Abstract
Background
Hepatocellular carcinoma (HCC) is the fifth most common cancer, and it is the second most common cancer-related mortality globally. The prognostic value of high mobility group box 1 (HMGB1) remains controversial. The purpose of this study is to conduct a meta-analysis and literature review to evaluate the association of HMGB1 expression with the prognosis of patients with HCC.
Methods
A detailed literature search was made in Medline, Google Scholar and others for related research publications. The data were extracted and assessed by two reviewers independently. Analysis of pooled data were performed, Hazard Ratio (HR) and mean difference with corresponding confidence intervals (CIs) were calculated and summarized respectively.
Results
10 relevant articles were included for this meta-analysis study. HMGB1 mRNA levels in HCC were significantly higher than those in normal (p<0.00001) and para-tumor tissues (p = 0.002) respectively. The protein levels of HMGB1 in HCC were significantly higher than those in para-tumor tissues (p = 0.005). Two studies reported the serum HMGB1 levels in patients with HCC of TNM stages, and indicating significantly different between stage I and II, stage II and III, as well as stage III and IV (two studies showed p<0.01 and p<0.001 respectively). The overall survival (OS) was significantly shorter in HCC patients with high HMGB1 expression compared those with low HMGB1 expression and the pooled HR was 1.31 with 95% CI 1.20–1.44, Z = 5.82, p<0.0001. Two additional studies showed that there were higher serum HMGB1 levels in patients with chronic hepatitis than those in healthy people (p<0.05).
Conclusions
The results of this meta-analysis suggest that HMGB1 mRNA and protein tissue levels in the patients with HCC are significantly higher than those in para-tumor and normal liver tissues respectively. Tissue HMGB1 overexpression is a potential biomarker for HCC diagnosis, and it is significantly associated with the prognosis of patients with HCC.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer in men, worldwide, and seventh among women, and it is the second leading cause of cancer-related mortality globally [1]. HCC commonly occurs in Asia and Africa, and its incidence rate is starting to increase in Western countries [2], [3]. Due to lack of effective biomarkers for diagnosis and prognosis, HCC is frequently found in late stages, when curative therapy approaches like resection, liver transplantation, radio frequency ablation (RFA), and transarterial chemoembolization (TACE) don't produce satisfactory clinical outcomes. HCC patients present with high recurrence and metastasis [4], [5], the 5-year survival rate after surgery for HCC has been only 25%–39% [6]. Therefore, it is critical to identify the biomolecular markers for the diagnosis and prognosis to monitor recurrence of HCC.
The serum a-fetoprotein (AFP) has been widely used as a risk assessment factor in patients with cirrhosis and a screening method for early detection of HCC patients, and a prognostic factor for prediction of tumor recurrence. AFP is the only HCC biomarker that has been studied through to phase 5 of biomarker development [7]. However, its sensitivity and specificity vary significantly ranging from 40%–65% and 76%–96%, respectively [8], [9], [10]. Recent studies have identified other potential biomarkers for early detection of HCC, including the circulating AFP isoform AFP-L3 [11], [12], des-gamma-carboxy prothrombin (DCP) [12], Golgi protein-73 (GP73) [13] and circulating miRNA [14]. However, most of them are only in phase 1 or 2 stages, further studies are needed to determine whether these biomarkers can be translated from bench works to clinical settings. The effects of other biomarkers to predict the prognosis for HCC such as vascular endothelial growth factor (VEGF), Cyclin-dependent Kinases(CDK), β-catenin/Wnt pathway and microRNAs [15] have been controversial and again none of them has been applied to the clinical settings.
HMGB1 was discovered in calf thymus in 1973 and named for its electrophoretic mobility in polyacrylamide gels [16], [17]. HMGB1 is encoded on human chromosome 13q12-13 and consists of 215 amino acids. It is a highly conserved protein containing two DNA binding domains A and B as well as a negatively charged C-terminal tail domain [18], [19]. HMGB1 has different functions depending on its subcellular locations. HMGB1 is normally located in the nucleus where it acts as a DNA chaperon by regulating transcription, replication, recombination, repair, and genome stability [20]. In 2011, Hanahan and Weinberg proposed a new model to define the ten hallmarks for the tumor requirements [21]. Recent findings indicate that HMGB1 dysfunction is associated with every hallmark of cancer and contributes to cancer initiation and development [22]. HMGB1 overexpression and cytoplasmic localization have been observed in some cancer cells such as in colon cancer [23], gastrointestinal stromal tumors [24], cervical carcinomas [25] and melanoma [26]. Moreover, recent studies demonstrate that HMGB1 expression is a useful prognostic maker for colorectal cancer and diffuse malignant peritoneal mesothelioma [27], [28], [29]. Higher HMGB1 expression has been observed in HCC compared to normal liver tissues [30], [31], [32]. HMGB1 could be a potential biomarker that may be used to detect HCC at early stage and predict its prognosis. However, the prognostic value of HMGB1 for survival remains controversial due to limited statistic power. There is no meta-analysis study to reveal the association between HMGB1 expression and clinicopathological parameters of HCC. Therefore, we performed a meta-analysis to evaluate the association of HMGB1 expression with the clinicopathological parameters and prognosis in HCC.
Methods
Search strategy and selection criteria
The following electronic databases were searched for relevant articles without any language restrictions: Web of Science (1945 ∼ 2014), the Cochrane Library Database (Issue 12, 2014), PubMed (1966 ∼ 2014), EMBASE (1980 ∼ 2014), CINAHL (1982 ∼ 2014), CNKI, Google scholar, and the Chinese Biomedical Database (CBM) (1982 ∼ 2014). We searched articles using the following terms: HMGB1 or high motility group box 1 and hepatocellular carcinoma, liver cancer or HCC. There were 27 articles identified from PubMed, 31 articles from Web Science, 33 articles from EMBASE, 262 articles from CNKI. 5950 articles from Google scholar were identified, however, only 500 of them were screened by titles and abstracts since the rest of them were not relevant. Finally total 853 articles were screened by titles and abstracts.
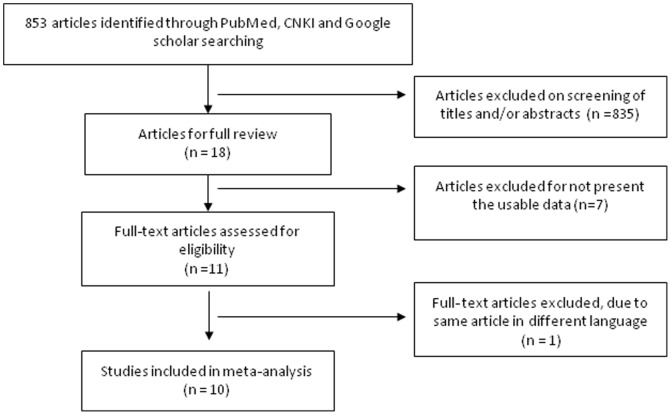
The inclusion criteria included: 1) The articles which the association between HMGB1 expression and the clinicopathological significance of HCC was evaluated; 2) The articles which the association of HMGB1 expression and prognosis in patients with HCC was evaluated; 3) The studies which utilized RT-PCR for detection of HMGB1 mRNA, ELISA for serum HMGB1 and Western Blot for tissue HMGB1 expression. The exclusion criteria included: 1) The studies which used the same population or overlapping database; 2) The studies of in vitro cell culture models (Figure 1). The search identified 18 articles of which 10 were eligible for quantitative analysis in this meta-analysis. The detailed information of 10 relevant citations was listed in Table 1.
Figure 1. Schematic flow diagram for selection of included studies.
Table 1. Main Characteristics of included studies.
| Author | Year | HCC Sample size | Comments |
| Liu et al. [31] | 2012 | 161 | High expression of HMGB1 predicts poor prognosis for HCC |
| Jiang et al. [30] | 2012 | 34 | HMGB1 is associated with clinicopathologic features in patients with HCC |
| Cheng et al. [70] | 2008 | 161 | Serum HMGB1 is associated with clinicopathologic features in patients with HCC |
| Xiao et al. [56] | 2014 | 208 | The association of HMGB1 Gene with the prognosis of HCC |
| Ueno et al. [32] | 2011 | 36 | Significance of HMGB1 expression in HCC |
| Han et al. [71] | 2014 | 13 | Changes of HMGB1 expression in HCC |
| Gou et al. [72] | 2013 | 82 | Detection and clinical significance of HMGB1 in HCC |
| Wang et al. [73] | 2013 | 39 | Increased HMGB1 expression in HCC and its mechanism |
| Feng et al. [74] | 2011 | 40 | The expression and clinical significance of HMGB1 in HCC |
| Wang et al. [57] | 2013 | 155 | Expression, subcellular location and prognostic relevance of HMGB1 in HCC |
Data extraction and study assessment
Two reviewers (Zhang and Han) independently extracted data and reviewed the contents of the manuscripts to determine whether they met the criteria for inclusion. Any discontent was discussed and resolved by a consensus including a third investigator (Wu). A data extract form was developed accordingly. One review author (Zhang) extracted the following data from the included studies: first author's name, year of publication, number of patients, stage of HCC, grade of HCC, and HMGB1 expression. The second author (Han) checked the extracted data, and disagreement was resolved by the discussion with the third author (Wu) for all issues.
Statistics analysis
All analysis was performed with Review Manager 5.2. Heterogeneity between studies was assessed using the Q-test and I 2 index. Mean differences with 95% confidence intervals were calculated by using a fixed or random effect model depending on heterogeneity (a fixed effect model for I 2<50%, a random effect model for I 2>50%). Meta-analysis was performed to compare HMGB1 mRNA expression between HCC and normal tissue, HMGB1 mRNA and protein expression levels between HCC and para-tumor tissue. Two studies investigated serum HMGB1 protein levels in TNM stages of HCC that were listed in Table 2. Another two studies detected serum HMGB1 levels in patients with hepatitis and normal controls, the data were listed in Table 3. The multivariate HRs were collected and the log HRs and its standard errors were calculated for individual study. Pooled hazard ratio (HR) with a 95% confidence interval was calculated for the association between HMGB1 expression and prognosis. All p values were two sided. Funnel plots were used for detection of publication bias. A sensitivity analysis, in which one study was removed at a time, was conducted to assess the result stability.
Table 2. Serum HMGB1 levels (ng/ml) in TNM stages of patients with HCC.
Table 3. Serum HMGB1 levels (ng/ml) in patients with hepatitis and normal controls.
Results
Total 853 articles were screened by titles and abstracts. The search identified 18 articles of which 10 were eligible for quantitative analysis in this meta-analysis. 7 articles were excluded due to different detect methods used. One article was excluded due to duplication. 10 relevant articles were included for full review in detail and 6 of them were conducted with meta-analysis (Figure 1). The following items were collected from each study: first author's name, year of publication, number of patients, stages of HCC, detect methods, HMGB1 expression and prognosis (Table 1 and 2).
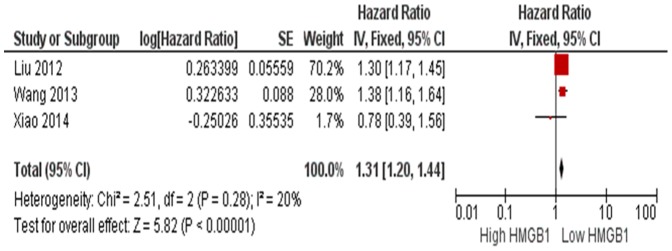
HMGB1 mRNA level in HCC was significantly higher than in normal and para-tumor tissues respectively. The pooled mean difference for HCC versus normal liver tissue was 0.38, (95% CI 0.22–0.54, Z = 4.59, p<0.00001, heterogeneity I 2 = 91%, Figure 2) and for HCC versus para-tumor, mean difference was 0.31, (95% CI 0.12–0.51, Z = 3.12, p = 0.002, heterogeneity I 2 = 93%, Figure 3). The protein level of HMGB1 in HCC was significantly higher than those in para-tumor tissues, the pooled mean difference was 0.51 with 95% CI 0.16–0.86, (Z = 2.83, p = 0.005, heterogeneity I 2 = 98%, Figure 4). Figure 5 showed the result of a meta-analysis of hazard ratios (HR) of HMGB1 high expression and low expression in the survival of patients with HCC. The overall survival (OS) was significantly shorter in HCC patients with high HMGB1 expression compared those with low HMGB1 expression, and the pooled HR was 1.31 with 95% CI 1.20–1.44, (Z = 5.82, p<0.0001, heterogeneity I 2 = 20%, Figure 5).
Figure 2. Forest plot for HMGB1mRNA/GADPH in HCC and normal liver tissue.
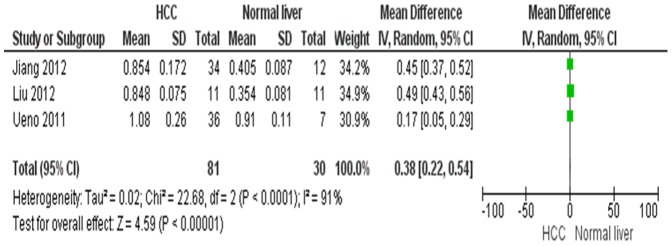
Figure 3. Forest plot for HMGB1mRNA/GADPH in HCC and para-tumor tissue.
Figure 4. Forest plot for HMGB1 protein/GADPH in HCC and para-tumor tissue.
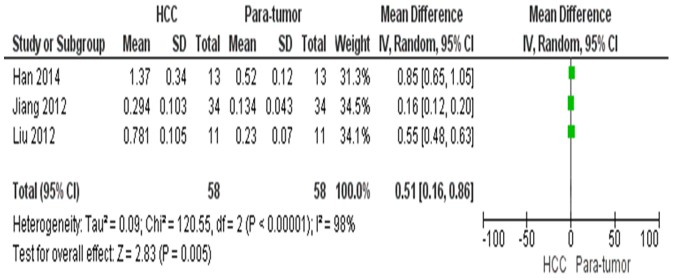
Figure 5. Forest plot for the association of HMGB1expression level and the risk of HCC.
Four of included studies were fully reviewed without performing meta-analysis due to less number of patients in the selected articles. Two studies reported the serum HMGB1 levels in patients with HCC of TNM stage I, II, III, IV (Table 2). The differences were significant between stage I and II, stage II and III, as well as stage III and IV groups (two studies showed p<0.01 and p<0.0001 respectively). Another two studies investigated the serum HMGB1 levels, suggesting there were higher serum HMGB1 levels in patients with chronic hepatitis than in healthy people (p<0.01 and p<0.05 respectively) (Table 3).
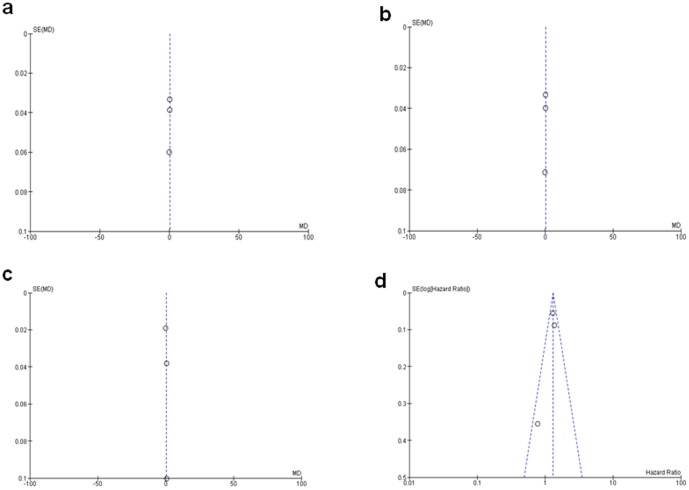
A sensitivity analysis, in which one study was removed at a time, was conducted to assess the result stability. The pooled HR was not significantly changed, indicating the stability of our analyses. The funnel plots were largely symmetric (Figure 6) suggesting there were no publication biases in the meta-analysis of HMGB1 expression and clinicopathological features.
Figure 6. Funnel plot for publication bias.
a: HMGB1 mRNA/GADPH in HCC and normal liver tissue; b: HMGB1 mRNA/GADPH in HCC and para-tumor tissue; c: HMGB1 protein/GADPH in HCC and para-tumor tissue; d: the association of HMGB1expression and the risk of HCC.
Discussion
For the first time, we conducted a meta-analysis and evaluated HMGB1 mRNA and protein expression levels in tissues from patients with HCC and normal liver or para-tumor tissues. The heterogeneity between studies was assessed using the Q-test and I 2 index. All I 2 indexes for comparisons of mRNA and protein expression between HCC and normal or para-tumor tissue were great than 75%, indicating that there was significant heterogeneity between studies. All p values for test of overall effect were equal or less than 0.005, indicating mRNA and protein expression levels in HCC were significantly increased compared to those in the normal and para-tumor tissues respectively. This suggests that HMGB1 plays an important role in the pathogenesis of HCC. HMGB1 is a potential tissue biomarker for HCC diagnosis.
Cancer development is a multistep process. Accumulated evidences indicate that HMGB1 is associated with ten functional capabilities of cancers as Hanahan and Weinberg described in 2011 [21]. Its function depends on the subcellular locations. Within the nucleus, HMGB1 acts as a DNA chaperon to bind to the minor groove of DNA, stabilizing nucleosides and facilitating the assembly of site-specific DNA-binding proteins such as the nuclear hormone/nuclear hormone receptor complex and p53, p73, as well as retinoblastoma protein (Rb) transcription complexes [33], [34]. When HMGB1 releases outside of the cell, it interacts with a wide range of proteins, such as the receptor of advanced glycation end products (RAGE), Toll-like receptors (TLRs: TLR2, TLR4 and TLR9), mitogen-activated protein kinase (MAPK), NF (nuclear factor)-κB, and coordinates immune system activation, cell migration, cell growth, angiogenesis, and tissue repair as well as regeneration [35], [36], [37], [38], [39], [40].
Recent studies report that the expression of HMGB1 is upregulated in several types of malignancies including gastric cancer [41], breast cancer [33], nasopharyngeal carcinoma [42], and squamous cell carcinoma of the head and neck [43], [44]. HMGB1 is not only constitutively expressed in the nucleus of cancer cells, but also is released by the tumor cells, macrophages, monocytes, dendritic cells (DCs), and T cells [45]. Released HMGB1 that acts as a proinflammatory cytokine from necrotic tumor cells creates a microenvironment and triggers signaling pathway, such as the NF-κB and inflammasome, to induce proinflammatory cytokine release. This loop accelerates inflammatory response and tumor formation [22], [46], [47]. Therefore, serum HMGB1 levels could be prediction for progression of patients with HCC.
Two studies reported the serum HMGB1 levels in patients with HCC of TNM stage I, II, III, IV. The differences were significant between stage I and II, stage II and III, as well as stage III and IV groups (two studies showed p<0.01 and p<0.0001 respectively). Higher HMGB1 expression was detected in advanced stages of HCC patients. This suggests that HMGB1 plays an important role in the invasion, metastasis and progression of HCC. In support of this, recent evidences indicated HMGB1 was co-localized with RAGE, suggesting their potential contribution to cellular migration and tumor invasion. The expression of RAGE was closely associated with the invasive and metastatic behavior in different types of cancer such as gastric [48] and colorectal cancer [49]. The interaction of HMGB1 with RAGE leads to activation of the NF-κB, MAPK, and type IV collagenase (MMP-2/MMP-9) signaling pathways, all of which degrade extracellular matrix protein and play a major role in tumor invasion and metastasis [50], [51], [52], [53]. Furthermore, tumor growth and metastasis were suppressed by blocking RAGE-HMGB1 complex in human and murine cancer cells [52], [54]. Knockdown of HMGB1 gene also inhibited liver cancer growth and metastasis [31], [55]. Therefore, the overexpression of HMGB1 is not only a potential biomarker for initial diagnosis of HCC, but it also could be a predictor of prognosis in patients with HCC. More studies are needed for a meta-analysis in the future to confirm the progression value of HMGB1 in the patients with HCC.
A few of studies reported controversial results of the association between HMGB1 expression and the prognosis of patients with HCC [31], [56], [57]. Those studies on prognostic significance of HMGB1 expression were limited by low statistical power. We performed a meta-analysis for the first time and pooled three studies together (n = 524). The low heterogeneity (I 2 = 20%) could be due to different methods to measure HMGB1 intensity in immunohistochemistry. Our pooled HR was 1.31 with 95% CI 1.20–1.44, test for overall effect p<0.0001, suggesting that high HMGB1 expression was significantly associated with the mortality of patients with HCC. Compared with low HMGB1 expression, HMGB1 high expression was associated with more than 1.3 times the risk of mortality. HMGB1 affects the prognosis of HCC through complicated pathways. HMGB1 has the following abilities to induce tumorigenesis: 1). tumor cells and tumor-infiltrating leukocytes secret HMGB1 in tumor microenvironment which activates NK-κB and inflammatory pathways, promoting tumor growth, invasion and metastasis [46], [47], [58]; 2). HMGB1 also has the ability to increase the production of ATP and provide more energy for tumor growth [59]; 3). In addition, HMGB1 promotes angiogenesis [60], [61], [62], [63] for tumor growth and metastasis; 4). Lastly, HMGB1 inhibits antitumor immunity for tumor survival [64], [65], [66]. All these activities contribute to poor prognosis of HCC patients with high expression of HMGB1. Therefore, HMGB1 overexpression predicts a poor prognosis in patients with HCC.
Interestingly, two studies investigated serum HMGB1 in patients with chronic hepatitis (the precursor of HCC) and healthy people. Significantly higher serum HMGB1 was detected in the patients with chronic hepatitis compared to those detected in healthy people. The difference of serum HMGB1 levels between the two studies could be due to that different methods were used to detect the serum concentration of HMGB1. HMGB1 lacks a classic signal sequence. During infection, HMGB1 is translocated from the nucleus to the cytoplasm and is subsequently released into the extracellular milieu by HCV or HBV infection [67], [68], [69]. HMGB1 is likely to be crucial in initiating and mediating hepatitis. Further studies are needed to verify its role in the pathogenesis of chronic hepatitis and subsequently in HCC.
We evaluated the HMGB1 expression in patients with HCC (both tumor and para-tumor tissues) and normal liver tissues. Further studies should be focused on finding cut-offs for the diagnosis of HCC and determination of its stages. In the future, more studies are needed to verify if HMGB1 blockage can be used in the development of new therapeutic agents for HCC. Finally, our study only selected the published articles, but it did not include some relevant unpublished papers which may result in certain publication bias. Thus the result should be interpreted carefully.
In conclusion, HMGB1 mRNA and protein tissue levels in the patients with HCC are significantly higher than in those of para-tumor and normal liver tissues respectively. HMGB1 overexpression is a potential biomarker for HCC diagnosis, and it significantly associated with the prognosis of patients with HCC.
Supporting Information
A PRISMA Checklist.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding from Jinan Municipal Science and Technology Development Plan (201401246) and Natural Science Foundation of Shandong Province (ZR2013H047). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55: 74–108. [DOI] [PubMed] [Google Scholar]
- 3. Bosch FX, Ribes J, Diaz M, Cleries R (2004) Primary liver cancer: worldwide incidence and trends. Gastroenterology 127: S5–S16. [DOI] [PubMed] [Google Scholar]
- 4. El-Serag HB, Marrero JA, Rudolph L, Reddy KR (2008) Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology 134: 1752–1763. [DOI] [PubMed] [Google Scholar]
- 5. Forner A, Bruix J (2008) Locoregional treatment for hepatocellular carcinoma: From clinical exploration to robust clinical data, changing standards of care. Hepatology 47: 5–7. [DOI] [PubMed] [Google Scholar]
- 6. Thomas MB, Zhu AX (2005) Hepatocellular carcinoma: the need for progress. J Clin Oncol 23: 2892–2899. [DOI] [PubMed] [Google Scholar]
- 7. Zhang BH, Yang BH, Tang ZY (2004) Randomized controlled trial of screening for hepatocellular carcinoma. J Cancer Res Clin Oncol 130: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trevisani F, D'Intino PE, Morselli-Labate AM, Mazzella G, Accogli E, et al. (2001) Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol 34: 570–575. [DOI] [PubMed] [Google Scholar]
- 9. Yoshida S, Kurokohchi K, Arima K, Masaki T, Hosomi N, et al. (2002) Clinical significance of lens culinaris agglutinin-reactive fraction of serum alpha-fetoprotein in patients with hepatocellular carcinoma. Int J Oncol 20: 305–309. [PubMed] [Google Scholar]
- 10. Khien VV, Mao HV, Chinh TT, Ha PT, Bang MH, et al. (2001) Clinical evaluation of lentil lectin-reactive alpha-fetoprotein-L3 in histology-proven hepatocellular carcinoma. Int J Biol Markers 16: 105–111. [DOI] [PubMed] [Google Scholar]
- 11. Yamamoto K, Imamura H, Matsuyama Y, Kume Y, Ikeda H, et al. (2010) AFP, AFP-L3, DCP, and GP73 as markers for monitoring treatment response and recurrence and as surrogate markers of clinicopathological variables of HCC. J Gastroenterol 45: 1272–1282. [DOI] [PubMed] [Google Scholar]
- 12. Nakamura S, Nouso K, Sakaguchi K, Ito YM, Ohashi Y, et al. (2006) Sensitivity and specificity of des-gamma-carboxy prothrombin for diagnosis of patients with hepatocellular carcinomas varies according to tumor size. Am J Gastroenterol 101: 2038–2043. [DOI] [PubMed] [Google Scholar]
- 13. Tian L, Wang Y, Xu D, Gui J, Jia X, et al. (2011) Serological AFP/Golgi protein 73 could be a new diagnostic parameter of hepatic diseases. Int J Cancer 129: 1923–1931. [DOI] [PubMed] [Google Scholar]
- 14. Qi J, Wang J, Katayama H, Sen S, Liu SM (2013) Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma 60: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinato DJ, Pirisi M, Maslen L, Sharma R (2014) Tissue biomarkers of prognostic significance in hepatocellular carcinoma. Adv Anat Pathol 21: 270–284. [DOI] [PubMed] [Google Scholar]
- 16. Goodwin GH, Johns EW (1973) Isolation and characterisation of two calf-thymus chromatin non-histone proteins with high contents of acidic and basic amino acids. Eur J Biochem 40: 215–219. [DOI] [PubMed] [Google Scholar]
- 17. Goodwin GH SC, Johns EW (1973) A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem 38: 14–19. [DOI] [PubMed] [Google Scholar]
- 18. Weir HM, Kraulis PJ, Hill CS, Raine AR, Laue ED, et al. (1993) Structure of the HMG box motif in the B-domain of HMG1. EMBO J 12: 1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Read CM, Cary PD, Crane-Robinson C, Driscoll PC, Norman DG (1993) Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res 21: 3427–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Prasad R, Wilson SH (2010) HMGB1: roles in base excision repair and related function. Biochim Biophys Acta 1799: 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 22. Tang D, Kang R, Zeh HJ 3rd, Lotze MT (2010) High-mobility group box 1 and cancer. Biochim Biophys Acta 1799: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Volp K, Brezniceanu ML, Bosser S, Brabletz T, Kirchner T, et al. (2006) Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut 55: 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi YR, Kim H, Kang HJ, Kim NG, Kim JJ, et al. (2003) Overexpression of high mobility group box 1 in gastrointestinal stromal tumors with KIT mutation. Cancer Res 63: 2188–2193. [PubMed] [Google Scholar]
- 25. Hao Q, Du XQ, Fu X, Tian J (2008) [Expression and clinical significance of HMGB1 and RAGE in cervical squamous cell carcinoma]. Zhonghua Zhong Liu Za Zhi 30: 292–295. [PubMed] [Google Scholar]
- 26. Poser I, Golob M, Buettner R, Bosserhoff AK (2003) Upregulation of HMG1 leads to melanoma inhibitory activity expression in malignant melanoma cells and contributes to their malignancy phenotype. Mol Cell Biol 23: 2991–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tabata C, Shibata E, Tabata R, Kanemura S, Mikami K, et al. (2013) Serum HMGB1 as a prognostic marker for malignant pleural mesothelioma. BMC Cancer 13: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fahmueller YN, Nagel D, Hoffmann RT, Tatsch K, Jakobs T, et al. (2013) Immunogenic cell death biomarkers HMGB1, RAGE, and DNAse indicate response to radioembolization therapy and prognosis in colorectal cancer patients. Int J Cancer 132: 2349–2358. [DOI] [PubMed] [Google Scholar]
- 29. Chiharu Tabata ES (2013) Rie Tabata Shingo Kanemura Koji Mikami Yoshitaka Nogi, (2013) et al. Serum HMGB1 as a prognostic marker for malignant pleural mesothelioma. BMC Cancer 13: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang W, Wang Z, Li X, Fan X, Duan Y (2012) High-mobility group box 1 is associated with clinicopathologic features in patients with hepatocellular carcinoma. Pathol Oncol Res 18: 293–298. [DOI] [PubMed] [Google Scholar]
- 31. Liu F, Zhang Y, Peng Z, Gao H, Xu L, et al. (2012) High expression of high mobility group box 1 (hmgb1) predicts poor prognosis for hepatocellular carcinoma after curative hepatectomy. J Transl Med 10: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ueno S HK, Sakoda M, Iino S, Kurahara H MK, Ando K, et al. (2011) Significance of High Mobility Group Box-1 (HMGB1) Expression in Hepatocellular Carcinoma. Med J Kago Shima Univ 62: 27–35. [Google Scholar]
- 33. Brezniceanu ML, Volp K, Bosser S, Solbach C, Lichter P, et al. (2003) HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. FASEB J 17: 1295–1297. [DOI] [PubMed] [Google Scholar]
- 34. Thomas JO, Travers AA (2001) HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci 26: 167–174. [DOI] [PubMed] [Google Scholar]
- 35. Ghavami S, Rashedi I, Dattilo BM, Eshraghi M, Chazin WJ, et al. (2008) S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol 83: 1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palumbo R, Galvez BG, Pusterla T, De Marchis F, Cossu G, et al. (2007) Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-kappaB activation. J Cell Biol 179: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dong Xda E, Ito N, Lotze MT, Demarco RA, Popovic P, et al. (2007) High mobility group box I (HMGB1) release from tumor cells after treatment: implications for development of targeted chemoimmunotherapy. J Immunother 30: 596–606. [DOI] [PubMed] [Google Scholar]
- 38. Scaffidi P, Misteli T, Bianchi ME (2002) Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418: 191–195. [DOI] [PubMed] [Google Scholar]
- 39. Tian J, Avalos AM, Mao SY, Chen B, Senthil K, et al. (2007) Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol 8: 487–496. [DOI] [PubMed] [Google Scholar]
- 40. Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, et al. (1999) HMG-1 as a late mediator of endotoxin lethality in mice. Science 285: 248–251. [DOI] [PubMed] [Google Scholar]
- 41. Akaike H, Kono K, Sugai H, Takahashi A, Mimura K, et al. (2007) Expression of high mobility group box chromosomal protein-1 (HMGB-1) in gastric cancer. Anticancer Res 27: 449–457. [PubMed] [Google Scholar]
- 42. Wu D, Ding Y, Wang S, Zhang Q, Liu L (2008) Increased expression of high mobility group box 1 (HMGB1) is associated with progression and poor prognosis in human nasopharyngeal carcinoma. J Pathol 216: 167–175. [DOI] [PubMed] [Google Scholar]
- 43. Liu Y, Xie C, Zhang X, Huang D, Zhou X, et al. (2010) Elevated expression of HMGB1 in squamous-cell carcinoma of the head and neck and its clinical significance. Eur J Cancer 46: 3007–3015. [DOI] [PubMed] [Google Scholar]
- 44. Lotze MT, Tracey KJ (2005) High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol 5: 331–342. [DOI] [PubMed] [Google Scholar]
- 45. Naglova H, Bucova M (2012) HMGB1 and its physiological and pathological roles. Bratisl Lek Listy 113: 163–171. [DOI] [PubMed] [Google Scholar]
- 46. Gebhardt C, Riehl A, Durchdewald M, Nemeth J, Furstenberger G, et al. (2008) RAGE signaling sustains inflammation and promotes tumor development. J Exp Med 205: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mittal D, Saccheri F, Venereau E, Pusterla T, Bianchi ME, et al. (2010) TLR4-mediated skin carcinogenesis is dependent on immune and radioresistant cells. EMBO J 29: 2242–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuniyasu H, Oue N, Wakikawa A, Shigeishi H, Matsutani N, et al. (2002) Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J Pathol 196: 163–170. [DOI] [PubMed] [Google Scholar]
- 49. Sasahira T, Akama Y, Fujii K, Kuniyasu H (2005) Expression of receptor for advanced glycation end products and HMGB1/amphoterin in colorectal adenomas. Virchows Arch 446: 411–415. [DOI] [PubMed] [Google Scholar]
- 50. Dumitriu IE, Baruah P, Manfredi AA, Bianchi ME, Rovere-Querini P (2005) HMGB1: guiding immunity from within. Trends Immunol 26: 381–387. [DOI] [PubMed] [Google Scholar]
- 51. Evans A, Lennard TW, Davies BR (2004) High-mobility group protein 1(Y): metastasis-associated or metastasis-inducing? J Surg Oncol 88: 86–99. [DOI] [PubMed] [Google Scholar]
- 52. Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, et al. (2000) Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 405: 354–360. [DOI] [PubMed] [Google Scholar]
- 53. Takada M, Hirata K, Ajiki T, Suzuki Y, Kuroda Y (2004) Expression of receptor for advanced glycation end products (RAGE) and MMP-9 in human pancreatic cancer cells. Hepatogastroenterology 51: 928–930. [PubMed] [Google Scholar]
- 54. Huttunen HJ, Fages C, Kuja-Panula J, Ridley AJ, Rauvala H (2002) Receptor for advanced glycation end products-binding COOH-terminal motif of amphoterin inhibits invasive migration and metastasis. Cancer Res 62: 4805–4811. [PubMed] [Google Scholar]
- 55. Dong YD, Cui L, Peng CH, Cheng DF, Han BS, et al. (2013) Expression and clinical significance of HMGB1 in human liver cancer: Knockdown inhibits tumor growth and metastasis in vitro and in vivo. Oncol Rep 29: 87–94. [DOI] [PubMed] [Google Scholar]
- 56. Xiao J, Ding Y, Huang J, Li Q, Liu Y, et al. (2014) The association of HMGB1 gene with the prognosis of HCC. PLoS One 9: e89097. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Wang Y (2013) Expression,Subcellular Location and Prognostic Relevance of High Mobility Group Box 1(HMGB1) in Hepatocellular Carcinoma. University of Science and Technology Graduation Thesis 2013: 11–14 Available: http://cdmd.cnki.com.cn/Article/CDMD-10487-1013273569.htm. [Google Scholar]
- 58. Jube S, Rivera ZS, Bianchi ME, Powers A, Wang E, et al. (2012) Cancer cell secretion of the DAMP protein HMGB1 supports progression in malignant mesothelioma. Cancer Res 72: 3290–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kang R, Tang D, Schapiro NE, Loux T, Livesey KM, et al. (2014) The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 33: 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yan W, Chang Y, Liang X, Cardinal JS, Huang H, et al. (2012) High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology 55: 1863–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tafani M, Schito L, Pellegrini L, Villanova L, Marfe G, et al. (2011) Hypoxia-increased RAGE and P2X7R expression regulates tumor cell invasion through phosphorylation of Erk1/2 and Akt and nuclear translocation of NF-{kappa}B. Carcinogenesis 32: 1167–1175. [DOI] [PubMed] [Google Scholar]
- 62. van Beijnum JR, Nowak-Sliwinska P, van den Boezem E, Hautvast P, Buurman WA, et al. (2013) Tumor angiogenesis is enforced by autocrine regulation of high-mobility group box 1. Oncogene 32: 363–374. [DOI] [PubMed] [Google Scholar]
- 63. Sasahira T, Kirita T, Bhawal UK, Ikeda M, Nagasawa A, et al. (2007) The expression of receptor for advanced glycation end products is associated with angiogenesis in human oral squamous cell carcinoma. Virchows Arch 450: 287–295. [DOI] [PubMed] [Google Scholar]
- 64. Kusume A, Sasahira T, Luo Y, Isobe M, Nakagawa N, et al. (2009) Suppression of dendritic cells by HMGB1 is associated with lymph node metastasis of human colon cancer. Pathobiology 76: 155–162. [DOI] [PubMed] [Google Scholar]
- 65. Liu Z, Falo LD Jr, You Z (2011) Knockdown of HMGB1 in tumor cells attenuates their ability to induce regulatory T cells and uncovers naturally acquired CD8 T cell-dependent antitumor immunity. J Immunol 187: 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. He Y, Zha J, Wang Y, Liu W, Yang X, et al. (2013) Tissue damage-associated “danger signals” influence T-cell responses that promote the progression of preneoplasia to cancer. Cancer Res 73: 629–639. [DOI] [PubMed] [Google Scholar]
- 67. Liu HB, Fan XG, Huang JJ, Li N, Peng JP, et al. (2007) [Serum level of HMGB1 in patients with hepatitis B and its clinical significance]. Zhonghua Gan Zang Bing Za Zhi 15: 812–815. [PubMed] [Google Scholar]
- 68. Jung JH, Park JH, Jee MH, Keum SJ, Cho MS, et al. (2011) Hepatitis C virus infection is blocked by HMGB1 released from virus-infected cells. J Virol 85: 9359–9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou RR, Zhao SS, Zou MX, Zhang P, Zhang BX, et al. (2011) HMGB1 cytoplasmic translocation in patients with acute liver failure. BMC Gastroenterol 11: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cheng BQ, Jia CQ, Liu CT, Lu XF, Zhong N, et al. (2008) Serum high mobility group box chromosomal protein 1 is associated with clinicopathologic features in patients with hepatocellular carcinoma. Dig Liver Dis 40: 446–452. [DOI] [PubMed] [Google Scholar]
- 71.Han F WT, Chen G (2014) Changes of high mobility group box 1 expression in hepatocellular carcinoma tissues. Journal of Medical Postgraduates 4.. [Google Scholar]
- 72.Gou H SL, Han L (2013) Detection and clinical significance of high mobility group box protein 1 in liver cancer. Journal of Preventive Medicine 40.. [Google Scholar]
- 73. Wang T HF, Liang J, Wang F, Chen G (2013) Increased HMGB1 expression in primary hepatic carcinoma and its mechanism Chinese Journal of Laboratory Diagnosis. 17: 1783–1785. [Google Scholar]
- 74.Feng H (2011) The Expression and Clinical Significance of High Mobility Group Protein B 1 in Hepatocellular Carcinoma Chongqing Medical University Graduation Thesis: 11–17. Available: http://cdmd.cnki.com.cn/Article/CDMD-10631-1011173410.htm
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A PRISMA Checklist.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.