 5 years of age) with congenital microcephaly. Because of these phenotypical differences, it is thus unclear whether this variation in HUWE1 is pathogenic.
In summary, our trio exome sequencing study identified deleterious DNMs in genes previously causally linked to ID in 12 cases out of the 41 studied herein, resulting in a molecular diagnostic yield of 29%. Recently, de Ligt et al. (2012) and Rauch et al. (2012) performed trio exome sequencing in individuals with severe ID and obtained a diagnostic yield, based on the presence of predicted-damaging point mutations in currently known ID genes, of 20% and 35%, respectively [2], [4], [21]. Overall, the contribution of inherited autosomal or X-linked recessive mutations appears limited in the three cohorts. The study of Rauch et al (2012) and ours were intentionally centered on sporadic cases, which might have created a bias against inherited mutations. However, it is important to emphasize that most cases with moderate or severe ID are sporadic, at least in Western societies. de Ligt et al. (2012) observed a proportionally smaller number of DNMs in their cohort when compared to that of Rauch et al. (2012) and ours. This difference may be related to the use of a different sequencing technology, which is associated with a lower depth, possibly accounting for the lower diagnostic yield observed in this study. Indeed, exploration of a subset of unexplained cases from this cohort using whole-genome sequencing revealed additional pathogenic DNMs in known ID genes, bringing the point mutation molecular diagnostic yield in this cohort to 34% [21].
Our study also provides evidence for the potential pathogenicity of 12 additional DNMs in as many genes. Some of these genes represent strong candidates. For instance, both HNRNPU and WAC map to small critical regions associated with ID, which were defined by a series of microdeletions. De novo truncating mutations in each of these genes were previously described in cases with severe ID. We now report additional truncating DNMs in these genes in cases with similar phenotypes as those already published, further supporting their involvement in ID. Similarly, we and others have identified damaging DNMs in RYR2 and MYH10 in patients with similar features. Finally, we discovered a truncating DNM in EGR1, the haploinsufficiency of which affects learning and memory in mice. Although the characterization of additional cases will be needed to confirm the involvement of these candidate genes in ID, these results indicate that the contribution of DNMs to the pathogenesis of moderate or severe ID could be even greater than that suggested by the diagnostic rate observed in this study.
In conclusion, our study suggests that DNMs represent a predominant cause of moderate or severe ID. High-depth trio-based exome sequencing is an effective method to establish molecular diagnosis in such cases.
Materials and Methods
Study subjects and ethics statement
The cases reported here (18 males, 23 females) with moderate (n = 12) or severe (n = 29) ID were recruited at the Sainte-Justine Hospital (Montreal, Canada), after the approval of the ethics committee, and informed consent was obtained from each participant or legal guardian. Inclusion criteria for the probands were: 1) absence of a history of ID, epilepsy or ASD in first or second-degree relatives; 2) moderate or severe ID with or without epilepsy or autistic features; 3) absence of pathogenic copy number variants as revealed by array comparative genome hybridization performed on a clinical basis (using a 135k-feature whole-genome microarray (SignatureChip OS2.0 manufactured for Signature Genomic Laboratories (Spokane, WA, USA) by Roche NimbleGen, Madison, WI, USA); 4) absence of specific changes on brain imaging. The clinical description of the 41 affected individuals is summarized in Table S4. For cases with likely or possibly pathogenic variants, a more detailed clinical description can be found in Text S1.
Exome capture and sequencing
Genomic DNA (3 µg) extracted from blood samples were used for exome capture and sequencing at the McGill University and Genome Quebec Innovation Center (Montreal, Quebec, Canada) using the Agilent SureSelect v4 exome capture kit, according to the manufacturer's recommendations, followed by 100 bp paired-end sequencing of each trio exomes on a single lane of the Illumina HiSeq2000.
Data analysis
Exome sequence data processing, alignment (using a Burrows-Wheeler algorithm, BWA-mem), and variant calling were done according to the Broad Institute Genome Analysis Tool Kit (GATK v4) best practices (http://www.broadinstitute.org/gatk/guide/topic?name=best-practices), and variant annotation was done using Annovar [61]. The median coverage of the target bases was 135× with 95% of the target bases being covered ≥10×. We focused on variants affecting the exonic regions and consensus splice site sequences (defined herein as intronic bases up to positions −3 and +6 from the exon boundaries). Only variants whose positions were covered at ≥10× and supported by at least 4 variant reads constituting ≥20% of the total reads for each called position were retained. This typically yielded an average of ∼22,000 variants. This variant list was subsequently reduced to an average of ∼500 rare variants by filtering out those that are present in ≥0.5% of in-house exome data sets (n = 600) from unrelated projects, as well as variants present in the 1000 Genome or in the Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/) with minor allele frequencies (MAF) ≥0.5%. Putative DNMs (typically <10/exome) were then extracted from the rare variant list by further excluding those that were present in the exomes of the parents. The sequencing reads carrying putative DNMs were inspected visually in each trio, using the Integrative Genomics Viewer (IGV) [62], to exclude obvious false positives. All putative DNMs were validated by bidirectional Sanger sequencing in the corresponding trio.
Supporting Information
Impact of the NANS synonymous de novo mutation in exon 4, c.603G>A (p. = ) (NM_018946.3), identified in patient 143.91 on exon splicing.
(DOC)
Confirmed DNMs identified in this study.
(XLSX)
Genes included in the physical protein-protein interaction network analysis.
(XLSX)
Inherited rare variations identified in the 41 probands of this study.
(XLSX)
Clinical phenotypes of the 41 affected individuals.
(XLSX)
Detailed clinical description of the patients with likely and possibly pathogenic DNMs identified in this study.
(DOCX)
Acknowledgments
We are grateful to the Génome Québec and McGill Innovation Center Massive Parallel Sequencing team for the exome sequencing service and to the bioinformatics analysis team of Réseau de Médecine Génétique Appliquée du Québec (RMGA) for the primary analysis of the sequence data. We thank Philippe Lemay for useful discussions. We are also grateful to all the families that participated in this study.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
JLM is a National Scientist of the Fonds de Recherche du Québec - Santé. MS holds a clinician-scientist award from the Canadian Institutes of Health Research (CIHR). This study was funded by a grant from CIHR (FRN 119440). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ropers HH (2010) Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet 11: 161–187. [DOI] [PubMed] [Google Scholar]
- 2. de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, et al. (2012) Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- 3. Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, et al. (2011) Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 88: 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, et al. (2012) Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380: 1674–1682. [DOI] [PubMed] [Google Scholar]
- 5. Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, et al. (2010) A de novo paradigm for mental retardation. Nat Genet 42: 1109–1112. [DOI] [PubMed] [Google Scholar]
- 6. Epi4K Consortium (2013) Epilepsy Phenome/Genome P, Allen AS, Berkovic SF, Cossette P, et al. (2013) De novo mutations in epileptic encephalopathies. Nature 501: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, et al. (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, et al. (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, et al. (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, et al. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, et al. (2012) De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet 44: 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoyer J, Ekici AB, Endele S, Popp B, Zweier C, et al. (2012) Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet 90: 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carvill GL, Heavin SB, Yendle SC, McMahon JM, O'Roak BJ, et al. (2013) Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet 45: 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kortum F, Das S, Flindt M, Morris-Rosendahl DJ, Stefanova I, et al. (2011) The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet 48: 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Willemsen MH, Nijhof B, Fenckova M, Nillesen WM, Bongers EM, et al. (2013) GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. J Med Genet 50: 507–514. [DOI] [PubMed] [Google Scholar]
- 16. Bonnet C, Ali Khan A, Bresso E, Vigouroux C, Beri M, et al. (2013) Extended spectrum of MBD5 mutations in neurodevelopmental disorders. Eur J Hum Genet 21: 1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Talkowski ME, Mullegama SV, Rosenfeld JA, van Bon BW, Shen Y, et al. (2011) Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am J Hum Genet 89: 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kleefstra T, Kramer JM, Neveling K, Willemsen MH, Koemans TS, et al. (2012) Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet 91: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Asadollahi R, Oneda B, Sheth F, Azzarello-Burri S, Baldinger R, et al. (2013) Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur J Hum Genet 21: 1100–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Haelst MM, Monroe GR, Duran K, van Binsbergen E, Breur JM, et al. (2014) Further confirmation of the MED13L haploinsufficiency syndrome. Eur J Hum Genet [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, et al. (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature 511: 344–347. [DOI] [PubMed] [Google Scholar]
- 22. Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, et al. (2010) De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet 42: 483–485. [DOI] [PubMed] [Google Scholar]
- 23. Hamdan FF, Daoud H, Patry L, Dionne-Laporte A, Spiegelman D, et al. (2013) Parent-child exome sequencing identifies a de novo truncating mutation in TCF4 in non-syndromic intellectual disability. Clin Genet 83: 198–200. [DOI] [PubMed] [Google Scholar]
- 24. Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, et al. (2012) Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet 49: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, et al. (2007) Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome). Am J Hum Genet 80: 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haack TB, Hogarth P, Kruer MC, Gregory A, Wieland T, et al. (2012) Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am J Hum Genet 91: 1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, et al. (2013) De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45: 445–449, 449e441. [DOI] [PubMed] [Google Scholar]
- 28. Filges I, Shimojima K, Okamoto N, Rothlisberger B, Weber P, et al. (2011) Reduced expression by SETBP1 haploinsufficiency causes developmental and expressive language delay indicating a phenotype distinct from Schinzel-Giedion syndrome. J Med Genet 48: 117–122. [DOI] [PubMed] [Google Scholar]
- 29. O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, et al. (2012) Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Palumbo O, Fichera M, Palumbo P, Rizzo R, Mazzolla E, et al. (2014) TBR1 is the candidate gene for intellectual disability in patients with a 2q24.2 interstitial deletion. Am J Med Genet A 164 A: 828–833. [DOI] [PubMed] [Google Scholar]
- 31. Endele S, Rosenberger G, Geider K, Popp B, Tamer C, et al. (2010) Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet 42: 1021–1026. [DOI] [PubMed] [Google Scholar]
- 32. Lemke JR, Hendrickx R, Geider K, Laube B, Schwake M, et al. (2014) GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann Neurol 75: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ballif BC, Rosenfeld JA, Traylor R, Theisen A, Bader PI, et al. (2012) High-resolution array CGH defines critical regions and candidate genes for microcephaly, abnormalities of the corpus callosum, and seizure phenotypes in patients with microdeletions of 1q43q44. Hum Genet 131: 145–156. [DOI] [PubMed] [Google Scholar]
- 34. Thierry G, Beneteau C, Pichon O, Flori E, Isidor B, et al. (2012) Molecular characterization of 1q44 microdeletion in 11 patients reveals three candidate genes for intellectual disability and seizures. Am J Med Genet A 158A: 1633–1640. [DOI] [PubMed] [Google Scholar]
- 35. Isken O, Maquat LE (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856. [DOI] [PubMed] [Google Scholar]
- 36. Zhang F, Yu X (2011) WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell 41: 384–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wentzel C, Rajcan-Separovic E, Ruivenkamp CA, Chantot-Bastaraud S, Metay C, et al. (2011) Genomic and clinical characteristics of six patients with partially overlapping interstitial deletions at 10p12p11. Eur J Hum Genet 19: 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Venetucci L, Denegri M, Napolitano C, Priori SG (2012) Inherited calcium channelopathies in the pathophysiology of arrhythmias. Nat Rev Cardiol 9: 561–575. [DOI] [PubMed] [Google Scholar]
- 39. Van Petegem F (2012) Ryanodine receptors: structure and function. J Biol Chem 287: 31624–31632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. LaPage MJ, Russell MW, Bradley DJ, Dick M 2nd (2012) Novel ryanodine receptor 2 mutation associated with a severe phenotype of catecholaminergic polymorphic ventricular tachycardia. J Pediatr 161: 362–364. [DOI] [PubMed] [Google Scholar]
- 41. Johnson JN, Tester DJ, Bass NE, Ackerman MJ (2010) Cardiac channel molecular autopsy for sudden unexpected death in epilepsy. J Child Neurol 25: 916–921. [DOI] [PubMed] [Google Scholar]
- 42. Lehnart SE, Mongillo M, Bellinger A, Lindegger N, Chen BX, et al. (2008) Leaky Ca2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J Clin Invest 118: 2230–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takano K, Liu D, Tarpey P, Gallant E, Lam A, et al. (2012) An X-linked channelopathy with cardiomegaly due to a CLIC2 mutation enhancing ryanodine receptor channel activity. Hum Mol Genet 21: 4497–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma X, Bao J, Adelstein RS (2007) Loss of cell adhesion causes hydrocephalus in nonmuscle myosin II-B-ablated and mutated mice. Mol Biol Cell 18: 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma X, Kawamoto S, Hara Y, Adelstein RS (2004) A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Mol Biol Cell 15: 2568–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tuzovic L, Yu L, Zeng W, Li X, Lu H, et al. (2013) A human de novo mutation in MYH10 phenocopies the loss of function mutation in mice. Rare Dis 1: e26144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muto S, Senda M, Akai Y, Sato L, Suzuki T, et al. (2007) Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc Natl Acad Sci U S A 104: 4285–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Minakuchi M, Kakazu N, Gorrin-Rivas MJ, Abe T, Copeland TD, et al. (2001) Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem 268: 1340–1351. [DOI] [PubMed] [Google Scholar]
- 49. Leung JW, Leitch A, Wood JL, Shaw-Smith C, Metcalfe K, et al. (2011) SET nuclear oncogene associates with microcephalin/MCPH1 and regulates chromosome condensation. J Biol Chem 286: 21393–21400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, et al. (2002) Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet 71: 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Poirier R, Cheval H, Mailhes C, Garel S, Charnay P, et al. (2008) Distinct functions of egr gene family members in cognitive processes. Front Neurosci 2: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bozon B, Davis S, Laroche S (2002) Regulated transcription of the immediate-early gene Zif268: mechanisms and gene dosage-dependent function in synaptic plasticity and memory formation. Hippocampus 12: 570–577. [DOI] [PubMed] [Google Scholar]
- 53. Jones MW, Errington ML, French PJ, Fine A, Bliss TV, et al. (2001) A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci 4: 289–296. [DOI] [PubMed] [Google Scholar]
- 54. Dickey AS, Strack S (2011) PKA/AKAP1 and PP2A/Bbeta2 regulate neuronal morphogenesis via Drp1 phosphorylation and mitochondrial bioenergetics. J Neurosci 31: 15716–15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Munton RP, Vizi S, Mansuy IM (2004) The role of protein phosphatase-1 in the modulation of synaptic and structural plasticity. FEBS Lett 567: 121–128. [DOI] [PubMed] [Google Scholar]
- 56. Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, et al. (2007) ESCRT-III recognition by VPS4 ATPases. Nature 449: 740–744. [DOI] [PubMed] [Google Scholar]
- 57. McCullough J, Colf LA, Sundquist WI (2013) Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem 82: 663–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, et al. (2010) Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci U S A 107: 12889–12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, et al. (2013) GeneMANIA prediction server 2013 update. Nucleic Acids Res 41: W115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, et al. (2008) Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet 82: 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Impact of the NANS synonymous de novo mutation in exon 4, c.603G>A (p. = ) (NM_018946.3), identified in patient 143.91 on exon splicing.
(DOC)
Confirmed DNMs identified in this study.
(XLSX)
Genes included in the physical protein-protein interaction network analysis.
(XLSX)
Inherited rare variations identified in the 41 probands of this study.
(XLSX)
Clinical phenotypes of the 41 affected individuals.
(XLSX)
Detailed clinical description of the patients with likely and possibly pathogenic DNMs identified in this study.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.