Abstract
Members of the plant NUCLEAR FACTOR Y (NF-Y) family are composed of the NF-YA, NF-YB, and NF-YC subunits. In Brassica napus (canola), each of these subunits forms a multimember subfamily. Plant NF-Ys were reported to be involved in several abiotic stresses. In this study, we demonstrated that multiple members of thirty three BnNF-Ys responded rapidly to salinity, drought, or ABA treatments. Transcripts of five BnNF-YAs, seven BnNF-YBs, and two BnNF-YCs were up-regulated by salinity stress, whereas the expression of thirteen BnNF-YAs, ten BnNF-YBs, and four BnNF-YCs were induced by drought stress. Under NaCl treatments, the expression of one BnNF-YA10 and four NF-YBs (BnNF-YB3, BnNF-YB7, BnNF-YB10, and BnNF-YB14) were greatly increased. Under PEG treatments, the expression levels of four NF-YAs (BnNF-YA9, BnNF-YA10, BnNF-YA11, and BnNF-YA12) and five NF-YBs (BnNF-YB1, BnNF-YB8, BnNF-YB10, BnNF-YB13, and BnNF-YB14) were greatly induced. The expression profiles of 20 of the 27 salinity- or drought-induced BnNF-Ys were also affected by ABA treatment. The expression levels of six NF-YAs (BnNF-YA1, BnNF-YA7, BnNF-YA8, BnNF-YA9, BnNF-YA10, and BnNF-YA12) and seven BnNF-YB members (BnNF-YB2, BnNF-YB3, BnNF-YB7, BnNF-YB10, BnNF-YB11, BnNF-YB13, and BnNF-YB14) and two NF-YC members (BnNF-YC2 and BnNF-YC3) were greatly up-regulated by ABA treatments. Only a few BnNF-Ys were inhibited by the above three treatments. Several NF-Y subfamily members exhibited collinear expression patterns. The promoters of all stress-responsive BnNF-Ys harbored at least two types of stress-related cis-elements, such as ABRE, DRE, MYB, or MYC. The cis-element organization of BnNF-Ys was similar to that of Arabidopsis thaliana, and the promoter regions exhibited higher levels of nucleotide sequence identity with Brassica rapa than with Brassica oleracea. This work represents an entry point for investigating the roles of canola NF-Y proteins during abiotic stress responses and provides insight into the genetic evolution of Brassica NF-Ys.
Introduction
Abiotic stress, such as salinity, drought or dramatic temperature change, adversely affects plant growth and productivity. Plants, being sessile, have evolved a range of strategies to adapt to detrimental environmental conditions. Plants respond to unfavorable conditions through two pathways, i.e., the abscisic acid (ABA)-dependent and ABA-independent pathway [1]–[3]. Transcription factors (TFs) play vital roles in mediating stress tolerance in both of these pathways.
NUCLEAR FACTOR Y (NF-Y) TFs are ubiquitous in higher eukaryotes and belong to the CCAAT-binding factor (CBF) family, also known as the Heme Activator Protein (HAP) family in yeast. The NF-Y heterocomplex consists of at least three subunits, namely NF-YA (also named CBF-B or HAP2), NF-YB (CBF-A or HAP3), and NF-YC (CBF-C or HAP5) [4]. In planta, each NF-Y subunit is encoded by a multi-member family, and there are at least 10 annotated members in each NF-Y subfamily in Arabidopsis [5]–[7]. Rice harbors at least 10 NF-YA genes, 12 NF-YB genes, and 8 NF-YC genes [8]. In a previous study, we identified 14 NF-YA genes, 14 NF-YB genes, and 5 NF-YC genes in Brassica napus L. [9]. NF-Y family members are extensively involved in plant development and stress responses, as summarized in a recent review [10].
Increasing evidence suggests that NF-Y subunits are important regulators of abiotic stress responses [11]–[14]. Overexpression of NF-YA1 in Arabidopsis was shown to result in hypersensitivity to salt stress during early postgerminative growth [15]. Real-time PCR analysis revealed that Glycine max (soybean) GmNF-YA3 was induced by ABA and other abiotic stresses, and that overexpression of GmNF-YA3 in Arabidopsis enhanced drought resistance [16]. A microarray analysis identified Arabidopsis NF-YB2 as one of the most strongly induced genes under multiple treatments, including cold, mannitol (dehydration stress), and NaCl (salinity) [17]. Further analysis confirmed that AtNF-YB2 functions in early flowering under osmotic stress [12]. AtNF-YB1 was found to improve plant performance under drought conditions and its ortholog in maize, ZmNF-YB2, was shown to resist drought stress in the field [11]. Several drought-related NF-YB genes were identified in Triticum aestivum (wheat), Populus euphratica (poplar), and Hordeum vulgare (barley) [18]–[20]. In the NF-YC subfamily, the transcript level of AtNF-YC2 was highly induced by light, oxidative, heat, cold, and drought stress [21]. Overexpression of PwHAP5 (homolog of Arabidopsis NF-YC2) from the conifer (Picea wilsonii) partially rescued the increased sensitivity of nf-yc2 to salt, drought, and ABA treatments [22].
In addition, several members of the NF-Y family were shown to be regulated by the microRNA169 (miR169) family, suggesting that a complex regulatory cascade is activated under stress conditions [13], [14], [23]. Li et al. (2008) demonstrated that AtNF-YA5 is a target of miRNA169a or miRNA169c [13]. At least six other miR169 targets were predicted to exist in the AtNF-YA family [24]. Of those six NF-YAs, NF-YA2, 3, 7, and 10 were found to be involved in multiple abiotic stresses, especially those involving low nutrient availability [25]. In rice, an NF-YA gene (Os03g29760) known to be a target of the miR169 family was found to be induced by high salinity [14]. The above results suggest that plant NF-Ys play a crucial role in the plant’s response to abiotic stress. However, the roles of BnNF-Ys under stress have not been examined in detail.
Canola (B. napus) constitutes the third largest vegetable oil source in the world. However, salinity and drought stress have a major limiting impact on its production and yield stability. Drought accounts for at least a 30% yield loss in canola every year [26]. In this study, we examined the expression patterns of BnNF-Ys under drought, high salinity, and ABA conditions. Our results suggest that 27 of 33 putative BnNF-Ys were induced by salt or drought stresses. Furthermore, 20 BnNF-Ys may be involved in the ABA-related pathway. We cloned the promoters of 28 BnNF-Ys from canola, which mostly included regions ∼1000bp upstream of the coding sequences, and identified their stress-related cis-elements. Taken together, our results suggest that most BnNF-Y members are regulated by abiotic stresses in an abscisic acid (ABA)-dependent or ABA-independent manner and serve as an entry point to investigate the roles of these genes in the plant’s response to environmental stresses.
Materials and Methods
Plant materials and growth conditions
Seeds of oilseed rape (Brassica napus ‘Nanyanyou 1’, kindly provided by Professor Zhaopu Liu at Nanjing Agricultural University) were kept in darkness for 2 days and germinated in Hoagland nutrient solution. The plants were grown for 3 weeks in the greenhouse with a light intensity of 392∼415µmol m−2 s−1 during a daily cycle consisting of 16h of light at 25°C and 8h of darkness at 18°C. For dehydration and salt stress treatments, plants were grown hydroponically in solution containing 15% (w/v) PEG6000 and 150mM NaCl plus ½ Hoagland nutrient solutions for 1 or 3h, respectively. For ABA treatments, 100µM ABA was added to the ½ Hoagland nutrient solution and the plants were treated for 1 or 3h. Seedlings growing on ½ Hoagland nutrient solution for various periods of time were used as the control. The harvested samples were immediately frozen in liquid nitrogen and stored at −80°C until used for gene expression analysis. All experiments were performed in biological triplicate.
Genomic DNA extraction, total RNA isolation, and primary cDNA synthesis
Genomic DNA was extracted from oilseed rape using a method from Rogers and Bendich’s CTAB-based protocol [27]. Total RNA of each sample was extracted using an E.Z.N.A. Plant RNA Kit (Omega Biotek, Cat#R6827, USA). The quality and quantity of extracted DNA and RNA from all samples were confirmed by both agarose gel electrophoresis and spectrophotometry (Thermo Scientific, NanoDropTM 1000, USA). Total RNA samples were pretreated with an RNase-Free DNase Set (Omega Biotek, Cat#E1091, USA) to eliminate residual genomic DNA. Primary cDNA was synthesized using a PrimeScript RT-PCR Kit (TaKaRa Code: D6110A, Japan), according to the manufacturer’s instructions.
Analysis of Gene Expression by Quantitative real-time PCR (qRT-PCR)
Quantitative real-time PCR was performed on an Applied Biosystems 7500 real-time PCR system using the SYBR Premix ExTaqTM Kit (TaKaRa Code: DRR041A, Japan). Sequence data, primers, and the PCR procedure used in this study were provided in our previous study [9]. All sequences of qRT-PCR amplification products were verified by sequencing. 18S rRNA was used as an internal control for expression analysis. Data were processed using the 2−ΔΔCT method [28], [29].
The transcript levels of each BnNF-Y gene in each tissue were first normalized to those of the housekeeping gene 18S and then compared to the transcript levels of each time-point control to obtain the relative gene expression level (i.e., the levels of expression in the same untreated tissue at the corresponding time point). Each data point represents the mean ±SE of three independent experiments. For the figures S1-S5, the transcript levels of each BnNF-Y genes were first normalized to those of the housekeeping gene 18S and then compared to the levels in the 0-h leaf control.
Statistical Analysis and Correlation Analysis
Significant differences between three treated samples and three untreated controls (same tissue and time-point only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS 18.0 (SPSS Corp., Chicago, IL, USA). Figures were created using Sigma Plot 10.0 (Systat Software, Inc. Germany). Correlation analysis of expression patterns was determined using relative gene expression levels from Fig. S1, Fig. S2, and Fig. S3. The regression coefficient was calculated in Microsoft Excel.
Measurements of Relative Water Content (RWC) from Drought-Stressed Plants
RWC measurements under PEG treatments were conducted as previously described [20].
Isolation of NF-Y promoters and Identification of their regulatory elements
To identify the promoter regions of NF-Ys from canola, each BnNF-Y coding region was used as a query in a BLASTN search against the Brassica database (http://brassicadb.org/brad/index.php), which includes B. rapa and B. oleracea sequences, and the highest hits were identified. Based on BnNF-Y coding sequences and the promoter sequences of the retrieved orthologous Brassica NF-Ys, most promoter regions of BnNF-Y sequences could be amplified with the primers listed in Table S1. Others promoter regions were amplified using a Genome Walking Kit (TaKaRa Code: D316, Table S1). In addition, the promoters of Arabidopsis NF-Y sequences were downloaded from the Plant Promoter Database (http://133.66.216.33/ppdb/cgi-bin/index.cgi). Sequences ∼1000bp upstream of the ATG start codon of each gene were retrieved from the genome, and known elements were identified using Plant Cis-element Regulatory DNA Elements (PLACE) (http://www.dna.affrc.go.jp/PLACE/) and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Phylogenetic tree analysis
The phylogenetic tree based on the promoter regions of plant NF-Ys was constructed using the neighbor-joining method implemented in MEGA software (version 4.1) with the following parameters: Jones–Taylor–Thornton (JTT) model and 1,000 bootstrap replicates.
Results
Expression Patterns of BnNF-Y Genes in the Leaves and Roots of Canola in Response to Salinity Stress
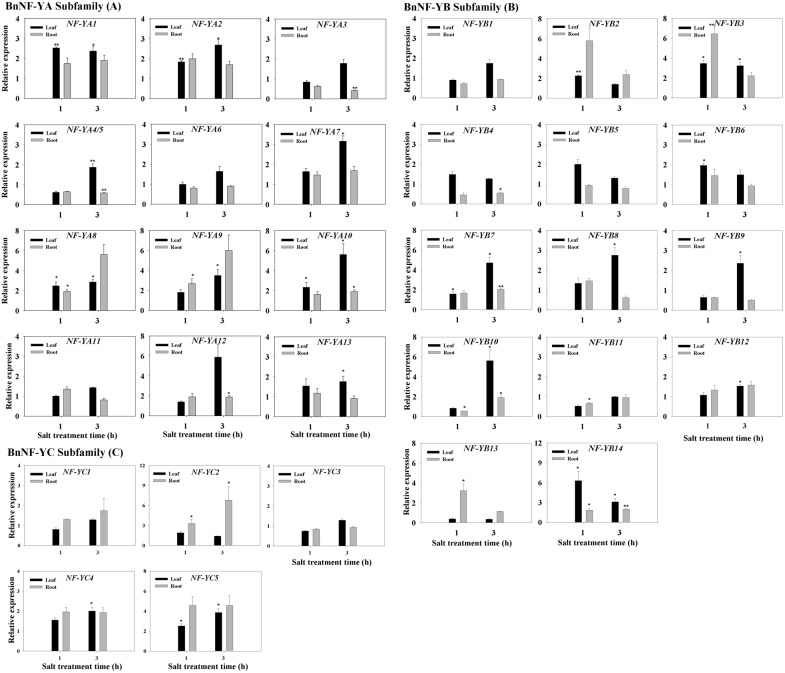
We previously identified BnNF-Y TF families in canola (B. napus L.) [9]. Since numerous reports have demonstrated that plant NF-Ys function in abiotic stress responses [13], [15], we examined whether BnNF-Ys had similar roles. To investigate whether BnNF-Y genes were responsive to salt stress, we monitored the transcript levels of all 33 BnNF-Ys in both the leaves and roots up to 3h of exposure to high salinity. First, we quantified the transcript levels of each BnNF-Y gene in different tissues and expressed these levels relative to those in 0-h untreated leaves. In this study, a gene was considered to be induced or down-regulated if its expression levels were both significantly different from those of an untreated control and >2-fold higher or lower than the control values, respectively. As shown in Fig. S1A, S2A and S3A, in the absence of any stress, the expression levels of most BnNF-YA subfamily members were not significantly altered during a 3-h observation period, except for BnNF-YA4/5, BnNF-YA11, BnNF-YA12, and BnNF-YA13. The transcription levels of BnNF-YA4/5 (BnNF-YA4 and BnNF-YA5 are alternative splice variants, with BnNF-YA4 being 11 amino acids shorter than BnNF-YA5) [9], and BnNF-YA11 were up-regulated in the 3-h root samples, while levels of BnNF-YA12 and BnNF-YA13 were down-regulated in 1-h leaf samples. This implies that these BnNF-YAs are subject to circadian clock regulation. To minimize the effect of circadian-mediated changes, we recalculated the expression levels by dividing the expression level of each gene to the corresponding gene expression level in the untreated control at each time point and comparing values in the same tissues. As shown in Fig. 1A, the transcript levels of BnNF-YA1, BnNF-YA2, BnNF-YA8 and BnNF-YA10 in leaves were higher than those of the control after 1h of NaCl treatment and remained elevated up to 3h of treatment, whereas those of, BnNF-YA7 and BnNF-YA9 in leaves were higher than those of the control up to 3h of treatment. In roots, two BnNF-YA subfamily members (BnNF-YA9 and BnNF-YA10) were induced at 1h or 3h. Most salt-responsive BnNF-YAs were only moderately up-regulated (2∼4 fold) upon exposure to salt stress, whereas BnNF-YA3 and BnNF-YA4/5 were slightly down-regulated in roots upon NaCl treatment.
Figure 1. Expression pattern of the BnNF-Y genes in the leaves and roots of plants subjected to salinity stress.
The expression of BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) genes in the leaves and roots of plants exposed to 150mM NaCl. Three-week-old canola seedlings were treated with 150mM NaCl for 1 and 3h. Total RNA was extracted from leaves (L) and roots (R) for quantitative PCR (qRT-PCR) analysis. Transcript levels of each BnNF-Y were first normalized to those of the housekeeping gene 18S and then compared to each time-point control (i.e., the level of untreated sample at the corresponding time point). Each data point represents the mean ±SE of three independent experiments. Significant differences between treated samples and untreated controls (same tissue only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS. Expression levels of untreated samples (C, 1-h or 3-h leaf and root samples) were arbitrarily set to 1.0.
We found that the expression of only two members of the BnNF-YB subfamily (i.e., BnNF-YB10, and BnNF-YB13) increased during a 3-h period in the absence of any treatments (Fig. S1B, S2B, and S3B). In leaves, the transcript levels of six BnNF-YBs (i.e., BnNF-YB3, BnNF-YB7, BnNF-YB8, BnNF-YB9, BnNF-YB10, and BnNF-YB14) were elevated after 1h or 3h of NaCl stress (Fig. 1B). In roots, NaCl treatments increased the transcript levels of BnNF-YB3, BnNF-YB13, and BnNF-YB14. Our results show that the levels of BnNF-YB1, BnNF-YB5, BnNF-YB6, BnNF-YB9, BnNF-YB11, and BnNF-YB12 transcripts were mostly not affected by salinity. Only the expression level of BnNF-YB4 in the roots was decreased at 3h.
None of the five BnNF-YC subunits was regulated by the biological clock, since the expression levels of these genes remained constant in the absence of any treatments (Fig. S1C, S2C, and S3C). Only BnNF-YC5 was induced in the leaf samples both at the 1- and 3-h time-points under salinity (Fig. 1C). BnNF-YC2 transcripts were more abundant in roots subjected to NaCl treatments than in untreated roots.
Expression Patterns of BnNF-Y Genes in Leaves and Roots of Canola in Response to drought Stress
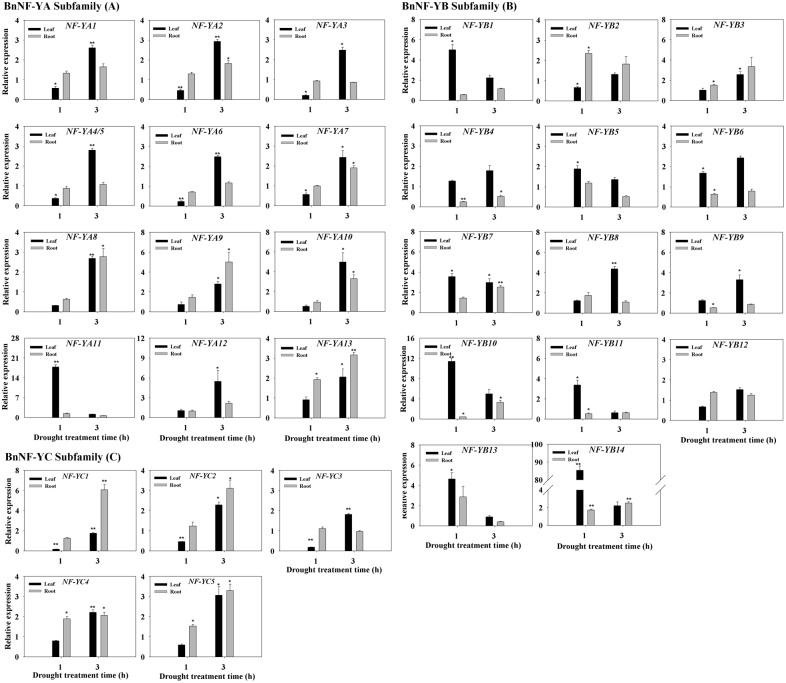
As shown in Fig. 2, all BnNF-YA members were responsive to PEG6000 treatments, which mimic drought stress, in leaf tissues. A 15% PEG treatment generally caused a 72% relative water content loss within 24h (Table S2). The expression levels of BnNF-YA10, BnNF-YA11 and BnNF-Y12 in leaves were dramatically up-regulated to more than four-fold the levels observed in the untreated control (Fig. 2A). BnNF-YA8, BnNF-YA9, BnNF-YA10 and BnNF-YA13 were induced in roots under drought stress. Four BnNF-YAs (BnNF-YA8, -YA9, -YA10, and -YA13) responded to drought stress both in leaves and roots, particularly up to 3h of treatment. The expression levels of five BnNF-YAs (BnNF-YA1, BnNF-YA2, -YA3, -YA4/5, and -YA6) decreased in leaves at 1h and increased at 3h.
Figure 2. Expression pattern of the BnNF-Y genes in plants subjected to drought stress.
The expression of BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) genes in the leaves and roots of plants exposed to 15% (w/v) PEG6000. Three-week-old canola seedlings were treated with 15% (w/v) PEG-6000 for 1 and 3h. Total RNA was extracted from leaves (L) and roots (R) for quantitative PCR (qRT-PCR) analysis. Transcript levels of each BnNF-Y were first normalized to those of the housekeeping gene 18S and then compared to each time-point control. Each data point represents the mean ±SE of three independent experiments. Significant differences between treated samples and untreated controls (same tissue only) are indicated by single (P<0.05) or double (P<0.01) asterisks, according to Dunnett’s method of one-way ANOVA in SPSS. Expression levels in untreated samples (CK, 1-h or 3-h leaf and root samples) were arbitrarily set to 1.0.
Most members of the BnNF-YB subfamily also responded to PEG treatments, except for BnNF-YB12, while the expression level of BnNF-YB14 increased 80-fold in leaves after 1h of treatment (Fig. 2B). As in the BnNF-YA subfamily, the transcripts of BnNF-YB were induced more strongly in leaves than in roots, since only 3 of 14 BnNF-YB members (BnNF-YB7, BnNF-YB10, and BnNF-YB14) were induced in roots subjected to drought stress. Four BnNF-YBs (BnNF-YB4, -B9, -B10 and -B11) exhibited decreased expression in the roots after 1h of treatment.
The transcript levels of four BnNF-YC members (BnNF-YC1, BnNF-YC2, BnNF-YC4, and BnNF-YC5) were elevated in both the leaf and root samples up to 3h of drought treatment, while those of three BnNF-YCs (BnNF-YC1, BnNF-YC2, and -YC3) were initially inhibited in leaves at the 1-h time point (Fig. 2C).
Expression Patterns of BnNF-Y Genes of Canola in Leaves and Roots in Response to ABA Stress
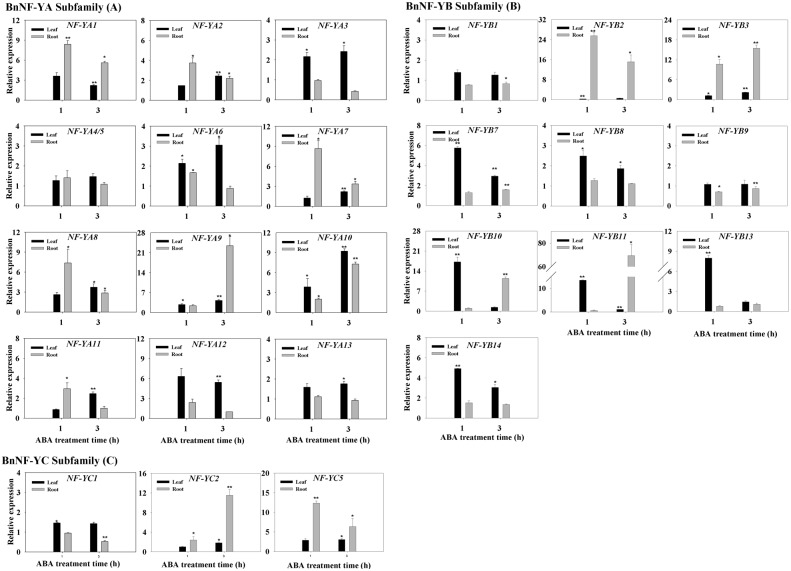
Plants respond to abiotic stresses via both the ABA-dependent and ABA-independent pathways. Some NF-Ys were reported to be involved in ABA-dependent pathways, whereas others were not [11], [13], [15]. To determine whether canola NF-Ys function via the ABA pathway, we further examined the expression profiles of BnNF-Ys that were found to be responsive to salinity or drought stress (i.e., all BnNF-YA members and most BnNF-YBs and BnNF-YCs; Figs 1 and 2) under ABA treatments. All BnNF-YAs, except for BnNF-YA4/5 and BnNF-YA13, were responsive to 3-h of ABA treatment (Fig. 3A). The transcript levels of BnNF-YA1, -YA2, -YA3, -YA6, -YA7, -YA8, -YA9, -YA10 and BnNF-YA11 were elevated at 1h. Also, most NaCl- or PEG- responsive BnNF-YB members were induced by ABA treatments, with the exception of BnNF-YB1 and BnNF-YB9 (Fig. 3B). The transcript level of BnNF-YB2 was inhibited in leaves after 1h of ABA treatment. Interestingly, several BnNF-YBs (BnNF-YB2, -YB3, -YB7, -YB10, -YB11, -YB13, and -YB14) were greatly up-regulated by ABA treatment. Amongst the BnNF-YC members, only the transcripts of BnNF-YC2 and BnNF-YC5 were increased in response to ABA treatment, and the upregulation was greater in the roots than in the leaves (Fig. 3C). The transcript levels of BnNF-YC1 remained almost constant from 1h to 3h of ABA treatment in all tissues examined.
Figure 3. Expression pattern of the BnNF-Y genes in the leaves and roots of plants exposed to 100 µM ABA.
Transcript levels of BnNF-Y (A), BnNF-YB (B), and BnNF-YC (C) genes after exposure to 100µM ABA. Three-week-old canola plantlets were treated with 100µM ABA for 1h and 3h. Total RNA was extracted from leaves and roots for quantitative PCR (qRT-PCR) analysis. Transcript levels of each BnNF-Y were first normalized to those of the housekeeping gene 18S and then compared to levels at each time point in the control. Each data point represents the mean ±SE of three independent experiments. Significant differences between treated samples and untreated controls (same tissue only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS. Expression levels in untreated samples (CK, 1-h or 3-h leaf and root samples) were arbitrarily set to 1.0.
To compare the gene expression levels of different BnNF-Ys in canola, the expression level of each NF-Y gene in the 0-h leaf or root samples (arbitrarily chosen as baseline in the real-time PCR analysis) was also determined by semi-quantitative RT-PCR analysis (35 cycles; Fig. S4). The BnNF-Ys showed diverse expression profiles. Interestingly, most BnNF-Ys were expressed at similar levels in 0-h leaves and 0-h roots.
Correlation between the gene expression patterns of BnNF-Y genes
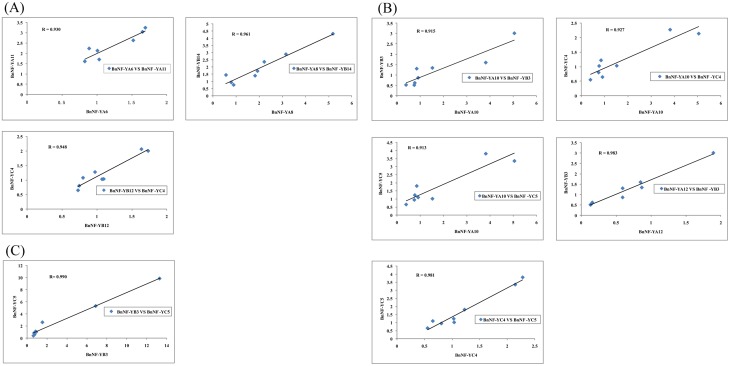
Since NF-Y TFs are known to form functional complexes in plants [30], [31], one would expect that NF-Ys with correlated expression profiles would be involved in the same regulatory processes under abiotic stress conditions. Extensive correlations were found to exist in the expression profiles of BnNF-Ys not only within the same subfamily, but also between the three subfamilies. The expression profiles of BnNF-YA6 and BnNF-YA11, of BnNF-YA8 and BnNF-YB14, and of BnNF-YC4 and BnNF-YB12 were highly correlated under NaCl treatment (R = 0.93, 0.948 and 0.961, respectively. Fig. 4A). Under drought stress, the expression of BnNF-YA10 and -YB3, -YC4, or -YC5, of -YA12 and -YB3, and of -YC4 and -YC5 were highly correlated (R = 0.915, 0.927, 0.913, 0.983, and 0981, respectively. Fig. 4B). Under ABA treatments, BnNF-YB3 and BnNF-YC5 showed very similar expression profiles (R = 0.99. Fig. 4C).
Figure 4. Correlation in expression levels of the three BnNF-Y subfamily members.
Correlated gene expression under salinity (A); drought (B); and ABA (C) treatments. Relative expression levels including 8 data points (relative NF-Y gene expression levels with and without treatments at 1h or 3h compared to the same 0-h samples) are used for each gene according to Fig. S1, S2, and S3. These data were fitted using linear regression analysis.
The Promoters of NF-Y Genes Contain Diverse Stress-responsive Elements
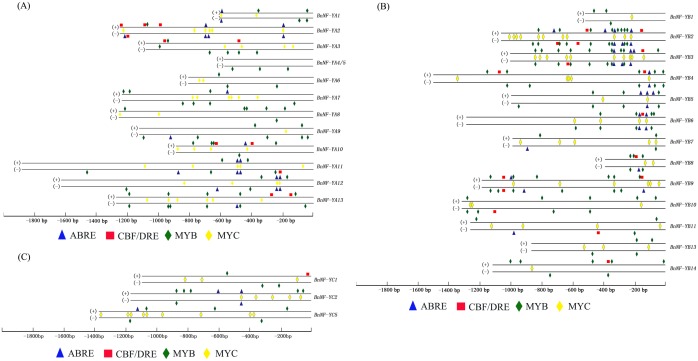
Since most BnNF-Y members were found to be regulated by abiotic stress, we sought to identify the promoter sequences of their corresponding genes and to identify stress-relative cis-elements. We cloned the promoters of 12 BnNF-YAs, 13 BnNF-YBs, and 3 BnNF-YCs that were responsive to one or more of the three treatments used above (i.e., NaCl, PEG6000, and ABA). Using a BLASTN search and genome walking, we successfully acquired all the promoter sequences, most of which consisted of around 1000bp upstream of the ATG start codon. However, we were unable to acquire the promoter sequences of two BnNF-YA members (BnNF-YA4/5 and BnNF-YA6) and two BnNF-YB members (BnNF-YB1 and BnNF-YB8) longer than 1000bp. We then identified cis-elements in these promoters using PLACE and PlantCARE software (Fig. 5), focusing on stress-related cis-elements, such as ABRE, CBF/DRE, MYB, and MYC. The promoters of each of the above-mentioned BnNF-Ys included at least two types of cis-elements, with the exception of the truncated BnNF-YA4/5 promoter sequences. All of the promoters, except for the promoter of BnNF-YA4/5, harbored MYB and MYC elements. More than half of the BnNF-YA and BnNF-YC promoters harbored at least one ABRE element, while only a few BnNF-Ys had DRE elements. We also constructed Arabidopsis cis-elements in the homologous NF-Y family and compared the arrangement of cis-elements in canola and Arabidopsis. Most canola NF-Y members had analogous cis-elements in Arabidopsis, but the number and arrangement of cis-elements differed (Fig. S5).
Figure 5. Diagrammatic representation of stress-related elements in the BnNF-Y promoters. Elements in the BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) promoters.
Symbols above the top line indicate elements that are in the forward orientation, and those below the bottom line indicate elements in the reverse orientation. MYC(CANNTG) sequences are between the double lines, to indicate their palindromic nature. Numbers are the distance in nucleotides between the sequences and ATG start codon.
Canola promoter sequences showed greater similarities with those of B. rapa than with those of Arabidopsis thaliana
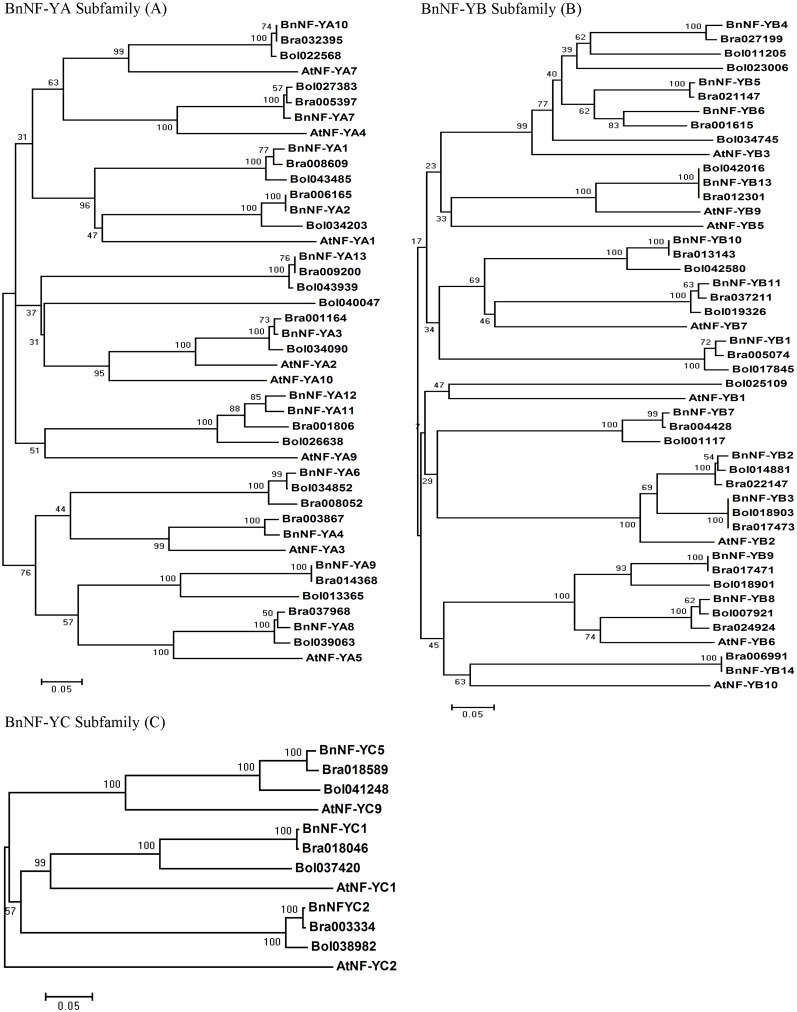
We previously showed that canola NF-Ys were more similar to those of the dicot Arabidopsis than to those of the monocot Oryza sativa (rice) based on putative amino acid sequences. In this study, we also used the promoter sequences to further explore the phylogenetic relationship amongst members of this family. All canola promoters grouped with promoters of their own homologs and formed clades with the corresponding subfamily members in the other species (Fig. 6). The promoter sequence of each canola NF-Y usually had the highest level of identity with B. rapa, and the lowest with Arabidopsis, with the exceptions of BnNF-YA6, BnNF-YB2 and BnNF-YB8. The promoter regions of these three BnNF-Ys were more similar to those of B. oleracea than to those of B. rapa.
Figure 6. Phylogenetic trees of Brassica and Arabidopsis NF-Y families based on the nucleotide sequences in the promoter regions.
The phylogenetic trees were constructed by the neighbor-joining method implemented by MEGA software, version 4.1. The numbers at each branch point represent the bootstrap scores (1,000 replicates). A branch with a bootstrap score of below 50 was usually considered unreliable. a Phylogenetic tree of Brassica and Arabidopsis NF-YA promoters. Due to low levels of nucleotide similarity, BnNF-YA4 homologues in B. oleracea were not included here. b Phylogenetic tree of Brassica and Arabidopsis NF-YB promoters. Due to low levels of nucleotide similarity, BnNF-YB14 homologues in B. oleracea were not included here. c Phylogenetic tree of Brassica and Arabidopsis NF-YC promoters. Bol and Bra represent B. oleracea and B. rapa, respectively. The numbers after the species abbreviation correspond to the individual gene name. Most promoters were at least 1000bp in length, except for those of BnNF-YA1 (595bp), BnNF-YA4/5 (577), BnNF-YA6 (806), BnNF-YA10 (892), BnNF-YB1 (519), BnNF-YB7 (988), BnNF-YB8 (391), and BnNF-YB13 (867) and their homologues in B. oleracea, B. rapa, and Arabidopsis.
Discussion
The fact that multiple members of the NF-Y family exist in plant genomes implies that these proteins have redundant or differentiated functions. In this study, we extensively explored the response of BnNF-Ys to abiotic stresses through qRT-PCR analysis. The expression profiles of the BnNF-Y TF family provide insight into their roles during the abiotic stress response.
It is not surprising that most identified BnNF-YAs are involved in salinity, drought, or ABA treatments, as most previously reported stress-responsive plant NF-Y members belong to the NF-YA subfamily [15]. Quantitative RT-PCR analysis indicated that transcripts of Arabidopsis NF-YA1 were increased by salinity, PEG, and ABA treatments. It was previously found that overexpression of Arabidopsis NF-YA1 rendered the transgenic plants more sensitive than the control to a 200mM NaCl treatment, whereas the corresponding Arabidopsis RNAi plants grew better than the control under these conditions [15]. BnNF-YA1 and BnNF-YA2, which showed the highest level of similarity with Arabidopsis NF-YA1 [9], were induced by salinity, PEG, and ABA treatments. Overexpression of Arabidopsis NF-YA2, NF-YA7, and NF-YA10 caused a dwarf phenotype and enhanced the plant’s resistance to multiple abiotic stresses [25]. We previously showed that BnNF-YA3, BnNF-YA13, and BnNF-YA14 formed a clade with AtNF-YA2 and AtNF-YA10 [9]. Expression of Arabidopsis NF-YA5 was up-regulated up to 14 days of drought treatment or within 24h of ABA treatment [13]. The ectopic expression of NF-YA5 in Arabidopsis increased the plant’s drought tolerance [13]. BnNF-YA8 or BnNF-YA9, which clustered with Arabidopsis NF-YA5, exhibited similar expression patterns up to 3h of PEG treatment. Using an inducible estradiol system, the overexpression of NF-YA1, 5, 6, or 9 was shown to result in increased ABA sensitivity during seed germination [32]. One NF-YA homolog, GmNF-YA3, from soybean was also shown to be involved in the abiotic stress response. Transcripts of GmNF-YA3 were induced after as little as 0.5h of NaCl, ABA, or cold treatment, and GmNF-YA3 showed a high level of similarity with Arabidopsis NF-YA3, 5, 6, and 8. Of the reported members of the plant NF-YA subfamily, the transcript levels of several members (including NF-YA1, 5, 6, and 9) were affected by ABA, implying that these genes regulate the plant’s response to abiotic stress via an ABA-dependent pathway. This may also be the case for BnNF-YA1, BnNF-YA8, and BnNF-YA9, which exhibit similar expression patterns under salinity, drought, or ABA stress.
AtNF-YB1 and its maize homologue ZmNF-YB2 were shown to enhance drought resistance, and maize plants overexpressing ZmNF-YB2 performed well in the field [11]. Barley HvNF-YB5, similar to Arabidopsis NF-YB1, showed around a 70-fold expression increase under salinity treatment [20]. Triticum aestivum (wheat) TaNF-YB2, homologous to Arabidopsis NF-YB1, was also upregulated in response to drought stress in the leaves [18]. BnNF-YB1, which is homologous to Arabidopsis NF-YB1, was induced only by drought in our study. Arabidopsis NF-YB2 was first found to be up-regulated by NaCl, mannitol, or cold (4°C) treatment [17]. Interestingly, NF-YB2 was identified as the top third gene to respond to osmotic stress. The expression of poplar NF-YB7 increased during a 15-d PEG or ABA treatment [33]. Transgenic Arabidopsis plants overexpressing poplar NF-YB7 exhibited an increased germination rate, root length, and photosynthetic rate under drought stress. Poplar NF-YB7 showed a high level of amino acid sequence identity with Arabidopsis NF-YB3, while Arabidopsis nf-yb3 mutant plants were more sensitive than the wild type to environmental stimuli. Canola BnNF-YB2 and BnNF-YB3, similar to Arabidopsis NF-YB2, were found to be responsive to NaCl and ABA treatments in this study. The expression of several poplar NF-YBs, such as NF-YB6, NF-YB11, and NF-YB13, was altered in the leaves of plants treated with PEG-6000 [19], while the expression of BnNF-YB7 and BnNF-YB14 (high identity with Poplar NF-YB11 and NF-YB13 in terms of the conserved core domain) was also up-regulated in the leaves under PEG or NaCl treatment (Fig. 1 and Fig. 2).
Previous studies revealed that a few NF-YC members were involved in the stress response [21], [22]. One NF-YC member, NF-YC2, from both Arabidopsis and Nicotiana tabacum (tobacco), was found to be activated by photooxidative stress [21]. This gene responded to multiple stresses, including heat, cold, and drought. In the same study, several Arabidopsis NF-YC members were shown to be induced by abiotic stress. The transcript levels of Arabidopsis NF-YC3 and NF-YC4 were increased under drought stress, while NF-YC4 was also induced by cold. Furthermore, Arabidopsis NF-YC2 formed a complex with NF-YB3, bZIP28, and NF-YA4 and was shown to play a role during endoplasmic reticulum stress [30]. PwHAP5 (Picea wilsonii) was up-regulated by NaCl, dehydration, and ABA treatment [22]. Ectopic expression of PwHAP5, which is homologous to Arabidopsis NF-YC2, improved salinity tolerance in plants. In our previous study, we showed that BnNF-YC2 and BnNF-YC5 grouped with NF-YC2 and NF-YC3 [9] and, like these homologs in Arabidopsis, responded to salinity and drought stress.
Since plants have so many NF-Ys, functional redundancy seems inevitable. Arabidopsis NF-YB2 and NF-YB3 function additively in the long-day flowering pathway [34]. LEAFY COTYLEDON 1 (LEC1) and LEAFY COTYLEDON1-LIKE (L1L) were shown to be involved in embryo development [35], [36]. Many of the BnNF-Ys characterized in our study were responsive to salinity, drought, or ABA treatment. Some of these members in the same subfamily were clustered in the same phylogenetic clade based on their promoter sequences, such as BnNF-YA1 and BnNF-YA2 (Fig. 6). Further evidence is needed to confirm the roles of these proteins during the abiotic stress response. It would also be interesting to explore whether these proteins from the three subfamilies combine to form trimeric complexes, as reported in yeast and animal systems [37]. It is not known whether the NF-Y complex always has fixed components or whether the components differ under different conditions. Even though extensive correlations in the expression patterns of TaNF-Y subfamily members were identified [18], no three members from the three different BnNF-Y subfamilies exhibited the same expression pattern under all three treatments in our study. This phenomenon implies that the canola BnNF-Y complex does not always consist of the same monomers under different conditions.
The plant’s response to abiotic stress involves the transcriptional regulation of genes via their cis-regulatory elements. ABRE, MYB, and MYC elements are known to be involved in the ABA-dependent stress pathway, while the DRE element plays a role in the ABA-independent pathway [38]–[40]. Two ABRE elements were identified in the promoter region of Arabidopsis drought-responsive NF-YA5, which functioned in an ABA-dependent manner [13]. Interestingly, GmNF-YA3, a homolog of NF-YA5, harbored an additional DRE cis-acting element in its promoter, which suggests that soybean NF-YA3 may be involved in both pathways [16]. A previous study of the global expression patterns of rice plants subjected to various abiotic stresses identified more ABRE and DRE elements in the promoter regions of genes responsive to both drought and salinity than in those specifically responsive to drought or salinity stress [41]. In our study, the promoter regions of BnNF-YA11 and BnNF-YA12, which were strongly induced by salinity and drought stress, each harbored 4 ABRE elements. Canola NF-YB2 and NF-YB3 each possessed at least 5 ABRE elements and were strongly up-regulated by ABA treatments. BnNF-YC2, which contains 2 ABRE elements in its promoter region, was strongly induced by ABA stress. In contrast, the promoter regions of canola NF-YB11 and NF-YB14, which were also strongly induced by ABA or drought treatment, had few DRE or ABRE elements, but several MYB or MYC elements, suggesting that MYB or MYC play roles in the abiotic stress response.
According to the well-known triangle theory [42], [43], canola, an allopolyploid, originated from the hybridization of B. rapa (the A genome) and B. oleaacea (the C genome), while all Brassica species basically arose from common Arabidopsis ancestors. Based on our NF-Y stress-related cis-element analysis and promoter sequence alignments, the upstream regulatory regions of NF-Y sequences of canola were found to be similar to those of Arabidopsis (Fig. 5 and 6). Through extensive comparisons based on nucleotide sequences, homoeologous segments conserved in canola and Arabidopsis were found to exhibit perfect collinearity [44]. Our study revealed that the promoters of canola NF-Ys were more similar to those of B. rapa than to B. oleracea. Canola (B. napus) was proposed to have multiple origins, and natural canola species were found to be more closely related to B. rapa species than to B. oleracea species, according to an Restriction fragment length polymorphisms (RFLP) analysis of nuclear, chloroplastic, and mitochondrial DNA [45]. A recent study found that two canola self-incompatibility genes (S-locus glycoproteins, SLGs) and an S-locus receptor kinase (SRK) had higher levels of amino acid sequence identity with their B. rapa homologues than with those from B. oleracea [46], supporting the notion that a subset of canola sequences are more closely related to B. rapa than to B. oleracea.
In conclusion, this study represents an extensive evaluation of BnNF-Y family members under salinity, drought, or ABA stress. The results presented here offer a useful foundation for further studies of BnNF-Y proteins under abiotic stress conditions. Several BnNF-Y members in each subfamily showed similar expression patterns, indicating that these genes may have redundant functions. Members of different families were found to have similar expression patterns, suggesting that BnNF-Ys form a heterocomplex. Our BnNF-Y promoter analysis shows that multiple BnNF-Y members contain abiotic stress-responsive elements and provides clues as to the evolution of BnNF-Ys in Brassica species.
Supporting Information
Expression pattern of BnNF-Y genes in the leaves and roots of plants subjected to salinity stress for 1 or 3 h. The expression of BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) in the leaves and roots of plants exposed to 150mM NaCl for the indicated periods of time. The transcript levels of each BnNF-Y gene were first normalized to those of the housekeeping gene 18S and then compared to the control (0-h level in the leaf). The expression levels of untreated samples (C, 0-h leaf samples) were arbitrarily set to 1.0. L, leaves; R, roots. CK, no treatment; Salt, NaCl treatment. Significant differences between different samples and 0-h samples (same tissue only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS.
(DOC)
Expression pattern of BnNF-Y genes exposed to osmotic stress. The expression of BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) genes in the leaves of plants exposed to treatment with 15% (w/v) PEG-6000 for the indicated periods. The transcript levels of each BnNF-Y gene were first normalized to those of the housekeeping gene 18S and then compared to the levels in the 0-h leaf control. Expression levels in untreated samples (C, 0-h leaf samples) were arbitrarily set to 1.0. L, leaves; R, roots. CK, no treatment; Drought, PEG6000 treatment. Significant differences between different samples and 0-h samples (same tissue only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS.
(DOC)
Expression pattern of the BnNF-Y genes after exposure to 100 µM ABA. The expression of BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) genes in the leaves and roots of plants exposed to 100µM ABA. Transcript levels of each BnNF-Y were first normalized to those of the housekeeping gene 18S and then compared to levels at 0h in the control (untreated) leaves. Expression levels in untreated samples (CK, 0-h leave samples) were arbitrarily set to 1.0. L, leaves; R, roots. CK, no treatment; ABA, ABA treatment. Significant differences between different samples and 0-h samples (same tissue only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS.
(DOC)
Semi-quantitative RT-PCR analysis of BnNF-Y expression in control leaves and roots. RT-PCR analysis was performed on untreated leaf (L) and root (R) tissue samples. The 18S housekeeping gene and BnNF-Ys were amplified for 28 cycles and 35 cycles, respectively.
(DOC)
Diagrammatic representation of NF-Y promoter regions. Arabidopsis NF-YA (A), NF-YB (B), and NF-YC (C) promoter regions. Symbols above the top line indicate elements in the forward orientation, and those below the bottom line are in the reverse orientation. MYC(CANNTG) sequences are between the double lines, to indicate their palindromic nature. Numbers are the distance in nucleotides between the sequences and ATG start codon.
(DOC)
Primers for BnNF-Y promoters. F indicates the PCR forward primer and R indicates the PCR reverse primer. Particularly, SP1, SP2, SP3 are three specific primers for Genome Walking.
(DOC)
Relative water content (RWC) of drought-stressed plants. Plants were treated with 10%, 15% and 20% (w/v) PEG6000 solution for 24 h.
(DOC)
Acknowledgments
We thank Prof. Zhaopu Liu from Nanjing Agricultural University for kindly supplying the canola (‘Nanyanyou 1’) seeds. We also thank Kathleen Farquharson for valuable comments on the manuscript revision. We also express our sincere thanks to the anonymous reviewers.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by grants from the Natural Science Foundation of Jiangsu province (BK2011635), the Doctoral Program of Higher Education of China (20120097120015), Fundamental Research Funds for the Central Universities (KYZ201206), the Scientific Research Foundation of the State Human Resource Ministry, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (RAPD program (809001)). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chen WJ, Zhu T (2004) Networks of transcription factors with roles in environmental stress response. Trends Plant Sci 9: 591–596. [DOI] [PubMed] [Google Scholar]
- 2. Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291. [DOI] [PubMed] [Google Scholar]
- 3. Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim IS, Sinha S, de Crombrugghe B, Maity SN (1996) Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol Cell Biol 16: 4003–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gusmaroli G, Tonelli C, Mantovani R (2001) Regulation of the CCAAT-binding NF-Y subunits in Arabidopsis thaliana . Gene 264: 173–185. [DOI] [PubMed] [Google Scholar]
- 6. Gusmaroli G, Tonelli C, Mantovani R (2002) Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283: 41–48. [DOI] [PubMed] [Google Scholar]
- 7. Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, et al. (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N (2008) Identification, characterization and interaction of HAP family genes in rice. Mol Gen Genet 279: 279–289. [DOI] [PubMed] [Google Scholar]
- 9. Liang M, Yin X, Lin Z, Zheng Q, Liu G, et al. (2014) Identification and characterization of NF-Y transcription factor families in Canola (Brassica napus L.). Planta 239: 107–126. [DOI] [PubMed] [Google Scholar]
- 10. Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, et al. (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci U S A 104: 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen NZ, Zhang XQ, Wei PC, Chen QJ, Ren F, et al. (2007) AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J Biochem Mol Biol 40: 1083–1089. [DOI] [PubMed] [Google Scholar]
- 13. Li WX, Oono Y, Zhu J, He XJ, Wu JM, et al. (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao B, Ge L, Liang R, Li W, Ruan K, et al. (2009) Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li YJ, Fang Y, Fu YR, Huang JG, Wu CA, et al. (2013) NFYA1 Is Involved in Regulation of Postgermination Growth Arrest Under Salt Stress in Arabidopsis. Plos One 8: e61289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni Z, Hu Z, Jiang Q, Zhang H (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol: 1–17. [DOI] [PubMed]
- 17. Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, et al. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stephenson TJ, McIntyre CL, Collet C, Xue GP (2007) Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol 65: 77–92. [DOI] [PubMed] [Google Scholar]
- 19.Yan DH, Xia X, Yin W (2013) NF-YB Family Genes Identified in a Poplar Genome-wide Analysis and Expressed in Populus euphratica Are Responsive to Drought Stress. Plant Mol Biol Report: 1–8.
- 20.Liang M, Hole D, Wu J, Blake T, Wu Y (2012) Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta: 1–13. [DOI] [PubMed]
- 21. Hackenberg D, Keetman U, Grimm B (2012) Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int J Mol Sci 13: 3458–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Yu Y, Wei J, Huang G, Zhang D, et al.. (2013) Homologous HAP5 subunit from Picea wilsonii improved tolerance to salt and decreased sensitivity to ABA in transformed Arabidopsis. Planta: 1–12. [DOI] [PubMed]
- 23. Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, et al. (2006) MtHAP2–1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula . Genes Dev 20: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- 25. Leyva-González MA, Ibarra-Laclette E, Cruz-Ramírez A, Herrera-Estrella L (2012) Functional and Transcriptome Analysis Reveals an Acclimatization Strategy for Abiotic Stress Tolerance Mediated by Arabidopsis NF-YA Family Members. PloS one 7: e48138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farooq M, Wahid A, Kobayashi N, Fujita D, Basra SMA (2009) Plant drought stress: effects, mechanisms and management. Agron Sustain Dev 29: 185–212. [Google Scholar]
- 27. Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5: 69–76. [DOI] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 29. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 30. Liu JX, Howell SH (2010) bZIP28 and NF-Y Transcription Factors Are Activated by ER Stress and Assemble into a Transcriptional Complex to Regulate Stress Response Genes in Arabidopsis. Plant Cell 22: 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, et al. (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis . Plant Cell 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mu J, Tan H, Hong S, Liang Y, Zuo J (2013) Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant 6: 188–201. [DOI] [PubMed] [Google Scholar]
- 33.Han X, Tang S, An Y, Zheng DC, Xia XL, et al.. (2013) Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J Exp Bot: ert262. [DOI] [PMC free article] [PubMed]
- 34. Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, et al. (2008) The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis . Planta 228: 709–723. [DOI] [PubMed] [Google Scholar]
- 35. Lotan T, Ohto M, Yee KM, West MAL, Lo R, et al. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- 36. Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, et al. (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dolfini D, Gatta R, Mantovani R (2012) NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol 47: 29–49. [DOI] [PubMed] [Google Scholar]
- 38. Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, et al. (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10: 88–94. [DOI] [PubMed] [Google Scholar]
- 41. Zhou J, Wang X, Jiao Y, Qin Y, Liu X, et al. (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63: 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jules J (2009) Plant Breeding Reviews Wiley.
- 43. Nagaharu U (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan J Bot 7: 389–452. [Google Scholar]
- 44. Cheung F, Trick M, Drou N, Lim YP, Park JY, et al. (2009) Comparative Analysis between Homoeologous Genome Segments of Brassica napus and Its Progenitor Species Reveals Extensive Sequence-Level Divergence. Plant Cell 21: 1912–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Song K, Osborn TC (1992) Polyphyletic Origins of Brassica Napus: New Evidence Based on Organelle and Nuclear Rflp Analyses. Genome 35: 992–1001. [Google Scholar]
- 46. Zhang XG, Yin DM, Ma CZ, Fu TD (2011) Phylogenetic Analysis of S-Locus Genes Reveals the Complicated Evolution Relationship of S Haplotypes in Brassica. Plant Mol Biol Report 29: 481–488. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression pattern of BnNF-Y genes in the leaves and roots of plants subjected to salinity stress for 1 or 3 h. The expression of BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) in the leaves and roots of plants exposed to 150mM NaCl for the indicated periods of time. The transcript levels of each BnNF-Y gene were first normalized to those of the housekeeping gene 18S and then compared to the control (0-h level in the leaf). The expression levels of untreated samples (C, 0-h leaf samples) were arbitrarily set to 1.0. L, leaves; R, roots. CK, no treatment; Salt, NaCl treatment. Significant differences between different samples and 0-h samples (same tissue only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS.
(DOC)
Expression pattern of BnNF-Y genes exposed to osmotic stress. The expression of BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) genes in the leaves of plants exposed to treatment with 15% (w/v) PEG-6000 for the indicated periods. The transcript levels of each BnNF-Y gene were first normalized to those of the housekeeping gene 18S and then compared to the levels in the 0-h leaf control. Expression levels in untreated samples (C, 0-h leaf samples) were arbitrarily set to 1.0. L, leaves; R, roots. CK, no treatment; Drought, PEG6000 treatment. Significant differences between different samples and 0-h samples (same tissue only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS.
(DOC)
Expression pattern of the BnNF-Y genes after exposure to 100 µM ABA. The expression of BnNF-YA (A), BnNF-YB (B), and BnNF-YC (C) genes in the leaves and roots of plants exposed to 100µM ABA. Transcript levels of each BnNF-Y were first normalized to those of the housekeeping gene 18S and then compared to levels at 0h in the control (untreated) leaves. Expression levels in untreated samples (CK, 0-h leave samples) were arbitrarily set to 1.0. L, leaves; R, roots. CK, no treatment; ABA, ABA treatment. Significant differences between different samples and 0-h samples (same tissue only) are indicated by a single (P<0.05) or double (P<0.01) asterisk, according to Dunnett’s method of one-way ANOVA in SPSS.
(DOC)
Semi-quantitative RT-PCR analysis of BnNF-Y expression in control leaves and roots. RT-PCR analysis was performed on untreated leaf (L) and root (R) tissue samples. The 18S housekeeping gene and BnNF-Ys were amplified for 28 cycles and 35 cycles, respectively.
(DOC)
Diagrammatic representation of NF-Y promoter regions. Arabidopsis NF-YA (A), NF-YB (B), and NF-YC (C) promoter regions. Symbols above the top line indicate elements in the forward orientation, and those below the bottom line are in the reverse orientation. MYC(CANNTG) sequences are between the double lines, to indicate their palindromic nature. Numbers are the distance in nucleotides between the sequences and ATG start codon.
(DOC)
Primers for BnNF-Y promoters. F indicates the PCR forward primer and R indicates the PCR reverse primer. Particularly, SP1, SP2, SP3 are three specific primers for Genome Walking.
(DOC)
Relative water content (RWC) of drought-stressed plants. Plants were treated with 10%, 15% and 20% (w/v) PEG6000 solution for 24 h.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.