Abstract
Conventional methods for the isolation of cancer-related circulating cell-free (ccf) DNA from patient blood (plasma) are time consuming and laborious. A DEP approach utilizing a microarray device now allows rapid isolation of ccf-DNA directly from a small volume of unprocessed blood. In this study, the DEP device is used to compare the ccf-DNA isolated directly from whole blood and plasma from 11 chronic lymphocytic leukemia (CLL) patients and one normal individual. Ccf-DNA from both blood and plasma samples was separated into DEP high-field regions, after which cells (blood), proteins, and other biomolecules were removed by a fluidic wash. The concentrated ccf-DNA was detected on-chip by fluorescence, and then eluted for PCR and DNA sequencing. The complete process from blood to PCR required less than 10 min; an additional 15 min was required to obtain plasma from whole blood. Ccf-DNA from the equivalent of 5 µL of CLL blood and 5 µL of plasma was amplified by PCR using Ig heavy-chain variable (IGHV) specific primers to identify the unique IGHV gene expressed by the leukemic B-cell clone. The PCR and DNA sequencing results obtained by DEP from all 11 CLL blood samples and from 8 of the 11 CLL plasma samples were exactly comparable to the DNA sequencing results obtained from genomic DNA isolated from CLL patient leukemic B cells (gold standard).
Keywords: Biomarkers, Cancer, Circulating cell-free (ccf)-DNA, Chronic lymphocytic leukemia (CLL), Dielectrophoresis
1 Introduction
Circulating cell-free (ccf) DNA is an important biomarker for early detection of cancer [1–4], residual disease [5, 6], monitoring chemotherapy [7], and other aspects of cancer management [1,8–15]. The isolation of cancer-related ccf-DNA from plasma may allow “liquid biopsies” to replace more invasive tissue biopsies for detecting and analyzing cancer mutations [1, 8, 10–14, 16–19]. However, the present methods for isolating ccf-DNA from plasma are complex, time-consuming, and relatively expensive processes that rule out use for point-of-care (POC) diagnostic applications. Conventional sample preparation processes have many other limitations that include (i) requirement of at least one or more milliliters of plasma, (ii) the processing of blood to plasma, (iii) a large number of manipulations that increases the chance for technician errors, (iv) decrease of recovery efficiency with decrease in sample size and concentration, (v) degradation of ccf-DNA by mechanical sheering during the processing steps, and (vi) limiting PCR analysis to shorter target DNA sequences due to the degradation of ccf-DNA. Finally, other potentially important cancer-related biomarkers, such as ccf-RNA, exosomes, and microvesicles, also require relatively long and involved processes for their isolation from plasma. With regard to hematological cancers, such as chronic lymphocytic leukemia (CLL) and lymphomas, DNA for PCR and sequencing can be obtained from transformed cells [20, 21], as well as from ccf-DNA isolated from plasma [22]. In the case of CLL, B cells from patients can be segregated into one of at least two major subsets on the basis of whether or not the Ig variable region has somatic mutations [23]. Patients with CLL cells that express unmutated Ig heavy-chain variable region genes (IGHV genes) tend to have an aggressive clinical course relative to that of patients who have CLL cells that express IGHV with somatic mutations [24–26]. For CLL diagnostics and management, genomic DNA is isolated from the peripheral blood mononuclear cells (PBMCs). The PBMCs are usually purified from the CLL patient blood samples by density centrifugation using Ficoll-Hypaque 1077. This is a long and labor-intensive process that adds considerable cost to patient management and precludes any POC applications. To assess the unique patient-specific IGHV expressed by the CLL B cells, PCR and DNA sequencing are performed on the isolated genomic DNA to determine the mutation status for the expressed IGHV gene [27–29].
Electrokinetic technologies, such as AC DEP, have long been known to provide effective separation of cells, nanoparticles, DNA, and other biomolecules [30–36]. However, until recently, DEP techniques remained impractical for use with high-conductance solutions (5–15 mS/cm), as well as with whole blood, plasma, and serum [33–36]. In earlier work, sample dilution to low-conductance conditions (<1 mS/cm) was required before effective DEP separations could be carried out [32,35–49]. While some progress was made using DEP under high-conductance conditions, these efforts have been limited to separations of cells and micron-sized entities by negative DEP forces using hybrid electrokinetic devices [37,50–54]. Such devices still could not be used with whole blood samples, and more importantly they did not provide efficient isolation of DNA from the sample. More recently, we have been able to develop electrokinetic techniques that allow nanoscale entities, including high molecular weight DNA and nanoparticles, to be isolated from high-conductance (>10 mS/cm) buffer solutions [55–57] and whole blood samples [58]. We were also able to demonstrate isolation of virus from blood and fluorescent detection of ccf-DNA from CLL patient blood samples [59]. Most recent, we were able demonstrate PCR and Sanger DNA sequencing results for ccf-DNA biomarkers isolated by DEP using only 25 µL samples of unprocessed CLL patient blood [60]. The PCR and Sanger sequencing results for the DEP process were equivalent to results obtained using conventional sample preparation of ccf-DNA from 1 mL of CLL patient plasma, and to the “gold standard” DNA sequencing results obtained using an established method for isolating DNA from the leukemic cells of CLL patients that requires 15–20 mL of blood. In the present study, we now compare the DEP isolation of ccf-DNA from CLL patient blood with CLL patient plasma.
2 Materials and methods
2.1 Sample acquisition
Blood samples were collected from CLL patients and healthy volunteers (IRB#: 080918) in collection tubes containing lithium heparin (Becton Dickinson). For the DEP experiments, 300 µL of blood was taken from the top of each undisturbed blood sample within 4–5 h of collection. Plasma was obtained by centrifuging the blood for 10 min at 1100 RCF. The supernatant (plasma) was pipetted into a microcentrifuge tube and either frozen or used directly for DEP experiments.
2.2 Dielectrophoretic (DEP) isolation of ccf-DNA from blood and plasma
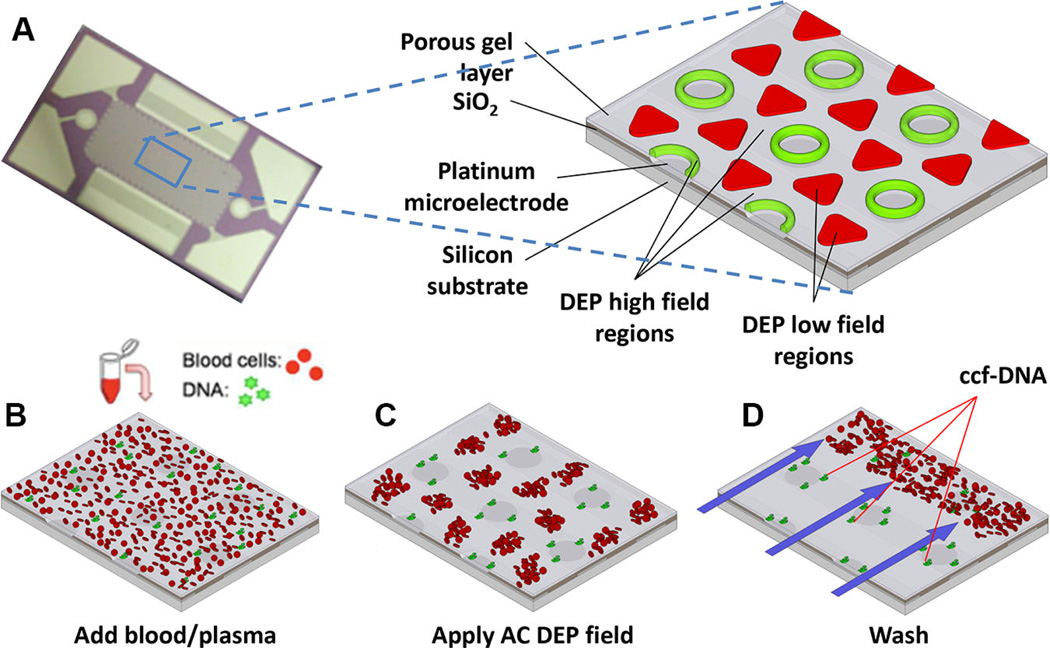
New AC DEP microarray devices (Biological Dynamics, La Jolla, CA) allow the rapid isolation of ccf-DNA and other nanoparticulate biomarkers (ccf-RNA, exosomes, etc.) directly from a small volume of blood and plasma. Figure 1A shows the alternating current electrokinetic microarray device (chip), which is approximately 10 mm × 20 mm and contains 1000 microelectrodes that are 60 µm in diameter. The expanded view shows a section of the chip which is fabricated on a silicon base with platinum microelectrodes insulated by SiO2 and over-coated with a porous hydrogel layer. The expanded view also shows the location of the DEP high-field regions (green) and the DEP low-field regions (red) which form when the AC field is applied. In the first step of the process, a blood or plasma sample containing the ccf-DNA is placed into the microarray device (chip) and an AC electric field is then applied (Fig. 1B). At a specific AC frequency and voltage level, the ccf-DNA, which is more polarizable than the surrounding medium, experiences positive DEP that causes it to concentrate into the DEP high-field regions over the circular microelectrode structures. In the case of blood samples, the blood cells that are less polarizable experience negative DEP that causes them to move into the DEP low-field regions between the microelectrodes (Fig. 1C). Concentration of the ccf-DNA into the DEP high-field regions requires only 3 min, after which a fluid wash removes the blood cells and other blood components from the microarray (Fig. 1D). This is possible because the ccf-DNA in the DEP high-field regions is held more strongly than the blood cells in the DEP low-field regions. Generally, proteins and lower molecular weight biomolecules in the blood are not affected by the AC fields and they are also removed by the washing procedure. After the washing step, the AC field is turned off, at which point the ccf-DNA, if fluorescently stained, can be analyzed on-chip by fluorescence and then the sample can be eluted for subsequent PCR and DNA sequencing analysis.
Figure 1.
AC DEP microarray device and scheme for isolation of ccf-DNA from blood and plasma. (A) Upper image shows the alternating current electrokinetic microarray device (chip) used to carry out the isolation of ccf-DNA directly from blood. Expanded view shows the device materials composition: porous gel, platinum microelectrodes, SiO2 layer, and silicon base; and the location of the DEP high-field (green) and the DEP low-field (red) regions when an AC field is applied. Lower figure shows (B) microarray with whole blood (red circles) containing fluorescent DNA (green dots); (C) application of the AC electric field causing the fluorescent DNA (green dots) to be concentrated in the DEP high-field regions on the microelectrodes, while the blood cells (red circles) move into the DEP low-field regions between the microelectrodes; and (D) the AC field remains on while a fluidic wash removes the blood cells from the microarray with DNA remaining concentrated in the DEP high-field regions.
The microarray chip itself is contained in a printed circuit board (PCB) cartridge that forms the sides of the fluidic chamber and is covered with an acrylic window, forming a flow cell with a 25 µL sample volume. A custom-built instrument system (Biological Dynamics) provides the electronic, optical, and fluidic functions under MATLAB software control. Each chip was pretreated by adding 25 µL of 0.5 × PBS (Lonza) to the flow cell and applying a 2 VRMS, 5 Hz sinusoidal waveform for 15 s to improve the hydrogel porosity. The 0.5 × PBS was then removed and 25 µL of either blood or plasma was added to the flow cell. An 11 V peak-to-peak (Vp-p), 10 kHz sinusoidal waveform was then applied to the chip for 3 min with no fluid flow. The same electric field was maintained while the chip was washed for 5 min at 200 µL/min with 1 × TE (Sigma-Aldrich). The electric field was then turned off, allowing captured DNA to diffuse into the 1 × TE solution. The 25 µL of fluid was removed within 30 s and stored in a microcentrifuge tube. For each CLL patient and healthy donor, this process was repeated four times, each time on a new microelectrode device. The 25 µL of eluted sample from each of the four runs was combined into a single microcentrifuge tube (100 µL total volume) and stored at 4°C for later analysis.
In order to visualize collection on the microelectrode array, the CLL and healthy donor blood and plasma samples were stained with SYBR Green I fluorescent dsDNA dye (Life Technologies, Carlsbad, CA). One and a half microliters of 100x SYBR Green I was added to 28.5 µL of blood and allowed to incubate at room temperature for 5 min. A total of 25 µL of this solution was added to the device and run as described above. After the 3 min of electric field collection and 5 min of washing, bright field and fluorescence images of the microelectrode pads were acquired using a CCD camera with a 10× objective, FITC filter, and a 470 nm LED excitation source. DNA with SYBR Green I from these imaged devices was not eluted or used in subsequent analysis.
2.3 DNA quantification
After DEP, the isolated ccf-DNA from the blood samples and the plasma samples was eluted and quantified using Quanit-iT PicoGreen (Life Technologies), a dsDNA dye. Each sample was diluted and combined with the PicoGreen reagent, and the resulting fluorescence was measured with a plate reader (Tecan). Standard curve used with PicoGreen assay is included in Supporting Information Fig. 1.
2.4 PCR, gel electrophoresis, and DNA sequencing
In order to verify that the collected ccf-DNA was from leukemic B-cells, it was amplified by PCR using Phusion High-Fidelity DNA Polymerase (New England Biolabs). The forward primers used were specific to the VH1, VH3, and VH4 regions, and the reverse primer was specific for the JH region. PCR thermal cycling conditions were a 5-min initial denaturation at 98°C followed by 40 cycles of 98°C denaturation for 15 s, 66°C annealing for 15 s, and 72°C extension for 15 s. While CLL blood samples were PCR amplified with each set of VH primers (VH1, VH3, and VH4), the plasma samples were only amplified with the correct VH primer. The PCR product was then analyzed by gel electrophoresis on a 2% agarose gel containing ethidium bromide (Life Technologies). The gels were viewed in a transilluminator and images were captured using a CCD camera. The images were analyzed with ImageJ software to determine the fluorescence in the region where the main 500–550 bp CLL target fragments should appear, regardless of whether or not a discrete band was observed. In ImageJ, the red channel was separated and used, while the blue and green channel data were discarded. The background fluorescence of the image was then removed using the “Subtract Background” tool. A 40-pixel wide by 22-pixel tall region was selected around the 500–550 bp region of each gel lane, and an “Integrate Density” measurement was taken. Remaining PCR product was cleaned up with the QIAquick PCR purification kit (Qiagen, Valencia, CA) and sequenced using the Sanger DNA sequencing method [60].
2.5 CLL PBMC genomic DNA extraction and IGHV analysis
PBMCs from CLL patients were isolated by density centrifugation using Ficoll-Hypaque 1077 (Sigma-Aldrich, St. Louis, MO) and suspended in fetal calf serum containing 10% DMSO (Sigma-Aldrich) for storage in liquid nitrogen [61]. DNA was extracted using a Qiagen kit (QIAamp DNA Mini kit, Qiagen) according to the manufacturer’s instructions and eluted in 30 µL of nuclease-free water. The IGHV gene characterization and mutation status was assessed as previously described [62, 63]. Most PCR products were sequenced directly, although in some cases, amplified products were cloned into pGEM-T (Promega, Madison, WI). Nucleotide sequences were analyzed using the ImMunoGenetic directory (European bioinformatics Institute ImMunoGeneTics Informations System available at http://imgt.cines.fr Leukemia 2011 Langerak, Davi, ERIC guidelines) [64]. Sequences with less than 98% homology with the corresponding germline IGHV gene were considered mutated. The heavy chain complementarity-determining region (HCDR3) was determined by the method of Kabat and Wu [29] as defined by the number of amino acids between codon 94 at the end of framework 3 and the conserved Trp of position 102 at the beginning of framework 4.
3 Results
3.1 DEP isolation of ccf-DNA
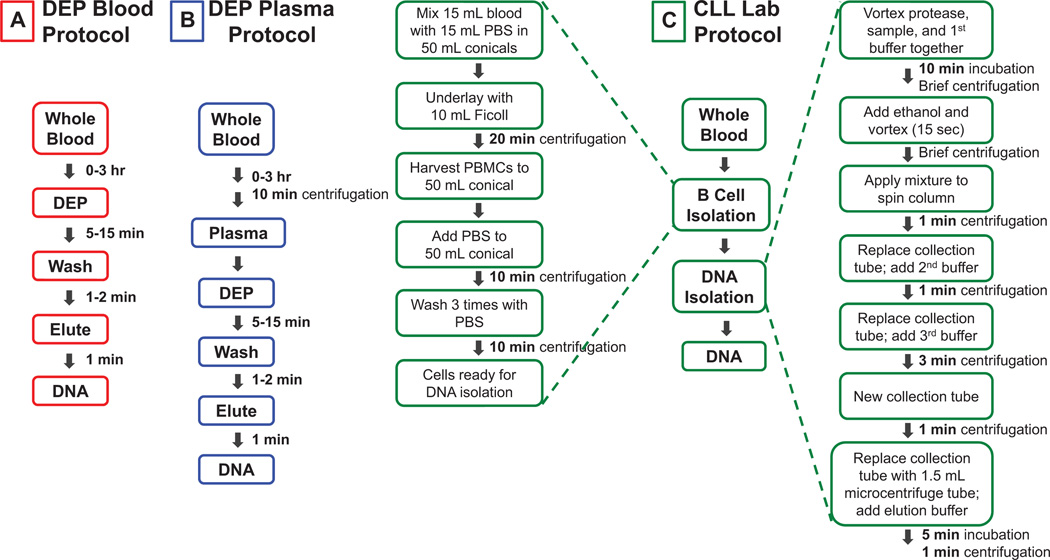
In this study, an AC DEP microarray was used to isolate ccf-DNA from 11 CLL patient blood and plasma samples and one normal blood and plasma sample. The study shows that the AC DEP technique and microarray allows (i) ccf-DNA to be isolated directly from 25 µL of unprocessed blood or plasma, (ii) on-chip fluorescence analysis of the ccf-DNA in less than 5 min, and (iii) elution of the ccf-DNA from the microarray chip for subsequent PCR and DNA sequencing analysis in less than 10 min. The manipulations for the DEP process comprise two simple steps: the addition of the blood or plasma sample into the microarray device and removal of the eluted sample upon completion of the process. Plasma has the required extra steps of pipetting and centrifugation before the sample is applied to the DEP microarray. Fluorescence analysis to determine the concentration of ccf-DNA was also carried out after elution from the DEP chip. The results from the DEP process for isolation ccf-DNA from both blood and plasma were then compared to conventional sample preparation process for isolation of genomic DNA from the leukemic B cells of CLL patients. This process requires a much larger blood sample and several hours to complete before fluorescence analysis, PCR, and DNA sequencing of the genomic DNA can be carried out. Figure 2 shows a comparison of the processing time and number of manipulations required for the DEP procedure for blood (Fig. 2A) and for plasma (Fig. 2B), with the CLL Lab procedure for isolating genomic DNA from patient B-lymphocyte cells (Fig. 2C). With regard to the DEP process, the blood sample can be applied to the microarray immediately. However, for this study the DEP isolation was generally carried from 0 to 3 h after the blood draw. For the CLL Lab procedure, the processing times include only the actual time necessary to run a specific processing step, that is, 10 min for centrifugation. Additional time is necessary for setting up, carrying out transfers such as pipetting, and for many other manipulations. When this process is performed manually, the additional manipulations can add at least another hour to the total time required for CLL Lab process, and they also add significantly to the overall cost of the assays.
Figure 2.
Processing times and steps for DEP isolation of ccf-DNA from blood and plasma compared with isolation of genomic DNA from CLL patient B-lymphocytes. (A) The DEP procedure used to isolate ccf-DNA directly from 25 µL samples of unprocessed CLL patient blood. (B) The DEP procedure used to isolate ccf-DNA directly from 25 µL samples of CLL patient plasma. (C) The procedure used to isolate genomic DNA from CLL patient B-lymphocyte cells starting with 15–20 mL of patient blood.
3.2 On-chip fluorescence detection of ccf-DNA
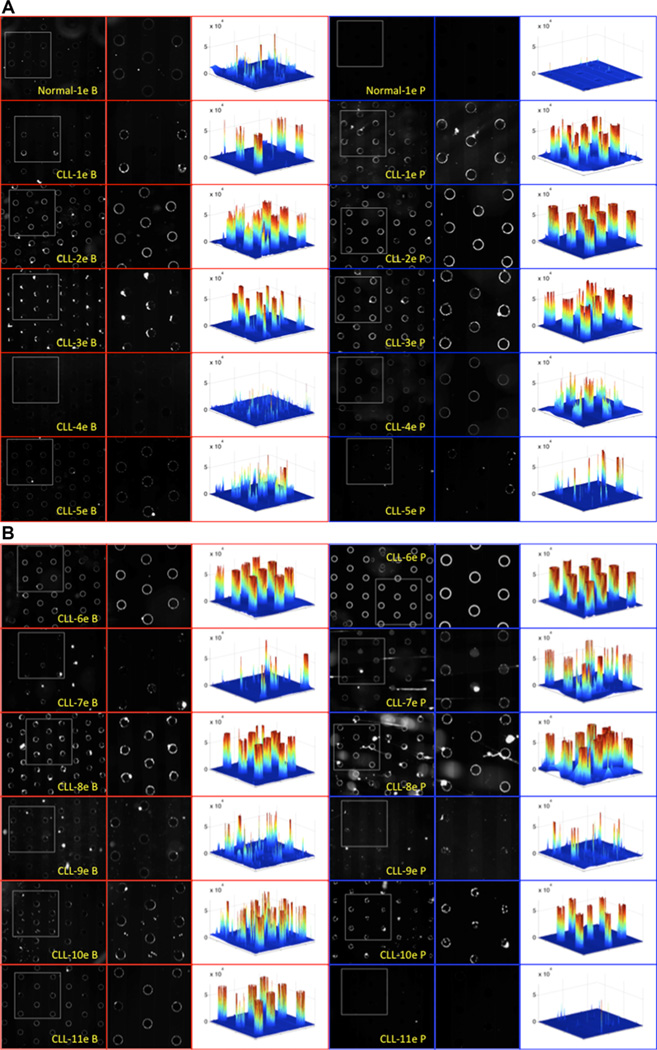
For on-chip fluorescence detection of the ccf-DNA, SYBR® Green I (Invitrogen) stain is added to the blood samples and plasma samples before the application of the DEP field. After DEP is carried out for 3 min and blood cells (blood samples) and plasma are removed by a fluidic wash, the fluorescently stained ccf-DNA, which is concentrated in the DEP high-field regions (on the microelectrodes), is detected. Figure 3A shows the fluorescent image results for ccf-DNA isolated by DEP from a normal blood (Normal-1e B) and plasma sample (Normal-1e P), and five CLL blood (CLL-1e B, CLL-2e B, etc.) and plasma samples (CLL-1e P, CLL-2e P, etc.). Figure 3B shows images from six more CLL blood and plasma patient samples, which were also run in this study. On the far right of each fluorescence image is a 3D fluorescence intensity image created by MATLAB, which provides better visualization of the relative amounts of ccf-DNA that were isolated. Overall, the fluorescent DNA levels (3D intensity) were higher in most of the CLL patient samples when compared to the fluorescent DNA levels obtained for the normal blood (Normal-1e B) and plasma (Normal-1e P) sample. The only exception being the plasma sample CLL-11e P, which has a very low level of fluorescence. The fluorescent DNA levels (3D intensity) were somewhat comparable between the CLL blood samples and CLL plasma samples, with the main exception being the plasma sample CLL-11e P.
Figure 3.
On-chip fluorescence imaging results from 25 µL blood and plasma samples showing SYBR Green stained ccf-DNA that was concentrated into the DEP high-field regions after the DEP field was applied for 3 min. (A) Fluorescence detection of ccf-DNA in five CLL patient blood and plasma samples and one normal blood and plasma sample. Images of one normal blood (Normal-1e B) and plasma sample (Normal-1e P) and five CLL blood (B) and plasma (P) samples (CLL-1e, CLL-2e, CLL-3e, CLL-4e, and CLL-5e). (B) Fluorescence detection of ccf-DNA in six additional CLL patient blood and plasma samples. Images of six additional CLL blood (B) and plasma (P) samples (CLL-6e, CLL-7e, CLL-8e, CLL-9e, CLL-10e, and CLL-11e). White dotted square areas in the images on the left side are enlarged in the center column images. The right side column shows 3D fluorescence intensity images created by MATLAB, which provide better visualization of the relative amounts of ccf-DNA that were isolated on the DEP high-field areas over the microelectrodes.
3.3 Concentration of eluted ccf-DNA from blood and plasma samples
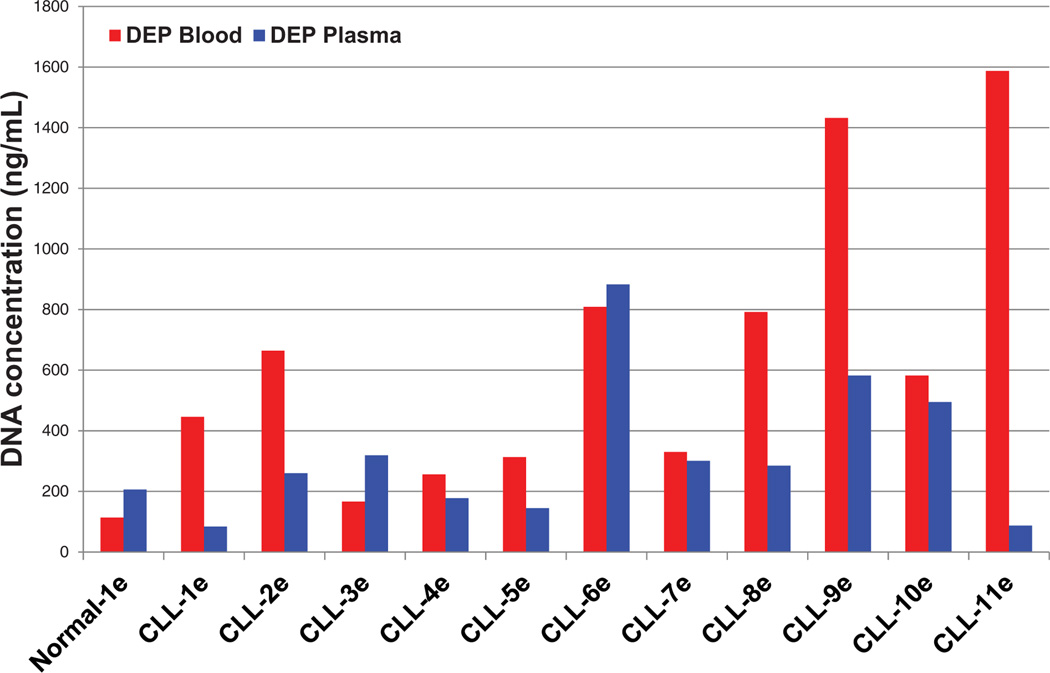
In additional experiments, Quant-iT PicoGreen (Invitrogen) fluorescence analysis was used to determine the concentration of the ccf-DNA in the CLL and normal blood and plasma samples after elution from the DEP microarray. For these experiments, SYBR Green I DNA dye was not added to the blood or plasma samples prior to DEP. Figure 4 shows the ccf-DNA concentration results (after DEP) for the eluted blood samples (red bars) and for the eluted plasma samples (blue bars). No significant correlation could be found between the DEP blood results and plasma results for the CLL samples. This is believed to be due to the elution process for removing the isolated ccf-DNA from the DEP device, which is only semiquantitative. Since the CLL laboratory procedure uses genomic DNA isolated from leukemic B cells, it does not provide meaningful gauge for ccf-DNA in the blood or plasma. Nevertheless, all of the CLL blood and most of the CLL plasma ccf-DNA concentrations were higher than the normal blood and plasma sample.
Figure 4.
Concentration of eluted ccf-DNA from blood and plasma samples. Bar graph of the ccf-DNA concentrations in the final eluted samples that were obtained directly from blood using the DEP process (red bars), and of the final eluted samples from plasma using the DEP (blue bars). The DNA concentrations were determined by fluorescence analysis using Quant-iT PicoGreen (Invitrogen) assay for dsDNA and normalized to the original sample volume.
3.4 PCR and DNA sequencing results
The eluted ccf-DNA isolated by DEP from both the CLL blood and plasma samples was amplified using primers for the IGHV1, IGHV3, and IGHV4 regions as described previously [60]. The equivalent of just 5 µL of the original CLL blood or plasma sample (25 µL) was amplified using IGHV specific primers. The correct IGHV PCR amplification products were obtained for all 11 CLL blood and plasma samples, although some bands were weak and secondary bands were also present. It should be pointed out that the CLL laboratory procedure does not require comparisons to blood or plasma samples from normal individuals; therefore, the ccf-DNA isolated by DEP from normal blood and plasma samples were not used for PCR or sequencing.
Once the IGHV regions for each of the CLL samples were analyzed using PCR, the resulting PCR products were sequenced and compared to the results previously obtained using an established method performed on genomic DNA obtained from CLL patient leukemic cells. Sanger sequencing was used to verify that the isolated ccf-DNA was coming from the leukemia cell population, and that the amplified IGHV regions matched the results obtained by PCR of genomic DNA isolated from B-lymphocytes. Supporting Information Table S1 shows the DNA sequencing results for all 11 CLL samples; the sequences obtained from the ccf-DNA isolated by DEP from the equivalent of 5 µL of blood and plasma are compared to the “gold standard” CLL patient sequences in the database, which were obtained from isolating genomic DNA from patient B-lymphocyte cells. All 11 CLL patient DNA sequences from the DEP blood samples show perfect homology with the genomic DNA “gold standard” results. Eight of the 11 CLL sequences for the DEP plasma samples show perfect homology with the genomic DNA “gold standard” results. The ccf-DNA from DEP plasma sample CLL-3e shows 98/100% homology, DEP plasma sample CLL-2e shows 84/100% homology, and DEP plasma sample CLL-4e did not sequence. Several possible reasons exist for these three CLL plasma sequencing results: (i) less ccf-DNA was present in the plasma samples; (ii) less DNA was eluted from the DEP microarray; and (iii) PCR is not fully optimized for amplification of very low levels of ccf-DNA.
4 Discussion
Ccf-DNA and ccf-RNA have the potential to become important biomarkers for cancer diagnostics and patient management. Ccf-DNA/RNA isolated from blood and/or plasma, constituting a “liquid biopsy,” may serve as an alternative to more invasive tissue biopsies in the detection and analysis of cancer mutations. Unfortunately, the time, complexity, and cost of employing conventional methods to isolate ccf-DNA/RNA from blood/plasma can limit the use of these procedures, especially for POC diagnostic applications. This study demonstrates the ability of a DEP microarray device to isolate ccf-DNA directly from a small amount (25 µL) of unprocessed blood or plasma samples. The DEP process comprises only two steps and can be completed in less than 10 min. In contrast, most conventional sample preparation processes typically involves obtaining 1–2 mL of plasma from 2–3 mL of blood and subsequently subjecting the plasma to a series of manipulations to obtain ccf-DNA over the course of 1–2 h [60]. The present CLL laboratory process for isolating genomic DNA from B-lymphocytes requires 15–20 mL of blood and is a complex and expensive process that takes several hours to complete. The DEP process enables the rapid use of unprocessed blood or plasma samples, which should lead to a significant reduction in the cost and complexity of ccf-DNA isolation relative to conventional methods for isolation of ccf-DNA. The DEP procedure for blood samples provides PCR and DNA sequencing results comparable to conventional sample preparation results [60], and to results obtained using an established method performed on genomic DNA. An additional advantage of using the DEP process is the ability to carry out fluorescence detection of ccf-DNA within minutes of application of the blood/plasma sample to the chip. In this study, CLL samples possessed higher fluorescence intensity levels for SYBR® Green stained ccf-DNA concentrated in DEP high-field regions than the normal blood samples. In many cases the fluorescence intensity levels of the CLL samples were substantially higher than those of the normal sample. Ultimately, the use of “on-chip” fluorescence to rapidly determine ccf-DNA levels in clinical blood samples could provide a first-level “alarm” for POC diagnostics. This could provide an indication of the presence or occurrence of an abnormality that could require further monitoring. In the case of solid tumors, researchers have demonstrated a correlation between ccf-DNA levels in patient plasma and survivability for lung and colon cancers [65,66]. However, the isolation of ccf-DNA in these studies required long and involved processes.
In summary, the DEP technique shows considerable potential for enabling rapid, simple, and cost-effective “liquid biopsy” and POC cancer diagnostics. In addition, the DEP technique may become a powerful tool for biomedical research. Currently, the true in vivo nature and actual levels of ccf-DNA/RNA, exosomes, and other nanoparticulate biomarkers in blood are not well known. The ability to rapidly isolate, in their unperturbed states, the cellular nanoparticulates released into the bloodstream by injured, necrotic, and transformed cells is critical to a better understanding of the disease process itself. Unquestionably, conventional sample preparation procedures, which involve processing plasma from blood and subsequently subjecting plasma to numerous time-consuming/labor-intensive physicalmanipulations, may lead to loss and degradation of the biomarkers. The use of this DEP technique for rapid isolation of ccf-DNA/RNA directly from blood samples promises to provide biomarkers in their unperturbed state and this may, in turn, enable researchers to deliver better diagnostic tools and research applications.
Supplementary Material
Acknowledgments
The authors thank Dr. Raj Krishan, Dr. David Charlot, and Gene Tu at Biological Dynamics (San Diego, CA) for their help and cooperation, and for allowing early access to the alternating current electrokinetic instrument and microarrays used in this study. The authors also thank Adam Wright for his help with image processing. The DEP technology used in the study was the result of original research carried out by the M. J. H. laboratory at the UCSD Moores Cancer Center under NIH NCI NanoTumor Center Grant (U54-CA119335). The work was also funded in part by NIH grant PO1-CA081534 for the CLL Research Consortium (T. J. K. PI).
With regard to financial conflict of interest, M. J. H. serves as an unpaid consultant to Biological Dynamics that provided the DEP devices for this study at no cost. The DEP technology used in the study was the result of original research carried out by the M. J. H. laboratory at the UCSD Moores Cancer Center under NIH NCI NanoTumor Center Grant (U54-CA119335). This work led to the filing of U.S. patent application (UCSD Docket # 2007-205-3MI; serial number 12/936,147; filed 4/3/2009). The technology was licensed to Biological Dynamics.
Abbreviations
- cf-DNA
circulating cell-free DNA
- CLL
chronic lymphocytic leukemia
- IGHV
Ig heavy-chain variable
- PBMC
peripheral blood mononuclear cell
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site
A. S. designed, carried out, and coordinated all experiments and prepared the manuscript. J. Y. M. performed PCR, DNA quantification, and prepared the manuscript. L. R. managed the CLL sample collection, storage, and distribution, provided patient clinical database and IGHV database/analysis, and edited the manuscript. E. M. G. performed IGHV gene sequence analysis, PCR, and IGHV sequencing, prepared tables and figures for the manuscript, and edited the manuscript. E. A. S. isolated ccf-DNA, carried out DNA quantification, contributed to and edited the manuscript. S. M. isolated ccf-DNA, and carried out DNA quantification and PCR. J. P. M. designed experiments, contributed material to the manuscript and edited the manuscript. G. F. W. designed PCR experiments, prepared background information for manuscript, and edited the manuscript. T. J. K. provided patient samples, clinical data, and edited the manuscript. M. J. H. designed experiments and prepared the manuscript. T. J. K. (UCSD Moores Cancer Center CLL laboratory) and M. J. H. (UCSD Departments Bioengineering and Nanoengineering) are the primary investigators and directed the study.
References
- 1.Schwarzenbach H, Hoon DS, Pantel K. Nat. Rev. Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 2.Fleischhacker M, Schmidt B. Biochim. Biophys. Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Ziegler A, Zangemeister-Wittke U, Stahel RA. Cancer Treat. Rev. 2002;28:255–271. doi: 10.1016/s0305-7372(02)00077-4. [DOI] [PubMed] [Google Scholar]
- 4.Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Rat-cliffe C, Roz E, Cirenei N, Bellomi M, Pelosi G, Pierotti MA, Pastorino U. J. Clin. Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Board RE, Knight L, Greystoke A, Blackhall FH, Hughes A, Dive C, Ranson M. Biomark. Insights. 2008;2:307–319. [PMC free article] [PubMed] [Google Scholar]
- 6.Sozzi G, Conte D, Mariana L, Lo Vullo S, Roz L, Lombardo C, Pierotti MA, Tavecchio L. Cancer Res. 2001;61:4675–4678. [PubMed] [Google Scholar]
- 7.Gautschi O, Bigosch C, Huegli B, Jermann M, Marx A, Chassé E, Ratschiller D, Weder W, Joerger M, Betticher DC, Stahel RA, Ziegler A. J. Clin. Oncol. 2004;22:4157–4164. doi: 10.1200/JCO.2004.11.123. [DOI] [PubMed] [Google Scholar]
- 8.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Sci. Transl. Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 9.Gormally E, Caboux E, Vineis P, Hainaut P. Mutat. Res. 2007;635:105–117. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Proc. Natl. Acad. Sci. USA. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA., Jr Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swisher EM, Wollan M, Mahtani SM, Willner JB, Garcia R, Goff BA, King MC. Am. J. Obstet. Gynecol. 2005;193:662–667. doi: 10.1016/j.ajog.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 13.Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, Donald E, Greystoke A, Ranson M, Hughes A, Dive C. Breast Cancer Res. Treat. 2010;120:461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 14.Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Clin. Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 15.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O’Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B, Diaz LA, Jr, Vel-culescu VE. Sci. Transl. Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee S, Kaye S. Eur. J. Cancer. 2011;47(Suppl 3):S116–S130. doi: 10.1016/S0959-8049(11)70155-1. [DOI] [PubMed] [Google Scholar]
- 17.Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, Milton DT, Strawn JR, Wakelee HA, Giaccone G. J. Clin. Oncol. 2011;29:2121–2127. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 18.Matulonis UA, Hirsch M, Palescandolo E, Kim E, Liu J, van Hummelen P, MacConaill L, Drapkin R, Hahn WC. PLoS ONE. 2011;6:e24433. doi: 10.1371/journal.pone.0024433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, Chirieac LR, Kaur R, Lightbown A, Simendinger J, Li T, Padera RF, Garc´ıa-Echeverr´ıa C, Weissleder R, Mahmood U, Cantley LC, Wong KK. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rassenti LZ, Huynh L, Toy TL, Chen L, Keating MJ, Gribben JG, Neuberg DS, Flinn IW, Rai KR, Byrd JC, Kay NE, Greaves A, Weiss A, Kipps TJ. N. Engl. J. Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 21.Ghia EM, Jain S, Widhopf GF, 2nd, Rassenti LZ, Keating MJ, Wierda WG, Gribben JG, Brown JR, Rai KR, Byrd JC, Kay NE, Greaves AW, Kipps TJ. Blood. 2008;111:5101–5108. doi: 10.1182/blood-2007-12-130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deligezer U, Erten N, Akisik EE, Dalay N. Exp. Mol. Pathol. 2006;80:72–76. doi: 10.1016/j.yexmp.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, Kipps TJ, Dighiero G, Schroeder HW, Jr, Ferrarini M, Chiorazzi N. J. Clin. Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 25.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 26.Kipps TJ, Rassenti LZ, Duffy S, Johnson T, Kobayashi R, Carson DA. Ann. N.Y. Acad. Sci. 1992;651:373–383. doi: 10.1111/j.1749-6632.1992.tb24638.x. [DOI] [PubMed] [Google Scholar]
- 27.Guarini A, Gaidano G, Mauro FR, Capello D, Mancini F, De Propris MS, Mancini M, Orsini E, Gentile M, Breccia M, Cuneo A, Castoldi G, Foa R. Blood. 2003;102:1035–1041. doi: 10.1182/blood-2002-12-3639. [DOI] [PubMed] [Google Scholar]
- 28.Giudicelli V, Chaume D, Jabado-Michaloud J, Le-franc MP. Stud. Health Technol. Inform. 2005;116:3–8. [PubMed] [Google Scholar]
- 29.Kabat EA, Wu TT. J. Immunol. 1991;147:1709–1719. [PubMed] [Google Scholar]
- 30.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Nat. Methods. 2006;3:369–375. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 31.Becker FF, Wang XB, Huang Y, Pethig R, Vykoukal J, Gascoyne PR. Proc. Natl. Acad. Sci. USA. 1995;92:860–864. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens M, Talary MS, Pethig R, Burnett AK, Mills KI. Bone Marrow Transplant. 1996;18:777–782. [PubMed] [Google Scholar]
- 33.Asbury CL, van den Engh G. Biophys. J. 1998;74:1024–1030. doi: 10.1016/s0006-3495(98)74027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes MP. In: Handbook of Nanoscience, Engineering, and Technology. 2nd edition. Goddard WA III, Brenner DW, Lyshevski SE, Iafrate GJ, editors. Boca Raton, FL: CRC Press; 2007. pp. 1–32. [Google Scholar]
- 35.Cheng J, Sheldon EL, Wu L, Uribe A, Gerrue LO, Carrino J, Heller MJ, O’Connell JP. Nat. Biotechnol. 1998;16:541–546. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 36.Cheng J, Sheldon EL, Wu L, Heller MJ, O’Connell JP. Anal. Chem. 1998;70:2321–2326. doi: 10.1021/ac971274g. [DOI] [PubMed] [Google Scholar]
- 37.Alazzam A, Stiharu I, Bhat R, Meguerditchian AN. Electrophoresis. 2011;32:1327–1336. doi: 10.1002/elps.201000625. [DOI] [PubMed] [Google Scholar]
- 38.Jaramillo Mdel C, Torrents E, Martínez-Duarte R, Madou MJ, Juárez A. Electrophoresis. 2010;31:2921–2928. doi: 10.1002/elps.201000082. [DOI] [PubMed] [Google Scholar]
- 39.Ermolina I, Milner J, Morgan H. Electrophoresis. 2006;27:3939–3948. doi: 10.1002/elps.200500928. [DOI] [PubMed] [Google Scholar]
- 40.Ramos A, Morgan H, Green NG, Castellanos A. J. Phys. D Appl. Phys. 1998;31:2338–2353. [Google Scholar]
- 41.Green NG, Ramos A, Morgan H. J. Phys. D Appl. Phys. 2000;33:632–641. [Google Scholar]
- 42.Cui L, Holmes D, Morgan H. Electrophoresis. 2001;22:3893–3901. doi: 10.1002/1522-2683(200110)22:18<3893::AID-ELPS3893>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Morgan H, Hughes MP, Green NG. Biophys. J. 1999;77:516–525. doi: 10.1016/S0006-3495(99)76908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ermolina I, Morgan H. J. Colloid Interface Sci. 2005;285:419–428. doi: 10.1016/j.jcis.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Asbury CL, Diercks AH, van den Engh G. Electrophoresis. 2002;23:2658–2666. doi: 10.1002/1522-2683(200208)23:16<2658::AID-ELPS2658>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 46.Washizu M, Kurusawa O, Arai I, Suzuki S, Shimamoto N. IEEE T. Ind. Appl. 1995;31:447–456. [Google Scholar]
- 47.Gagnon Z, Senapati S, Chang HC. Electrophoresis. 2010;31:666–671. doi: 10.1002/elps.200900473. [DOI] [PubMed] [Google Scholar]
- 48.Asokan SB, Jawerth L, Lloyd Carroll R, Cheney RE, Washburn S, Superfine R. Nano Lett. 2003;3:431–437. [Google Scholar]
- 49.Holzel R, Calander N, Chiragwandi Z, Willander M, Bier FF. Phys. Rev. Lett. 2005;95:128102. doi: 10.1103/PhysRevLett.95.128102. [DOI] [PubMed] [Google Scholar]
- 50.Han KH, Frazier AB. Lab Chip. 2008;8:1079–1086. doi: 10.1039/b802321b. [DOI] [PubMed] [Google Scholar]
- 51.Gao J, Sin ML, Liu T, Gau V, Liao JC, Wong PK. Lab Chip. 2011;11:1770–1775. doi: 10.1039/c1lc20054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pratt ED, Huang C, Hawkins BG, Gleghorn JP, Kirby BJ. Chem. Eng. Sci. 2011;66:1508–1522. doi: 10.1016/j.ces.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava SK, Artemiou A, Minerick AR. Electrophoresis. 2011;32:2530–2540. doi: 10.1002/elps.201100089. [DOI] [PubMed] [Google Scholar]
- 54.Kuczenski RS, Chang HC, Revzin A. Biomicrofluidics. 2011;5:032005. doi: 10.1063/1.3608135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krishnan R, Sullivan BD, Mifflin RL, Esener SC, Heller MJ. Electrophoresis. 2008;29:1765–1774. doi: 10.1002/elps.200800037. [DOI] [PubMed] [Google Scholar]
- 56.Krishnan R, Heller MJ. J. Biophotonics. 2009;2:253–261. doi: 10.1002/jbio.200910007. [DOI] [PubMed] [Google Scholar]
- 57.Krishnan R, Dehlinger DA, Gemmen GJ, Mifflin RL, Esener SC, Heller MJ. Electrochem. Commun. 2009;11:1661–1666. doi: 10.1016/j.elecom.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonnenberg A, Marciniak JY, Krishnan R, Heller MJ. Electrophoresis. 2012;33:2482–2490. doi: 10.1002/elps.201100700. [DOI] [PubMed] [Google Scholar]
- 59.Sonnenberg A, Marciniak JY, McCanna J, Krishnan R, Rassenti L, Kipps TJ, Heller MJ. Electrophoresis. 2013;34:1076–1084. doi: 10.1002/elps.201200444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonnenberg A, Marciniak JY, Rassenti L, Ghia EM, Skowronski EA, Manouchehri S, McCanna J, Widhopf GF, II, Kipps TJ, Heller MJ. Clin. Chem. 2014;60:500–509. doi: 10.1373/clinchem.2013.214874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen L, Huynh L, Apgar J, Tang L, Rassenti L, Weiss A, Kipps TJ. Blood. 2008;111:2685–2692. doi: 10.1182/blood-2006-12-062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agathangelidis A, Darzentas N, Hadzidimitriou A, Brochet X, Murray F, Yan X-J, Davis Z, van Gastel-Mol EJ, Tresoldi C, Chu CC, Cahill N, Giudicelli V, Tichy B, Pedersen LB, Foroni L, Bonello L, Janus A, Smedby K, Anagnostopoulos A, Merle-Beral H, Laoutaris N, Juliusson G, di Celle PF, Pospisilova S, Jurlander J, Geisler C, Tsaftaris A, Lefranc MP, Langerak AW, Oscier DG, Chiorazzi N, Belessi C, Davi F, Rosenquist R, Ghia P, Stamatopoulos K. Blood. 2012;119:4467–4475. doi: 10.1182/blood-2011-11-393694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Widhopf GF, 2nd, Rassenti LZ, Toy TL, Gribben JG, Wierda WG, Kipps TJ. Blood. 2004;104:2499–2504. doi: 10.1182/blood-2004-03-0818. [DOI] [PubMed] [Google Scholar]
- 64.Langerak AW, Davi F, Ghia P, Hadzidimitriou A, Murray F, Potter KN, Rosenquist R, Stamatopoulos K, Belessi C European Research Initiative on CLL (ERIC) Leukemia. 2011;25:979–984. doi: 10.1038/leu.2011.49. [DOI] [PubMed] [Google Scholar]
- 65.van der Drift MA, Hol BE, Klaassen CH, Prinsen CF, van Aarssen YA, Donders R, van der Stappen JW, Dekhuijzen PN, van der Heijden HF, Thunnissen FB. Lung Cancer. 2010;68:283–287. doi: 10.1016/j.lungcan.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 66.Guadalajara H, Dom´ınguez-Berzosa C, Garc´ıa-Arranz M, Herreros MD, Pascual I, Sanz-Baro R, Garc´ıa-Olmo DC, Garc´ıa-Olmo D. Cancer Detect. Prev. 2008;32:39–44. doi: 10.1016/j.cdp.2008.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.