Kif18A and Kif4A display a similar ability to attenuate the dynamics of microtubules but function to control the lengths of distinct subsets of spindle microtubules during mitosis. Kif18A and Kif4A are not functionally equivalent for chromosome alignment, and Kif18A's function in this process depends on its loop2 region.
Abstract
Microtubule length control is essential for the assembly and function of the mitotic spindle. Kinesin-like motor proteins that directly attenuate microtubule dynamics make key contributions to this control, but the specificity of these motors for different subpopulations of spindle microtubules is not understood. Kif18A (kinesin-8) localizes to the plus ends of the relatively slowly growing kinetochore fibers (K-fibers) and attenuates their dynamics, whereas Kif4A (kinesin-4) localizes to mitotic chromatin and suppresses the growth of highly dynamic, nonkinetochore microtubules. Although Kif18A and Kif4A similarly suppress microtubule growth in vitro, it remains unclear whether microtubule-attenuating motors control the lengths of K-fibers and nonkinetochore microtubules through a common mechanism. To address this question, we engineered chimeric kinesins that contain the Kif4A, Kif18B (kinesin-8), or Kif5B (kinesin-1) motor domain fused to the C-terminal tail of Kif18A. Each of these chimeric kinesins localizes to K-fibers; however, K-fiber length control requires an activity specific to kinesin-8s. Mutational studies of Kif18A indicate that this control depends on both its C-terminus and a unique, positively charged surface loop, called loop2, within the motor domain. These data support a model in which microtubule-attenuating kinesins are molecularly “tuned” to control the dynamics of specific subsets of spindle microtubules.
INTRODUCTION
During cell division, microtubule length control is critical for mitotic spindle positioning, spatial control of chromosome movements, and cytokinesis. However, it remains unclear how cells regulate the lengths of the different subpopulations of spindle microtubules necessary for these functions. For example, astral and nonkinetochore microtubules, which interact with the cell cortex and apply pushing forces to chromosome arms, display short half-lives, as well as fast polymerization and depolymerization rates (Zhai et al., 1995; Tirnauer et al., 2002; Stumpff et al., 2012). In contrast, the microtubules that bind to kinetochores—specialized protein structures that assemble at the centromeric regions of mitotic chromosomes—display relatively long half-lives with slower polymerization and depolymerization rates (Rieder and Salmon, 1994; Zhai et al., 1995). These kinetochore fibers (K-fibers) consist of 10–45 microtubules, and changes in their length facilitate the movement and alignment of chromosomes within the spindle (Rieder and Salmon, 1998). Whether cells use specialized or generalized mechanisms to control the lengths of dynamically diverse spindle microtubules is not understood.
Molecular motors of the kinesin-4 (Kif4A), kinesin-8 (Kif18A and Kif18B), and kinesin-13 (Kif2A, Kif2B, and MCAK) families play key roles in regulating spindle microtubule length to facilitate cell division in vertebrates (Kline-Smith and Walczak, 2002; Castoldi and Vernos, 2006; Manning et al., 2007; Mayr et al., 2007; Stumpff et al., 2008; Stout et al., 2011). The kinesin-13 motors function by inducing microtubule depolymerization, whereas Kif4A and Kif18A directly attenuate microtubule dynamic instability (Desai et al., 1999; Bringmann et al., 2004; Bieling et al., 2010; Du et al., 2010; Stumpff et al., 2011, 2012). Whereas individual kinesin-13 motors localize to and regulate both K-fibers and nonkinetochore microtubules, Kif4A, Kif18A, and Kif18B display specificity for particular microtubule subsets (Kline-Smith and Walczak, 2002; Moore et al., 2005; Lee et al., 2010; Stout et al., 2011; Tanenbaum et al., 2011; Stumpff et al., 2012; Wandke et al., 2012). Before anaphase, Kif4A localizes to mitotic chromatin, where it suppresses the lengths and growth rates of nonkinetochore microtubules (Lee et al., 2001; Stumpff et al., 2012; Wandke et al., 2012). In contrast, Kif18A localizes to and attenuates the dynamics of K-fibers, whereas Kif18B primarily localizes to and regulates the lengths of astral microtubules (Mayr et al., 2007; Stumpff et al., 2008; Lee et al., 2010; Stout et al., 2011; Tanenbaum et al., 2011).
Although the biochemical activity of Kif18B has not been determined, Kif18A and Kif4A similarly attenuate microtubule dynamics but display different motile behaviors. Kif18A and Kif4A are plus end–directed motors that accumulate at microtubule ends, where they suppress both polymerization and depolymerization (Bringmann et al., 2004; Bieling et al., 2010; Stumpff et al., 2012; Subramanian et al., 2013). Kif4A translocates along microtubules at a relatively fast velocity, where it forms a steady-state concentration at microtubule ends, indicating that individual Kif4A molecules readily dissociate from the microtubule tip (Bringmann et al., 2004; Bieling et al., 2010; Subramanian et al., 2013). In contrast, Kif18A is a slow, highly processive motor that stably associates with microtubule ends both in reconstituted systems and in mitotic cells (Du et al., 2010; Mayr et al., 2011; Stumpff et al., 2011). Stable microtubule end association is significantly reduced for Kif18A mutants lacking the C-terminal tail. The tail contains microtubule-binding domains that function to enhance the association of the protein with microtubules (Mayr et al., 2011; Stumpff et al., 2011; Su et al., 2011; Weaver et al., 2011). However, Kif18A truncation mutants still display some capacity to stably associate with microtubule ends, suggesting that regions within the Kif18A motor domain also contribute to microtubule tip binding (Stumpff et al., 2011). These differences in motor activity raise the question of whether Kif18A and Kif4A control microtubule lengths through a common mechanism or contain unique molecular attributes that facilitate their regulation of particular microtubule subsets.
To address this question, we tested the ability of the Kif4A, Kif18B, and Kif5B (kinesin-1) motor domains to functionally substitute for Kif18A during mitotic chromosome alignment. The kinesin-8 motor Kif18B, but not Kif4A or Kif5B, was capable of controlling K-fiber lengths to facilitate chromosome alignment, indicating that Kif4A and human kinesin-8 motors control microtubule lengths through distinct mechanisms. Furthermore, a positively charged surface loop within the motor domains of kinesin-8s, called loop2, is required for stable association with K-fiber ends, chromosome alignment, and spindle length control. These data support a model in which unique molecular structures within the motor domains of microtubule-attenuating kinesins facilitate their specific control of K-fiber versus nonkinetochore microtubule dynamics.
RESULTS
The Kif18A tail is not sufficient to facilitate accumulation of plus end–directed motors at K-fiber ends
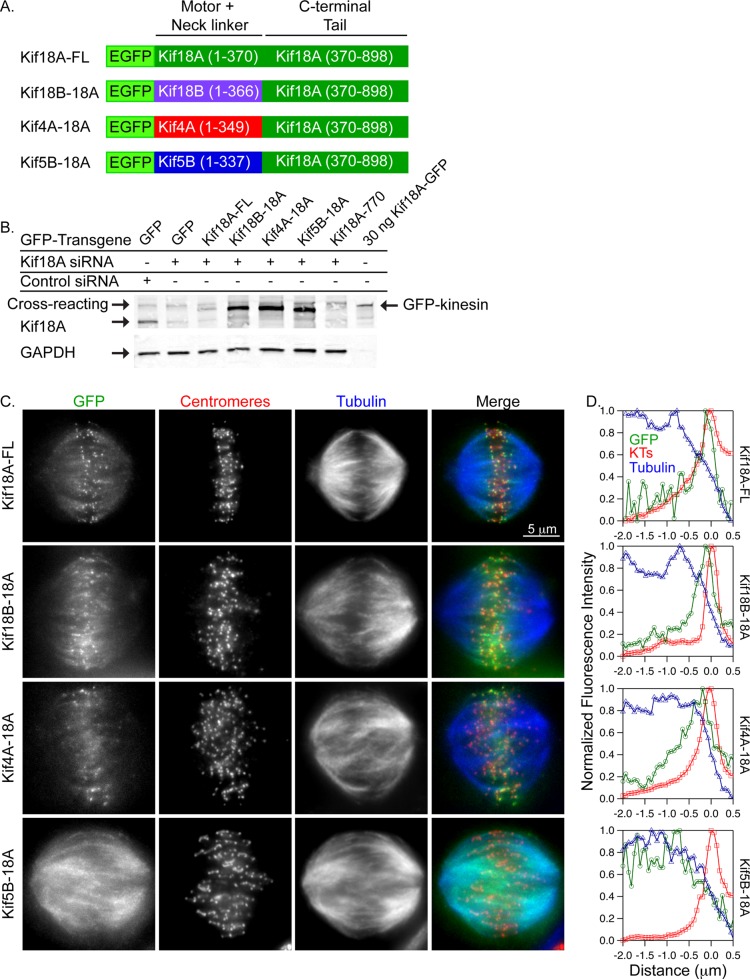
The Kif18A C-terminal tail is necessary for the motor's accumulation at K-fiber plus ends (Stumpff et al., 2011). To determine whether the Kif18A tail is sufficient to facilitate the concentration of other plus end–directed kinesins at K-fiber ends, we engineered chimeric kinesins consisting of the motor and neck-linker domains of Kif18B (kinesin-8), Kif4A (kinesin-4), and Kif5B (kinesin-1) fused to the Kif18A C-terminus (Figure 1A). Chimeric kinesins were expressed as enhanced green fluorescent protein (EGFP)-fusion proteins in HeLa cells depleted of endogenous Kif18A, and cells with similar GFP fluorescence levels were evaluated (Supplemental Figure S1). Kif18A small interfering RNA (siRNA)–treated cells expressed 9.8 ± 1.3% of the Kif18A detected by Western blot in control-treated cells (Figure 1B). Whereas all of the chimeras localized to K-fibers, only wild-type Kif18A and the Kif18B-18A chimera efficiently concentrated at K-fiber ends (Figure 1, C and D). Of interest, the Kif4A-18A chimera displayed accumulation at the ends of K-fibers on the spindle periphery but not at the ends of those on the spindle interior. In contrast, the Kif5B-18A chimera was uniformly distributed on the spindle (Figure 1, C and D). These data indicate that the Kif18A-tail can target plus end– directed kinesins to K-fibers; however, an element common to the kinesin-8 motors Kif18A and Kif18B is additionally required for efficient accumulation at K-fiber ends.
FIGURE 1:
The Kif18A C-terminus targets plus end–directed motors to K-fibers but is not sufficient for accumulation at K-fiber ends. (A) Schematic of Kif18A-tail chimeras. Numbers in parentheses indicate the amino acids used in each fragment. (B) Western blots of lysates transfected with the indicated siRNAs and GFP-tagged transgenes. Purified Kif18A-GFP (30 ng) was included for comparison. Blots were probed with anti-Kif18A (top) and anti-GAPDH (bottom) antibodies. The amount of Kif18A expressed in Kif18A siRNA–treated cells from three independent experiments is 9.8 ± 1.3% relative to controls. Note that Kif18A-770 does not contain the epitope recognized by the anti-Kif18A antibody. An anti-GFP blot of this sample is included in Supplemental Figure S2. (C) Fluorescence micrographs of the indicated GFP-tagged kinesins (green) in Kif18A-depleted HeLa cells immunostained with ACA (centromeres, red) and tubulin (blue). (D) Line scans of GFP-tagged kinesin (green trace) relative to tubulin (blue trace) and ACA fluorescence (red trace) along peripheral K-fibers.
A kinesin-8 motor domain specific activity facilitates stable binding of motors at K-fiber ends
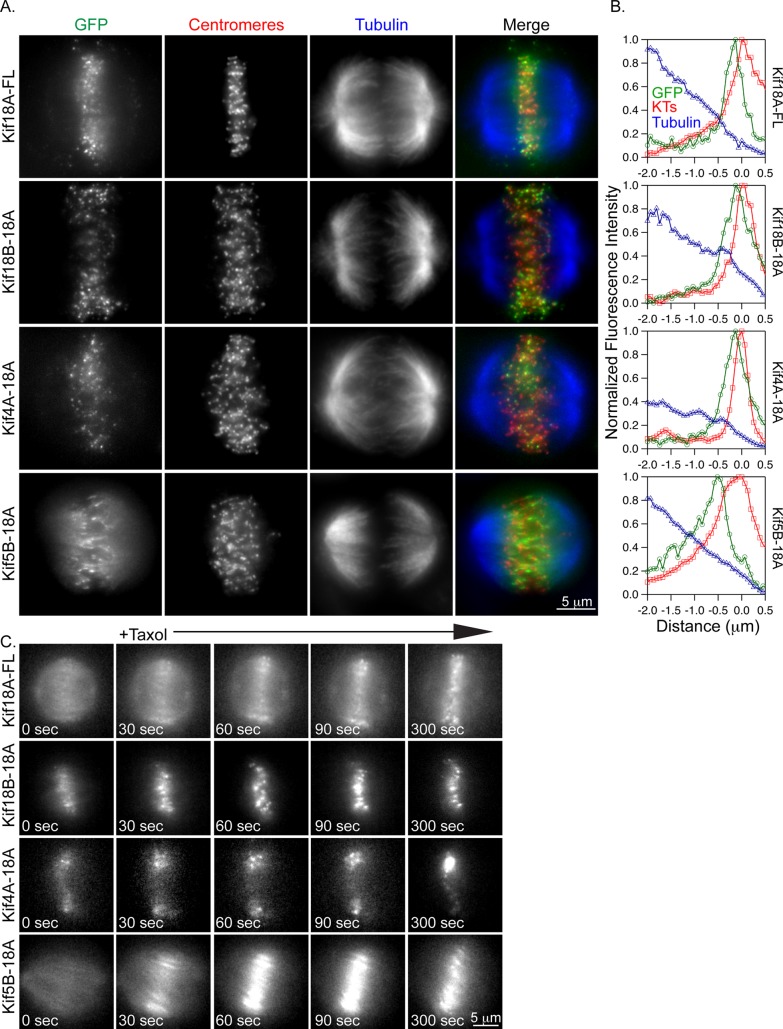
Given the ability of purified Kif4A to form a steady-state accumulation at the ends of stable microtubules in vitro, the accumulation of Kif4A-18A specifically at the ends of peripheral K-fibers could be explained if peripheral and interior K-fibers display differences in their dynamic instability (Subramanian et al., 2013). Consistent with this, previous studies demonstrated significant reduction in the oscillatory movements of kinetochores, which are closely coupled to K-fiber dynamic instability, on the periphery compared with those in the interior of the spindle (Canman et al., 2002; Cimini et al., 2004; Stumpff et al., 2008). Thus we hypothesized that peripheral K-fibers provide a stable track that allows a steady-state accumulation of Kif4A-18A. To test this idea, we briefly treated Kif18A-depleted HeLa cells expressing GFP-tagged chimeric kinesins with the microtubule-stabilizing drug Taxol (10 μM) and then analyzed kinesin localization. Similar to the previously observed behavior of Kif18A-FL in Taxol-treated cells, the concentration of each of the chimeric kinesins at the ends of K-fibers was increased after Taxol addition (Figure 2, A and B; Stumpff et al., 2011). GFP-chimera signal accumulated to K-fiber ends within 60–90 s of Taxol addition (Figure 2C). Of importance, this rapid accumulation is consistent with efficient plus end–directed movement of active motors to microtubule ends.
FIGURE 2:
Kif18A-tail chimeras rapidly accumulate at K-fiber ends in Taxol-treated cells. (A) Fluorescence micrographs of the indicated GFP-tagged kinesins (green) in Kif18A-depleted HeLa cells after a 5-min incubation with 10 μM Taxol. Cells were immunostained with ACA (centromeres, red) and tubulin (blue). (B) Line scans of GFP-tagged kinesins (green trace) relative to tubulin (blue trace) and ACA (red trace) along K-fibers in 10 μM Taxol–treated cells. (C) Still images from live-cell analyses of GFP-tagged kinesin relocalization after the addition of Taxol. Taxol was added immediately after the 0-s image was captured.
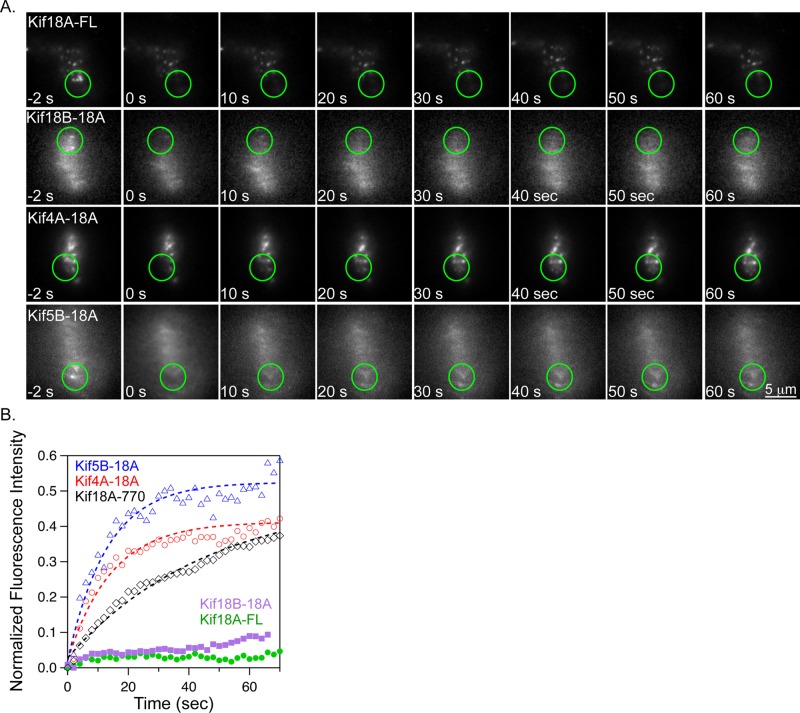
Although the localization of these chimeras was similar in Taxol-treated cells, it was not clear whether their behavior at K-fiber ends was comparable. To address this question, we used a fluorescence recovery after photobleaching (FRAP) assay to examine the kinetics of chimeric kinesins at K-fiber ends in Taxol-treated cells. For these experiments, a small region of interest containing GFP-kinesin was photobleached near a monomeric red fluorescent protein (mRFP)-CENP-B labeled centromere, and fluorescence recovery was visualized at 2-s intervals (Figure 3A). Wild-type Kif18A and the Kif18B-18A chimera displayed only a small mobile fraction at K-fiber ends under these conditions, consistent with previous studies of Kif18A turnover (Stumpff et al., 2011; Figure 3B and Table 1). In contrast, the Kif4A-18A and Kif5B-18A chimeras displayed significantly higher mobile fractions at K-fiber ends (Figure 3B and Table 1). A Kif18A truncation mutant (Kif18A-770) that lacks the microtubule-binding regions at the C-terminal end of the tail accumulates at K-fibers ends specifically in Taxol-treated cells (Supplemental Figure S2). Kif18A-770 displays an increase in the mobile fraction at K-fiber ends but does not fully recover (Figure 3B, Supplemental Figure S2, and Table 1). These data indicate that Kif18A's stable association at K-fiber ends requires both the C-terminal tail and a kinesin-8 motor–specific element that is shared between Kif18A and Kif18B. Furthermore, the behavior of Kif4A-18A and Kif5B-18A at the ends of Taxol-stabilized K-fibers is consistent with these motors forming a steady-state accumulation, similar to that observed for purified Kif4A (Subramanian et al., 2013).
FIGURE 3:
A kinesin-8–specific activity is required for stable association with K-fiber ends. (A) Stills from FRAP assays performed in live, Taxol-treated, Kif18A-depleted HeLa cells expressing the indicated GFP-tagged kinesin. Fluorescence intensity was measured before (−2 s) and after (0–60 s) a brief pulse with a 405-nm laser focused to the region indicated by the green circle. (B) Plot of relative GFP fluorescence as a function of time after a bleaching event for the indicated kinesins. Dashed lines are single-exponential fits to the data. Note that the Kif18B-18A and Kif18A-FL data did not fit a single exponential. Quantified data from FRAP experiments are reported in Table 1. Representative images of Kif18A-770 are included in Supplemental Figure S2.
TABLE 1:
FRAP measurements of kinesin behavior at K-fiber ends in Taxol-treated cells.
| GFP-kinesin | t1/2 (s) | Mobile fraction (%) | Fraction recovered 30 s after bleach (%) | Number of cells |
|---|---|---|---|---|
| Kif18A-FL | 45.0 ± 20.0 | 18.6 ± 8.8 | 6.6 ± 4.0 | 8 |
| Kif18B-18A | 19.5 ± 5.0** | 20.9 ± 8.2 | 12.2 ± 4.9 | 4 |
| Kif4A-18A | 10.4 ± 3.3** | 43.2 ± 21.5** | 40.4 ± 20.7** | 9 |
| Kif5B-18A | 8.3 ± 2.9** | 57.5 ± 16.3** | 51.9 ± 14.3** | 6 |
| Kif18A-770 | 20.4 ± 8.0** | 39.1 ± 17.0** | 23.5 ± 10.5** | 9 |
| Kif18A-L2-K6A | 16.2 ± 5.7** | 37.6 ± 17.0** | 29.4 ± 14.9** | 10 |
| Kif18A-ΔL2 | 13.7 ± 8.4** | 30.8 ± 10.7* | 22.9 ± 9.9** | 6 |
| Kif4A-L2-18A | 18.7 ± 9.9**,# | 25.9 ± 5.0*,# | 16.9 ± 6.1**,# | 9 |
*p ≤ 0.05 compared with Kif18A-FL, **p ≤ 0.01 compared with Kif18A-FL, #p ≤ 0.05 compared with Kif4A-18A.
The rates of fluorescence recovery in FRAP experiments, measured as the half-time to max recovery (t1/2) or the percentage of recovery after 30 s, also differed among the kinesins tested. The three kinesin-8 motor–containing constructs displayed relatively slow rates of recovery compared with the Kif4A-18A and Kif5B-18A chimeras (Figure 3B and Table 1). The rates of recovery correlate with the previously reported plus end–directed velocities measured for single molecules of full-length kinesin-8 (75–199 nm/s for Kif18A), tailless Kif18A (210–299 nm/s), kinesin-4 (800 nm/s for Xklp1), and Kif5B (780 nm/s for Kif5B-560), consistent with fluorescence recovery being facilitated by directional movement of motors into the bleached region (Lakamper et al., 2003; Bieling et al., 2010; Mayr et al., 2011; Stumpff et al., 2011).
Kif18A loop2 is required for its accumulation and stable binding at K-fiber ends
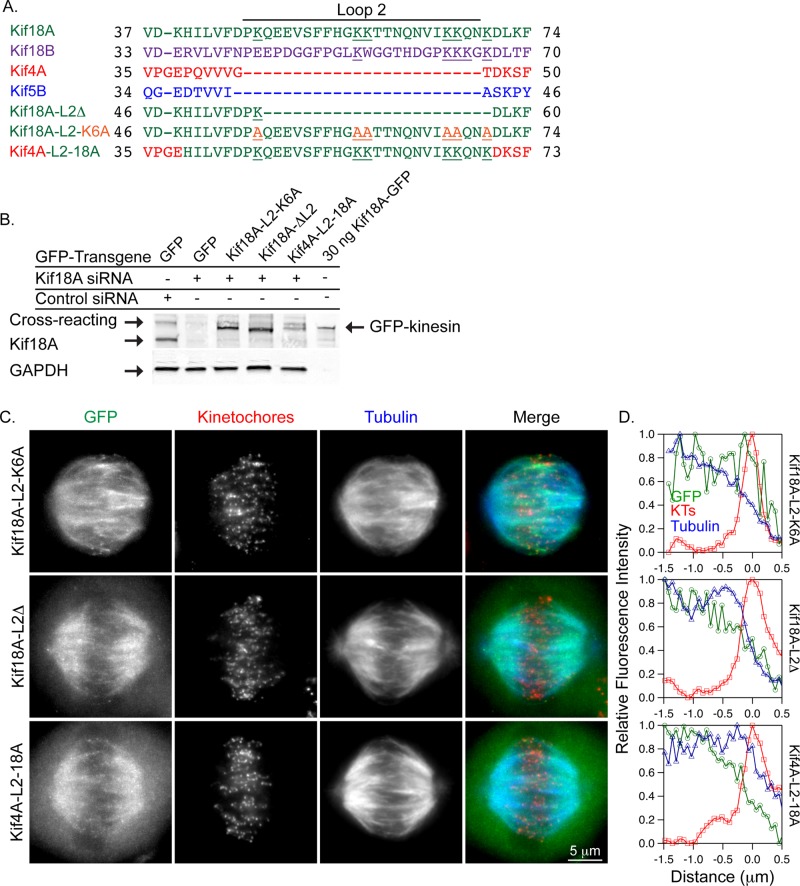
To understand the molecular basis of the differences in K-fiber end association displayed by kinesin-8 motors compared with Kif4A and Kif5B, we compared the sequences of these proteins to look for candidate structures in the kinesin-8s that might explain their ability to stably bind microtubule ends. This analysis revealed that Kif18A and Kif18B contain an extended loop2 region compared with Kif4A and Kif5B (Figure 4A). Kif18A loop2 contains six lysine residues, four of which are conserved in Kif18B (Figure 4A). Structural studies indicate that this positively charged extension in Kif18A directly contacts the negatively charged surface of α-tubulin (Peters et al., 2010). Furthermore, a similar loop2 structure is necessary for the microtubule depolymerization activity of kinesin-13 motors (Ogawa et al., 2004; Shipley et al., 2004). To test whether this unique surface loop contributes to kinesin-8–specific K-fiber end-binding activity, we engineered Kif18A mutants that contain a truncated loop2 (Kif18A-L2Δ) or alanine substitutions at each of the six lysine residues in loop2 (Kif18A-L2-K6A; Figure 4A).
FIGURE 4:
Kif18A's loop2 is necessary but not sufficient for motor accumulation at K-fiber ends. (A) Protein sequence alignment of the loop2 regions for the indicated kinesins. Kif18A's loop2 contains six lysine residues (underlined), which were mutated to alanine (orange, underlined) in Kif18A-L2-K6A. (B) Western blots of lysates transfected with the indicated siRNAs and GFP-tagged transgenes. Purified Kif18A-GFP (30 ng) was also loaded for comparison. Blots were probed with anti-Kif18A (top) and anti-GAPDH (bottom) antibodies. (C) Fluorescence micrographs of the indicated GFP-tagged kinesins (green) in cells immunostained with ACA (centromeres, red) and tubulin (blue). (D) Line scans of GFP-tagged kinesin (green trace) relative to tubulin (blue trace) and ACA fluorescence (red trace) along peripheral K-fibers.
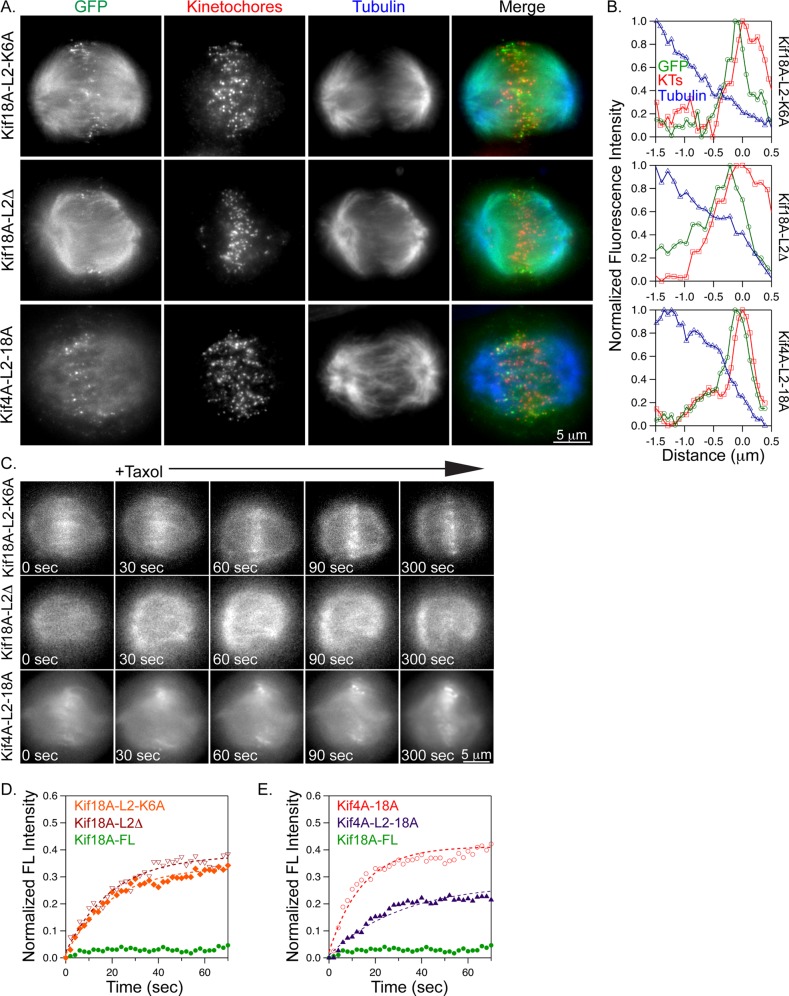
When expressed in mitotic cells depleted of endogenous Kif18A (Figure 4B), neither Kif18A-L2Δ nor Kif18A-L2-K6A efficiently accumulated at K-fiber plus ends (Figure 4, C and D). However, both loop2 mutants accumulated at K-fiber ends in cells briefly treated with 10 μM Taxol (Figure 5, A and B). The accumulation of Kif18A-L2-K6A occurred with similar kinetics to those displayed by the chimeric kinesins tested in this study; however, the Kif18A-L2Δ mutant accumulated more slowly and to a lesser extent, primarily on peripheral K-fibers (Figure 5C). FRAP studies revealed that both loop2 mutants displayed an increased mobile fraction at K-fiber ends in Taxol-treated cells compared with wild-type Kif18A (Figure 5D and Table 1). These data indicate that loop2 is necessary for Kif18A's stable association with K-fiber ends.
FIGURE 5:
Kif18A's loop2 is necessary but not sufficient for stable association with K-fiber ends. (A) Fluorescence micrographs of the indicated GFP-tagged loop2 mutant kinesins (green) in Kif18A-depleted HeLa cells after a 5-min incubation with Taxol. Cells were immunostained with ACA (centromeres, red) and tubulin (blue). (B) Line scans of GFP-tagged loop2 mutants (green trace) relative to tubulin (blue trace) and ACA (red trace) along K-fibers in 10 μM Taxol–treated cells. (C) Still images from live-cell analyses of GFP-tagged kinesin relocalization after the addition of 10 μM Taxol. Taxol was added immediately after the 0-s image was captured. (D, E) Plots displaying relative GFP fluorescence as a function of time after a bleaching event for the indicated kinesins. Dashed lines are single-exponential fits to the data. Quantified data from FRAP experiments are reported in Table 1.
To determine whether loop2 and the C-terminal tail of Kif18A are sufficient for the stable association of plus end–directed motors with K-fiber ends, we inserted the loop2 region of Kif18A into the Kif4A-18A chimera (Kif4A-L2-18A; Figure 4A). Kif4A-L2-18A failed to accumulate at K-fiber ends in cells with dynamic microtubules (Figure 4, C and D) but did concentrate efficiently at ends in Taxol-treated cells (Figure 5, A–C). In FRAP assays, Kif4A-L2-18A displayed a significantly reduced mobile fraction and a slower recovery rate compared with the Kif4A-18A chimera (Figure 5E and Table 1). Although the rate of recovery and mobile fraction for Kif4A-L2-18A were also significantly different from those measured for Kif18A-FL, they were comparable to the values obtained from FRAP analyses of Kif18B-18A. These data indicate that Kif18A's loop2 and C-terminal tail underlie the majority of the stable microtubule-end binding activity displayed by the two kinesin-8 motors.
The Kif4A motor domain cannot functionally substitute for Kif18A to control chromosome alignment and spindle length
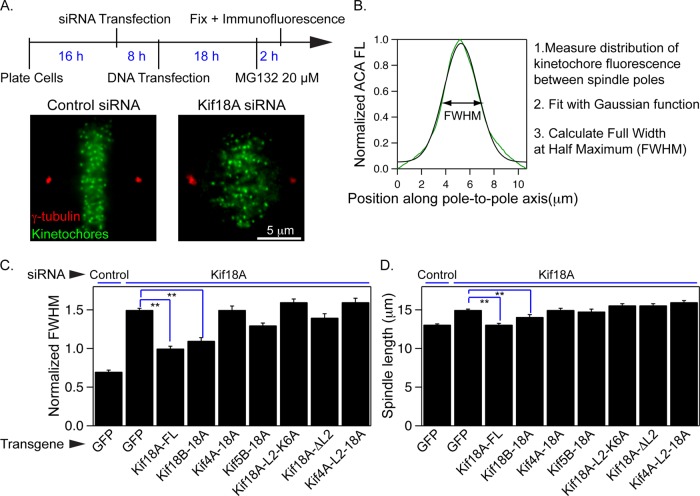
To determine whether loop2 is required for Kif18A's regulation of K-fiber lengths during mitosis, we assayed the ability of our chimeric kinesins and loop2 mutants to facilitate chromosome alignment and regulate spindle length in cells depleted of endogenous Kif18A. Cells depleted of Kif18A were transfected with GFP-tagged kinesins and then treated ∼18 h later with MG132 (20 μM), which prevents anaphase entry (Figure 6A). Cells were then fixed and immunofluorescently stained with antibodies against centromeric (ACA) and centrosomal (γ-tubulin) proteins. Under these conditions, the majority of cells transfected with scrambled control siRNAs and a GFP control plasmid had well-aligned kinetochores. In contrast, cells transfected with Kif18A siRNAs and a GFP control plasmid displayed unaligned kinetochores (Figure 6A).
FIGURE 6:
Kinesin-8 loop2 is required for mitotic chromosome alignment and spindle length regulation. (A) Schematic of experimental design. Representative images show control and Kif18A siRNA–treated cells immunostained with ACA (centromeres, green) and γ-tubulin (red) antibodies. (B) Representative plot of centromere (ACA) fluorescence distribution as a function of position along the pole-to-pole axis. Fluorescence data were fitted with a Gaussian function, and the full-width at half-maximum (FWHM) was calculated as a metric for centromere alignment. (C, D) Graphs of average FWHM (C) and average spindle length (D) calculated from cells transfected with the indicated siRNAs and transgenes. **p < 0.01.
Chromosome alignment was quantified in cells with similar GFP expression levels (Supplemental Figure S1) by measuring the distribution of ACA fluorescence along the pole-to-pole axis and computing the full-width at half-maximum of a Gaussian fit to the distribution (Figure 6B). Spindle length was measured as the distance between the two centrosomes. Kif18A-FL and the Kif18B-18A chimeras were able to facilitate chromosome alignment and reduce spindle length in the absence of endogenous Kif18A (Figure 6, C and D). In contrast, the Kif4A-18A, Kif4A-L2-18A, and Kif5B-18A chimeras were not capable of aligning chromosomes or reducing spindle length compared with GFP-expressing cells depleted of Kif18A (Figure 6, C and D). Similarly, the Kif18A-L2Δ and Kif18A-L2-K6A mutants failed to align chromosomes or reduce spindle length in the absence of endogenous Kif18A (Figure 6, C and D). These data indicate that loop2 provides an essential activity for Kif18A's function in regulating K-fiber lengths to align mitotic chromosomes. However, the inability of Kif4A-L2-18A to rescue chromosome alignment and spindle length in Kif18A-depleted cells suggests that these regions are not sufficient for Kif18A's mitotic functions.
DISCUSSION
The molecular control of microtubule dynamics is essential for diverse cellular processes, such as division, migration, and morphogenesis. Although microtubules are intrinsically dynamic, their growth and shortening are modulated in cells by polymerizers, depolymerizers, and attenuators. This regulation is complex within structures that comprise closely packed microtubule subpopulations with dramatically different dynamic properties, such as the mitotic spindle. The data presented here indicate that the Kif4A and Kif18A motor domains are not functionally equivalent for spindle microtubule length control and likely attenuate microtubule dynamics through distinct mechanisms. Furthermore, Kif18A's specific ability to control K-fiber lengths requires a unique, positively charged loop2 region common to kinesin-8 motors. This implies that the differential control of K-fibers and nonkinetochore microtubules relies in part on the specialized activities of microtubule-attenuating kinesins.
Kif4A controls the lengths and dynamics of nonkinetochore microtubules within the spindle to regulate polar ejection forces and spindle length (Stumpff et al., 2012; Wandke et al., 2012). Although Kif4A displays a similar attenuation of microtubule dynamics to Kif18A, it does not have an extended loop2 region or stably associate with microtubule ends (Bringmann et al., 2004; Bieling et al., 2010; Stumpff et al., 2012; Subramanian et al., 2013). Consistent with loop2 and stable end binding being required for K-fiber length control, the Kif4A-18A chimera was unable to regulate chromosome alignment or spindle length. These data suggest that Kif4A and Kif18A attenuate microtubule dynamics through distinct mechanisms. We speculate that Kif4A's fast velocity facilitates its steady-state accumulation at the ends of fast-growing nonkinetochore microtubules within the spindle. This concentration of motor could in turn inhibit addition and loss of tubulin. On the assumption that a threshold of motor accumulation is necessary to attenuate microtubule dynamics, such a mechanism could efficiently control the lengths of individual microtubules, but stable association with microtubule tips, as exhibited by the Kif18A and Kif18B motor domains, may be required to reach the motor concentrations necessary to attenuate the dynamics of the 15–20 microtubules in a HeLa cell K-fiber (McEwen et al., 2001).
Kif18A controls K-fiber lengths to spatially confine the movements of mitotic chromosomes to the spindle equator (Stumpff et al., 2008, 2012). This function depends on Kif18A's highly processive plus end–directed motility, stable association with microtubule ends, and direct suppression of microtubule dynamics (Mayr et al., 2011; Stumpff et al., 2011). Microtubule-binding regions found in the C-terminal tails of kinesin-8s promote targeting of these motors to the ends of microtubules by increasing motor processivity (Mayr et al., 2011; Stumpff et al., 2011; Su et al., 2011; Weaver et al., 2011). However, Kif18A's tail does not account for all of the motor's microtubule end–specific functions. For example, the ability of a Kif18A-truncation mutant lacking the tail to dwell at microtubule ends and suppress their dynamics is attenuated but not ablated, indicating that these properties are intrinsic to the Kif18A motor domain (Stumpff et al., 2011). Our data indicate that the loop2 region of kinesin-8s underlies these unique motor activities.
The contribution of Kif18A's loop2 to its function in controlling K-fiber lengths is particularly intriguing because an analogous region is required for kinesin-13–dependent microtubule depolymerization. Kinesin-13 loop2 forms an antiparallel β-sheet structure that ends with a KVD finger, which is essential for microtubule depolymerization activity but not microtubule binding (Ogawa et al., 2004; Shipley et al., 2004). These data suggest that kinesin-13 loop2 has a microtubule end–specific function. Consistent with this, loop2 is necessary for kinesin-13s to induce curvature between α-tubulin and β-tubulin subunits, an activity predicted to facilitate microtubule disassembly (Desai et al., 1999; Moores et al., 2002; Tan et al., 2008; Asenjo et al., 2013).
Our data suggest that, similar to kinesin-13 loop2, Kif18A's loop2 promotes microtubule tip association and functions in the regulation of K-fiber dynamics. However, in contrast to kinesin-13s, loop2 in Kif18A is longer, conformationally dynamic, and lacks a KVD finger (Peters et al., 2010). Cryo–electron microscopy studies indicate that the positively charged Kif18A loop2 directly associates with the negatively charged surface of α-tubulin (Peters et al., 2010). We speculate that the structural differences between the loop2 regions of kinesin-13s and Kif18A could explain the different effects of these motors on microtubule dynamics. Kif18A's loop2 could analogously modify the conformation of tubulin at microtubule tips in such a way that it inhibits loss or gain of tubulin dimers from the end rather than inducing terminal tubulin removal. Furthermore, stable association with microtubule tips may perturb the ability of other microtubule regulators to access microtubule ends. Our analyses of the Kif4A-L2-18A chimera indicate that although loop2 and the Kif18A tail facilitate Kif4A's stable association with K-fiber ends, they are not sufficient for K-fiber length control. Thus other, yet-to-be-identified differences between the structure and activity of the Kif4A and Kif18A motors likely contribute to the functional specificity of these kinesins.
Kif18B controls the lengths of astral microtubules during mitosis, but whether it functions as a microtubule attenuator or depolymerizer is unresolved (Stout et al., 2011; Tanenbaum et al., 2011). The similarity between the loop2 regions of Kif18A and Kif18B and the ability of the Kif18B-18A chimera to functionally substitute for Kif18A during chromosome alignment in our studies suggest that Kif18B attenuates microtubule dynamic instability similar to Kif18A. This implies that differentially localized kinesin-8s may use similar mechanisms to regulate distinct subsets of spindle microtubules. Although all kinesin-8 motors appear to have an extended loop2, the depolymerizing kinesin-8, Kip3p, has a longer (38 amino acids) and divergent loop2 compared with Kif18A and Kif18B (Gupta et al., 2006; Varga et al., 2006; Peters et al., 2010). Perhaps differences in the interactions of kinesin-8 loop2 structures with tubulin at the ends of microtubules can explain the diversity of effects on microtubule dynamics observed for kinesin-8 family members (Gupta et al., 2006; Varga et al., 2006; Du et al., 2010; Stumpff et al., 2011; Erent et al., 2012; Niwa et al., 2012).
MATERIALS AND METHODS
Plasmids and siRNAs
Chimeric kinesins were constructed from the codons indicated in Figures 1A and 4A by overlap extension PCR. Site-directed mutagenesis was used to make Kif18A-L2Δ and introduce silent mutations within the motor domains of Kif18A-FL and all Kif18A mutants to facilitate siRNA resistance (forward primer sequence, 5′-CGTCCGGAAAACACTAAAGAAAAAGCAGCaGGcTTcCAcAAAGTGGTTCATGTTGTGG-3′). The Kif18A-L2-K6A construct was synthesized (Life Technologies, Carlsbad, CA). Construction of mRFP-CENP-B was previously described (Stumpff et al., 2008). The scrambled control (Silencer Negative Control #1, Life Technologies) and validated Silencer Kif18A siRNAs (5′-GCUGGAUUUCAUAAAGUGG-3′; Life Technologies) used were also described previously (Stumpff et al., 2008, 2012).
Cell culture and transfections
HeLa cells were cultured at 37ºC and 5% CO2 in MEM-alpha (Life Technologies) containing 10% fetal bovine serum (FBS; Life Technologies) plus antibiotics. For siRNA transfections, (5–6.5) × 105 cells were plated in 60-mm2 dishes. Approximately 16 h later, cells were treated with 300 pmol of siRNA complexed with 12 μl of RNAiMax (Life Technologies) following the manufacturer's instructions. After 8 h, cells were trypsinized, pooled, and transfected with plasmid DNA using a Nucleofector 4D system (Lonza, Walkersville, MD). Electroporated cells were then plated on 12-mm glass coverslips (Electron Microscopy Sciences, Hatfield, PA) for fixed-cell assays or poly-l-lysine–coated, 35-mm2 glass-bottom dishes (MatTek, Ashland, MA) for live-cell studies. Cells were treated with 10 μM Taxol or 20 μM MG132 at 5 min or 2 h before fixation, respectively, as indicated in the text.
Western blot analysis
Cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40, and 0.1% SDS) 20 h after DNA transfection and 28 h after siRNA addition. Lysates were extracted on ice for 10 min. An equal volume of 2× Laemmli buffer was then added, and lysates were boiled for 10 min. Recombinant Kif18A-GFP was purified as previously described (Stumpff et al., 2011). Lysates and purified proteins were separated by electrophoresis on 4–15% Tris-glycine polyacrylamide gels (Bio-Rad, Hercules, CA) and transferred to polyvinylidene fluoride membrane (Bio-Rad). Membranes were blocked in Odyssey blocking buffer (1:1 ratio of Tris-buffered saline [TBS; 50 mM Tris-Cl, pH 7.4, and 150 mM NaCl] and Odyssey blocking reagent [Li-COR, Lincoln, NE]). Blocked membranes were probed with 1 μg/ml rabbit anti-Kif18A antibodies (Bethyl Laboratories, Burlington, ON, Canada), 4 μg/ml rabbit anti-GFP antibodies (Life Technologies), or 0.5 μg/ml mouse anti– glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies (Millipore, Billerica, MA) diluted in Odyssey blocking buffer with 0.1% Tween-20. Secondary DyLight 800 anti–rabbit immunoglobulin G (IgG) and DyLight 680 anti–mouse IgG antibodies (Thermo Scientific, Waltham, MA) were diluted to 0.03 μg/ml in Odyssey blocking buffer with 0.1% Tween-20 and 0.01% SDS. Secondary antibody fluorescence was detected with an Odyssey CLx (Li-COR). Kif18A knockdown was quantified from three independent experiments by densitometry using ImageJ software (National Institutes of Health, Bethesda, MD). The amount of Kif18A remaining in each Kif18A siRNA treated lysate was determined by comparing the Kif18A band density to that in the matched control lysate and correcting for protein load relative to the GAPDH signal.
Immunofluorescence microscopy
Cells were fixed on coverslips for 10 min in 1% paraformaldehyde (Electron Microscopy Sciences)/−20º C methanol, washed in 1× TBS, and blocked in 20% goat serum in antibody-diluting solution (1× TBS, 2% bovine serum albumin, 0.1% azide, 0.1% Triton-X 100) for 1 h. Cells were labeled with the following primary antibodies: 2.5 μg/ml human anti–centromere serum (Antibodies Incorporated, Davis, CA), 1 μg/ml mouse anti–α-tubulin (Sigma-Aldrich, St. Louis, MO), and 1 μg/ml mouse anti–γ-tubulin (Sigma-Aldrich). Cells were labeled with goat anti-human Alexa Fluor 594 or goat anti-mouse Alexa Fluor 647 secondary antibodies at 1 μg/ml (Life Technologies). Coverslips were mounted on glass slides in Prolong Gold antifade reagent plus 4′,6-diamidino-2-phenylindole (Life Technologies). Cells were imaged on a Nikon Ti-E inverted microscope controlled by NIS Elements software (Nikon Instruments, Melville, NY) with APO 100×/1.49 numerical aperture (NA) and Plan APO 60×/1.42 NA oil immersion objectives (Nikon Instruments), Spectra-X light engine (Lumencore, Beaverton, OR), and Clara cooled charge-coupled device (CCD) camera (Andor, South Windsor, CT). Optical sections were collected at 200-nm intervals through each spindle. Representative images are maximum intensity projections of five optical slices centered on the pole-to-pole axis. Brightness and contrast were adjusted using Photoshop CS6 (Adobe, San Jose, CA). Two-dimensional linear protein mapping was performed within single focal planes using the linescan tool in NIS Elements.
FRAP studies
Kif18A-depleted HeLa cells transfected with mRFP-CENP-B and GFP-tagged kinesin constructs were switched into CO2-independent medium containing 10% FBS (Life Technologies) just before imaging. For analyses of kinesin redistribution after Taxol addition, mitotic cells were imaged for 20 s at 10-s intervals, and then an equal volume of 37ºC CO2-independent medium containing Taxol was added so the final Taxol concentration was 10 μM. Cells were then imaged at 10-s intervals for 10 min. For FRAP assays, GFP-expressing cells were imaged at 2-s intervals for 10 s before bleaching. GFP foci near mRFP-CENP-B foci were then photobleached using a point-focused 405-nm laser, and GFP imaging was continued at 2-s intervals. GFP fluorescence was quantified in the bleached zone before and after laser activation using NIS Elements software. Fluorescence data were analyzed and plotted using Igor Pro (Wavemetrics, Portland, OR), and statistical comparisons were performed using a Student's two-tailed t test. Fluorescence intensities were normalized such that the prebleach value is set to 1 and the postbleach value is 0. Imaging was performed on the Nikon Ti-E inverted microscope described earlier, but images were captured with an iXon X3 electron-multiplying CCD camera (Andor) to minimize exposure times and photobleaching.
Quantification of chromosome alignment
Single focal plane images of MG132-arrested cells where both centrosomes were in the same focal plane were captured and rotated such that the pole-to-pole axis was horizontal. The Plot Profile command in ImageJ was used to measure the distribution of ACA fluorescence within a rectangular region of interest (ROI) with a length defined by the distance between the centrosomes and a set height of 17.5 μm. The average normalized ACA fluorescence in each pixel column within the ROI was plotted as a function of position along the pole-to-pole axis and fitted with a Gaussian in Igor Pro. The full-width at half-maximum of the Gaussian was calculated as a metric for chromosome alignment in a single cell. Statistical comparisons were performed using a Student's two-tailed t test.
Supplementary Material
Acknowledgments
We thank David Warshaw and Christopher Berger for insightful suggestions and critical feedback on the manuscript. This work was supported by a Leukemia and Lymphoma Society Special Fellow Award (3652-11), a Basil O'Connor Research Starter Scholar Award (#5-FY14-33), and a Vermont Cancer Center/LCCRO Program Award to J.S.
Abbreviations used:
- ACA
anti-centromere antibodies
- CENP-B
centromere protein-B
- EGFP
enhanced green fluorescent protein
- FL
full length
- FRAP
fluorescence recovery after photobleaching
- FWHM
full-width at half-maximum
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- K-fibers
kinetochore fibers
- Kif
kinesin family
- L2
loop2
- mRFP
monomeric red fluorescent protein
- siRNA
small interfering RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1132) on September 10, 2014.
REFERENCES
- Asenjo AB, Chatterjee C, Tan D, DePaoli V, Rice WJ, Diaz-Avalos R, Silvestry M, Sosa H. Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. Cell Rep. 2013;3:759–768. doi: 10.1016/j.celrep.2013.01.030. [DOI] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303:1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- Canman JC, Salmon ED, Fang G. Inducing precocious anaphase in cultured mammalian cells. Cell Motil Cytoskeleton. 2002;52:61–65. doi: 10.1002/cm.10032. [DOI] [PubMed] [Google Scholar]
- Castoldi M, Vernos I. Chromokinesin Xklp1 contributes to the regulation of microtubule density and organization during spindle assembly. Mol Biol Cell. 2006;17:1451–1460. doi: 10.1091/mbc.E05-04-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Erent M, Drummond DR, Cross RA. S. pombe kinesins-8 promote both nucleation and catastrophe of microtubules. PloS One. 2012;7:e30738. doi: 10.1371/journal.pone.0030738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Kline-Smith SL, Walczak CE. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol Biol Cell. 2002;13:2718–2731. doi: 10.1091/mbc.E01-12-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakamper S, Kallipolitou A, Woehlke G, Schliwa M, Meyhofer E. Single fungal kinesin motor molecules move processively along microtubules. Biophys J. 2003;84:1833–1843. doi: 10.1016/S0006-3495(03)74991-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Kim E, Park M, Moon E, Ahn SM, Kim W, Hwang KB, Kim YK, Choi W, Kim W. Cell cycle-regulated expression and subcellular localization of a kinesin-8 member human KIF18B. Gene. 2010;466:16–25. doi: 10.1016/j.gene.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Lee YM, Lee S, Lee E, Shin H, Hahn H, Choi W, Kim W. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem J. 2001;360:549–556. doi: 10.1042/0264-6021:3600549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Mayr MI, Storch M, Howard J, Mayer TU. A non-motor microtubule binding site is essential for the high processivity and mitotic function of kinesin-8 Kif18A. PloS One. 2011;6:e27471. doi: 10.1371/journal.pone.0027471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Chan GK, Zubrowski B, Savoian MS, Sauer MT, Yen TJ. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, Andrieux A, Job D, Wordeman L. MCAK associates with the tips of polymerizing microtubules. J Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores CA, Yu M, Guo J, Beraud C, Sakowicz R, Milligan RA. A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell. 2002;9:903–909. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- Niwa S, Nakajima K, Miki H, Minato Y, Wang D, Hirokawa N. KIF19A Is a Microtubule-depolymerizing kinesin for ciliary length control. Dev Cell. 2012;23:1167–1175. doi: 10.1016/j.devcel.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- Peters C, Brejc K, Belmont L, Bodey AJ, Lee Y, Yu M, Guo J, Sakowicz R, Hartman J, Moores CA. Insight into the molecular mechanism of the multitasking kinesin-8 motor. EMBO J. 2010;29:3437–3447. doi: 10.1038/emboj.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipley K, Hekmat-Nejad M, Turner J, Moores C, Anderson R, Milligan R, Sakowicz R, Fletterick R. Structure of a kinesin microtubule depolymerization machine. EMBO J. 2004;23:1422–1432. doi: 10.1038/sj.emboj.7600165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell. 2011;22:3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev Cell. 2012;22:1017–1029. doi: 10.1016/j.devcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Ti SC, Tan L, Darst SA, Kapoor TM. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 2013;154:377–390. doi: 10.1016/j.cell.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Rice WJ, Sosa H. Structure of the kinesin13-microtubule ring complex. Structure. 2008;16:1732–1739. doi: 10.1016/j.str.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Wandke C, Barisic M, Sigl R, Rauch V, Wolf F, Amaro AC, Tan CH, Pereira AJ, Kutay U, Maiato H, et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J Cell Biol. 2012;198:847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, LeBlanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21:1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.