By binding to both the E1-E2 core and the E3 subunit, Kgd4 acts as a molecular adaptor that is necessary to form a stable α-ketoglutarate dehydrogenase enzyme complex.
Abstract
The mitochondrial citric acid cycle is a central hub of cellular metabolism, providing intermediates for biosynthetic pathways and channeling electrons to the respiratory chain complexes. In this study, we elucidated the composition and organization of the multienzyme complex α-ketoglutarate dehydrogenase (α-KGDH). In addition to the three classical E1-E3 subunits, we identified a novel component, Kgd4 (Ymr31/MRPS36), which was previously assigned to be a subunit of the mitochondrial ribosome. Biochemical analyses demonstrate that this protein plays an evolutionarily conserved role in the organization of mitochondrial α-KGDH complexes of fungi and animals. By binding to both the E1-E2 core and the E3 subunit, Kgd4 acts as a molecular adaptor that is necessary to a form a stable α-KGDH enzyme complex. Our work thus reveals a novel subunit of a key citric acid–cycle enzyme and shows how this large complex is organized.
INTRODUCTION
The mitochondrial citric acid cycle is a central hub for cellular metabolism. It channels electrons from reduced substrates to the membrane-bound respiratory chain complexes for efficient energy conversion. In addition, intermediates of the citric acid cycle serve as precursors for synthesis of amino acids, nucleotides, lipids, and redox cofactors. A central enzyme of the citric acid cycle is α-ketoglutarate dehydrogenase (α-KGDH), which catalyzes the oxidative decarboxylation of α-ketoglutarate to succinyl-CoA and the concomitant formation of NADH and CO2. Succinyl-CoA is then converted to succinate by succinyl-CoA synthetase or used as a substrate in heme biosynthesis. In addition to this basic role in energy metabolism, KGDH is a major site of reactive oxygen species generation and plays an important role in metabolic reprogramming in cancers and during neurodegenerative diseases (Gibson et al., 2005; Calingasan et al., 2008; McLain et al., 2011; Trofimova et al., 2012; Soga, 2013; Quinlan et al., 2014).
The catalytic activities of KGDH are found in three polypeptides, in which E1 represents the α-KGDH, E2 is the dihydrolipoyl succinyltransferase, and E3 has dihydrolipoyl dehydrogenase activity (Reed and Hackert, 1990). The genes encoding the subunits of mitochondrial KGDH were identified first in yeast and were named KGD1 (E1), KGD2 (E2), and LPD1 (E3) (Roy and Dawes, 1987; Repetto and Tzagoloff, 1989, 1990). In an attempt to identify the composition and organization of the mitochondrial KGDH, we discovered a novel subunit we termed Kgd4. This subunit is part of a conserved family of proteins that were previously annotated to be components of the small subunit of the mitochondrial ribosome. Our data establish that these proteins instead play an important role in the organization of mitochondrial KGDH, because they are necessary for recruiting the E3 subunit to the E1-E2 core of the enzyme complex.
RESULTS
Kgd4 copurifies with α-KGDH
To analyze the composition and organization of mitochondrial KGDH, we designed a strategy to purify the enzyme from isolated mitochondria and identify copurified proteins by mass spectrometry. We created a yeast strain expressing C-terminally His7-tagged Kgd1. Mitochondria from this strain were isolated and lysed in Triton X-100, and KGDH was purified through metal affinity chromatography (Figure 1A). Bands that were enriched in the Kgd1-His7 purifications in comparison with the untagged control (wild type) were excised and analyzed by mass spectrometry (Figure 1B). This led to the identification of four proteins that specifically copurified: Kgd1, Kgd2, and Lpd1, the catalytic subunits of KGDH, and two bands containing peptides of Ymr31, suggesting this protein is a subunit of the complex. This was surprising because Ymr31 was previously described as a part of the mitochondrial ribosome (Matsushita et al., 1989; Cavdar Koc et al., 2001). However, purification of mitochondrial ribosomes typically uses sequential centrifugation steps that do not allow separating the small ribosomal subunit from the KGDH, because both have a similar size; the highly abundant KGDH is therefore found to contaminate ribosomal preparations (Koc et al., 2013). To confirm our finding that Ymr31 copurifies with KGDH, we equipped Ymr31 with a C-terminal His7-tag and purified complexes containing Ymr31. These experiments revealed that subunits of KGDH could be specifically recovered with Ymr31 (Figure 1C), but Lpd1 was more quantitatively enriched than Kgd1 and Kgd2, which likely reflects a partial dissociation of the complex during the rather harsh purification procedure. Importantly, proteins of either the large (Mrpl4) or the small (Mrps5) subunit of the mitochondrial ribosome were not copurified. We therefore concluded that Ymr31 is not part of the mitochondrial ribosome but likely a novel subunit of KGDH, and we renamed Ymr31 as α-KGDH 4 (Kgd4).
FIGURE 1:
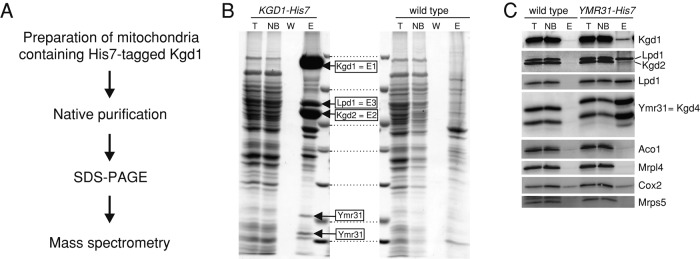
Kgd4 copurifies with α-KGDH. (A) Strategy to analyze the composition of KGDH. (B) Mitochondrial KGDH was purified using a C-terminal His7-tag on Kgd1. Proteins were separated on SDS–PAGE and analyzed by Coomassie staining and mass spectrometry. Boxes indicate proteins specifically identified in the elution fraction of the sample containing His7-tagged Kgd1. (C) Complexes of Ymr31 were purified as in B, and the purifications were analyzed by Western blotting using the indicated antibodies. T, total of the lysate; NB, unbound fraction; W, wash fraction; E, elution fraction.
Absence of Kgd4 decreases KGDH activity in vitro without destabilizing the catalytic subunits
We next set out to determine the impact of KGD4 deletion on KGDH activity and prepared lysates from mitochondria containing or lacking Kgd4. Robust KGDH activity was observed in lysates from wild-type cells, while KGDH activity was dramatically reduced in the absence of Kgd4 (Figure 2A). In contrast, malate dehydrogenase (MDH) activity was similar in both mitochondrial lysates (Figure 2B). A possible explanation for this strong KGDH activity defect in the absence of Kgd4 could be that a catalytic subunit is destabilized. To analyze which impact the deletion of an individual KGDH subunit has on the accumulation of the other subunits, we analyzed proteins from mitochondria isolated from cells lacking Kgd1, Kgd2, Kgd4, or Lpd1. Only Kgd1 levels were slightly decreased in the absence of Kgd2, while the steady-state levels of Lpd1 and Kgd2 were not affected by the absence of one of the other subunits (Figure 2C). Similarly, the mitochondrial ribosome (Mrpl4) or the mitochondrially encoded Cox2 protein accumulated normally in the absence of Kgd4, substantiating the notion that Kgd4 is not a ribosomal protein. We also tested for attachment of the essential cofactor lipoic acid to Kgd2 by Western blotting, but did not observe a decrease in lipoylation in the absence of Kgd4 (Figure 2D). In summary, the catalytic subunits of KGDH are not destabilized or nonfunctional in the absence of Kgd4.
FIGURE 2:
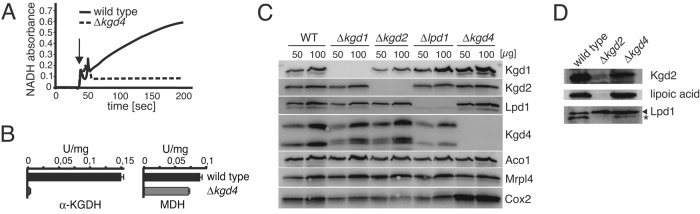
Absence of Kgd4 decreases KGDH activity in vitro without destabilizing the catalytic subunits. (A) Mitochondria from wild-type cells or cells lacking Kgd4 were lysed, and KGDH activity was photometrically determined. (B) Quantification of three independent enzyme assays for KGDH and MDH activity. (C) Total cellular protein was extracted from the indicated cells and analyzed by Western blotting. (D) Mitochondria of the indicated strains were analyzed by Western blotting using the indicated antibodies. The asterisk indicates the rest signal of the Western blotting with Kgd2 antibodies.
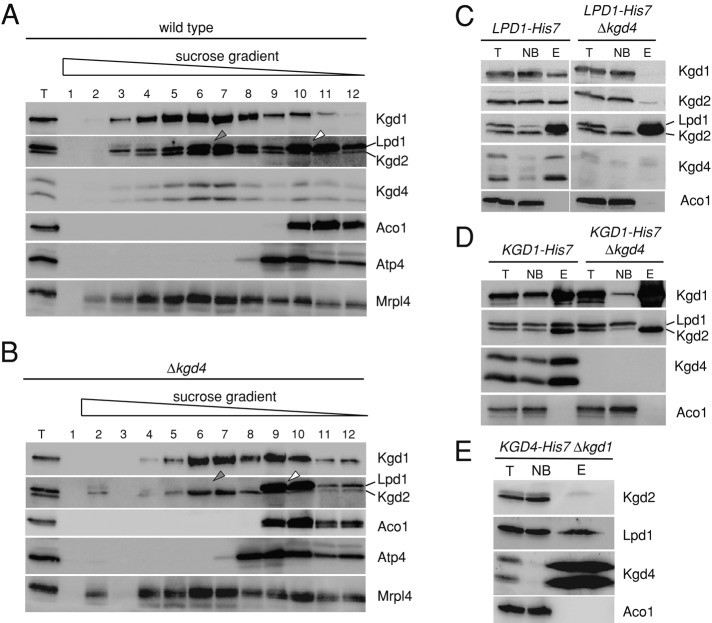
Kgd4 is necessary for a stable incorporation of the E3 subunit into the E1-E2 core of yeast α-KGDH
What could be the molecular function of Kgd4? Because the catalytic subunits are stable in the absence of Kgd4 (Figure 2C), we set out to analyze the composition of KGDH in Δkgd4 cells on linear sucrose gradients. In lysates from wild-type mitochondria, Kgd1, Kgd2, Lpd1, and Kgd4 comigrated (Figure 3A). Because Kgd4 comigrated quantitatively with the complex, these data support the notion that Kgd4 is a subunit of the KGDH. Lpd1 showed two main peaks, one comigrating with KGDH (gray arrow) and one in the top fractions (white arrow), confirming that a substantial amount of Lpd1 exists in a free form in the matrix (Repetto and Tzagoloff, 1991). When analyzing mitochondrial lysates from the Δkgd4 strain, we observed that Kgd2 migrated exclusively with Kgd1 (gray arrow), that a small pool of Kgd1 was additionally present in the top fractions, and that Lpd1 did not comigrate with Kgd2 but was exclusively recovered in the top fractions of the gradient (Figure 3B, white arrow). This observation indicates that a stable interaction of Lpd1 with Kgd1 and Kgd2 depends on Kgd4.
FIGURE 3:
Kgd4 is necessary for a stable incorporation of the E3 subunit into the E1-E2 core of yeast α-KGDH. (A) Mitochondria from the wild-type strain were lysed in Triton X-100 and subjected to centrifugation on a linear sucrose gradient. Fractions were collected and analyzed by Western blotting. (B) Mitochondria of the Δkgd4 strain were processed and analyzed as in A. (C) Mitochondria containing a C-terminally His7-tagged Lpd1 with or without Kgd4 were lysed, and proteins were purified on Ni-NTA. The fractions of this purification were analyzed by Western blotting. (D) Mitochondria containing Kgd1-His7 with or without Kgd4 were processed and analyzed as in C. (E) Mitochondria containing Kgd4-His7 but lacking Kgd1 were processed and analyzed as in C. T, total of the lysate; NB, unbound fraction; E, elution fraction.
We next set out to confirm this conclusion by two independent approaches and tested first for copurification of KGDH subunits when the enzyme was purified using His7-tagged subunits. We engineered yeast strains expressing a His7-tagged Lpd1 with or without Kgd4. Mitochondria of these strains were isolated, and His7-tagged Lpd1 was purified. In the presence of Kgd4, Kgd2 and Kgd1 were recovered together with Lpd1, while in the absence of Kgd4, only Lpd1 could be purified (Figure 3C), showing that the presence of Kgd4 is required to purify intact KGDH. Next we tested for copurification of KGDH subunits by using the His7-tagged Kgd1 strain with or without Kgd4. Kgd2, Lpd1, and Kgd4 were copurified together with Kgd1, as seen before (Figure 1B). Importantly, a stable complex of Kgd1 and Kgd2 that did not contain Lpd1 could be purified in the absence of Kgd4 (Figure 3D).
We next asked whether Kgd4 and Lpd1 would interact in the absence of Kgd1. We therefore purified complexes containing Kgd4 from mitochondria lacking Kgd1 and found that Lpd1 but not Kgd2 could be purified (Figure 3E). Taken together, these purification experiments confirmed that Lpd1 cannot stably incorporate into KGDH in the absence of Kgd4 and showed that Kgd4 directly binds to Lpd1.
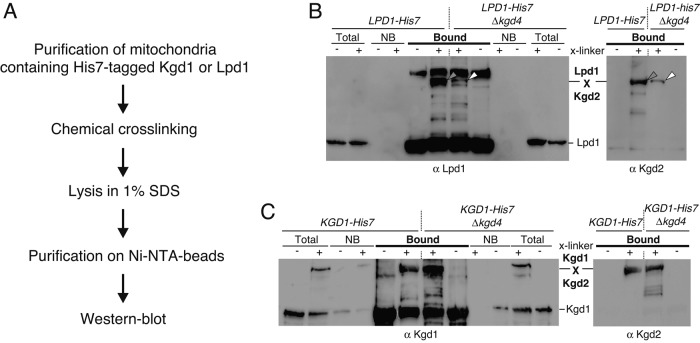
Kgd4 is required for efficient interaction of E3 with KGDH in intact organelles
Because the above-presented data were obtained from experiments using mitochondrial lysates, we used chemical cross-linking followed by purification under denaturing conditions (Figure 4A) to detect the interaction of Lpd1 with KGDH in intact organelles. We exposed isolated, intact mitochondria containing His7-tagged Lpd1 with or without Kgd4 to the membrane-permeable chemical cross-linker bismaleimidohexane. After the cross-linker was quenched, mitochondria were reisolated and boiled in SDS, and Lpd1 and its cross-linking products were purified and analyzed by Western blotting. These analyses revealed that a cross-linking product between Lpd1 and Kgd2 was formed in wild-type mitochondria, indicating a close contact of Lpd1 and Kgd2 (Figure 4B, gray arrows). Importantly, the efficiency with which this product appeared was dramatically decreased in the absence of Kgd4, demonstrating that Ldp1 and Kgd2 are not in similar proximity as in wild-type mitochondria (Figure 4B, white arrows). As a control, we tested for cross-linking between Kgd1 and Kgd2 in the presence or absence of Kgd4. The cross-linking product between the two proteins was similarly prominent in the presence or absence of Kgd4 (Figure 4C), confirming our fractionation experiments, which showed that both proteins interact stably with each other in the absence of Kgd4 (Figure 3B). Taken together, these data therefore confirm that Kgd4 is specifically necessary for stably recruiting the E3 subunit Lpd1 to the Kgd1-Kgd2 subcomplex.
FIGURE 4:
Kgd4 is required for efficient interaction of E3 with KGDH in intact organelles. (A) Cross-linking strategy to identify interactions between individual KGDH subunits. (B) Mitochondria prepared as in Figure 3C were exposed to the chemical cross-linker bismaleimidehexane, and Lpd1-His7 and its cross-linking products were purified under denaturing conditions on Ni-NTA beads. The fractions were analyzed by Western blotting. The gray arrow indicates the cross-linking product between Lpd1 and Kgd2 that decreases in the absence of Kgd4 (white arrow). (C) Mitochondria as in Figure 3D were processed and analyzed as in B. NB, unbound fraction.
Organization of α-KGDH
We next asked what interactions the KGDH subunits have with each other. To unravel these interactions, we analyzed complexes of KGDH subunits in mutants in which individual subunits were absent. We lysed mitochondria from Δkgd1, Δkgd2, or Δlpd1 cells and followed migration of the other subunits on linear sucrose gradients. In the absence of Kgd1, Kgd2 still migrated as a large complex, while Lpd1 or Kgd4 were recovered in the top fractions of these gradients (Figure 5A). These data confirm that Kgd2 forms a large, oligomeric complex (Reed and Hackert, 1990) and show that Lpd1 and Kgd4 do not stably interact with Kgd2 in the absence of Kgd1. In addition, these data also confirm that Kgd4 is not part of the mitochondrial ribosome, because Kgd4 did not migrate into the middle of the gradient in the absence of Kgd1 or Kgd2 (Figure 5A), while migration of the ribosome, as evidenced by proteins of the small (Mrps5) or large (Mrpl4) subunit, was unchanged.
FIGURE 5:
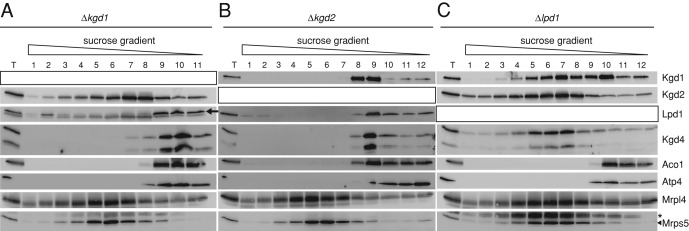
Organization of α-KGDH in mitochondria. (A) Mitochondria lacking Kgd1 were analyzed as in Figure 3A. The black arrow indicates the signal of Lpd1. (B) Mitochondria lacking Kgd2 were analyzed as in Figure 3A. (C) Mitochondria lacking Lpd1 were lysed and analyzed as in Figure 3A. T, total of the lysate. The asterisk indicates the signal of the previous Western blotting with Mrpl4 antibodies.
In contrast, Kgd1, Kgd4, or Lpd1 did not migrate as a similarly large complex in the absence of Kgd2 (Figure 5B), confirming that Kgd2 forms the core of KGDH around which the other subunits assemble (Reed and Hackert, 1990). In the absence of Lpd1, Kgd1 was partly shifted to the top fractions of these gradients, suggesting that the presence of Lpd1 partly stabilizes the interaction of Kgd1 with Kgd2 (Figures 5C and 3B). Importantly, Kgd4 comigrated with Kgd1 and Kgd2 in the absence of Lpd1 (Figure 5C). Because Kgd4 did not comigrate with Kgd2 without Kgd1 (Figure 5A), these data show that Kgd4 interacts with the Kgd1-Kgd2 complex and not with Kgd2. In summary, these data show that Kgd4 quantitatively associates with the core of KGDH formed by Kgd1 and Kgd2. Kgd4 is also present in the fully assembled complex and is necessary to stably incorporate Lpd1 into the complex.
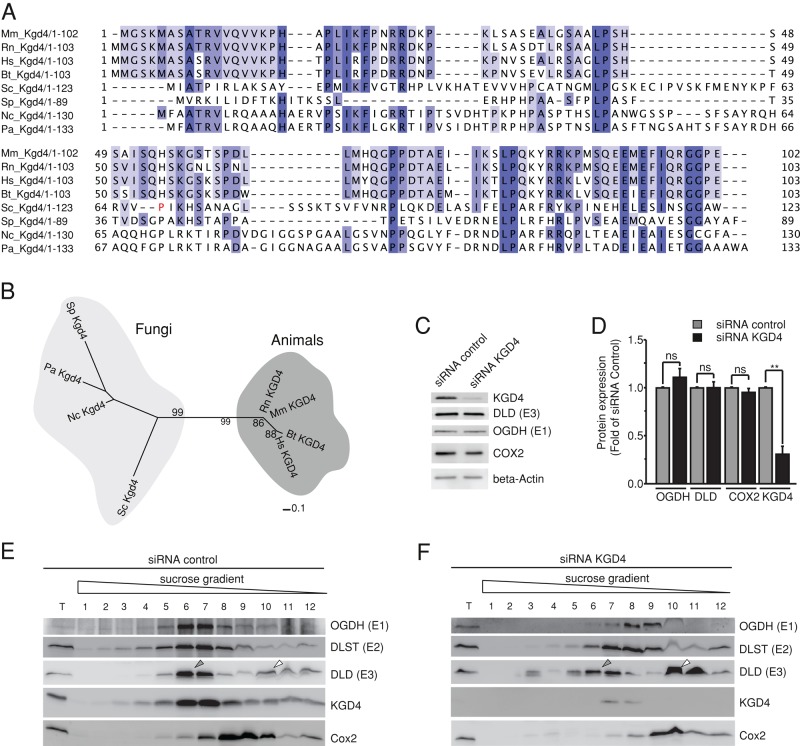
Kgd4 function is evolutionarily conserved
Phylogenetic analyses revealed that homologues of Kgd4 are present in fungi and animals (Figure 6, A and B). To functionally test the significance of a mammalian Kgd4 homologue for the organization of α-KGDH, we depleted KGD4 in a murine cell line using small interfering RNA (siRNA; Figure 6C). This knockdown was efficient and had no effect on the steady-state levels of the other KGDH subunits or the mitochondrially encoded COX2 (Figure 6, C and D). Cells were lysed and subjected to sucrose gradient centrifugation. E1, E2, E3, and KGD4 comigrated in lysates from control cells (Figure 6E, gray arrow). This also confirmed that KGD4 is part of the murine KGDH. On knockdown of KGD4, however, E1 and E2 were shifted upward in the gradient and no longer comigrated with E3, and increased amounts of E3 were detected in the top fractions of the gradient (Figure 6F, white arrow). The residual E3 signal in fractions 6 and 7 could derive from the branched-chain amino acid dehydrogenase, which also contains the identical E3 subunit and is similar in size to KGDH (Aevarsson et al., 1999). Importantly, these results demonstrate that knockdown of the murine Kgd4 homologue destabilizes the interaction between E3 and the other KGDH subunits in a similar way as in yeast. We therefore concluded that the members of Kgd4 protein family have an evolutionarily conserved role in recruiting the E3 subunits to KGDH.
FIGURE 6:
Kgd4 function is evolutionarily conserved. (A) Alignment of amino acid sequences of Kgd4 homologues. Sequences used for comparison were extracted from the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/protein): S. cerevisiae (Sc-gi45270840); Neurospora crassa (Nc-gi85092531), Schizosaccharomyces pombe (Sp-gi429240132), Podospora anserina (Pd-gi171678223), Homo sapiens (Hs-gi15150811), Rattus norvegicus (Rn-gi300797955), Mus musculus (Mm-gi13384742), and Bos taurus (Bt-gi78369264). (B) Phylogenetic tree of Kgd4 homologues from fungi and animals. Sequences used for comparison were extracted as in Figure 4A. (C) siRNA-mediated knockdown of KGD4 in murine glial cells. (D) Quantification of the knockdown efficiency. (E) Cells transfected with control RNA were lysed and subjected to density gradient centrifugation. The fractions were analyzed by Western blotting. (F) Cells in which KGD4 was depleted for 48 h were processed and analyzed as in D.
Kgd4 contains two separable domains to contact Lpd1 and the Kgd1-Kgd2 core
Having established that Kgd4 interacts with both the Kgd1-Kgd2 core and Lpd1 (Figures 3E and 5, A and C), we asked whether Kgd4 would contain N- or C-terminally located modules to establish interactions with the Kgd1-Kgd2 core or Lpd1, respectively. To identify such regions, we compared the Saccharomyces cerevisiae Kgd4 amino acid sequence with homologues from other species (Figure 6A). Typically, motifs engaged in protein–protein interactions exhibit a particularly high degree of conservation. Indeed, the N- and C-terminal parts of the sequences show regions of substantial conservation (Figure 6A). We therefore constructed two variants of Kgd4 that would allow the identification of specific interactions of the N- or of the C-terminus: Kgd4ΔC contains the N-terminal domain, encompassing amino acids 1–68, and Kgd4ΔN comprises the C-terminal amino acids 69–123 (Figure 7A). To allow detection and purification of these chimeric proteins, we added protein A-His7 tags (Figure 7A). In addition, a cleavable mitochondrial targeting signal (MTS) from the Oxa1 protein was included to allow import of the Kgd4ΔN construct (Figure 7A). These constructs were expressed in cells lacking wild-type Kgd4, mitochondria were isolated and lysed, and protein complexes were purified using the His7-tag. First, we tested which subunits of KGDH could be copurified with the N-terminal domain of Kgd4. These experiments revealed that Lpd1 but not Kgd1 or Kgd2 could be copurified with Kgd4ΔC (Figure 7B). This indicated that Kgd4 contacts Lpd1 via an N-terminal domain and suggests that the C-terminus is implicated in binding to the Kgd1-Kgd2 core.
FIGURE 7:
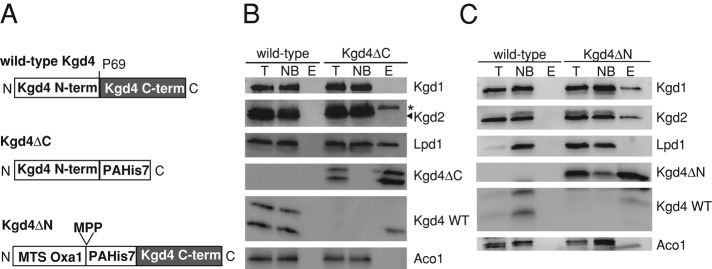
Kgd4 contains two separable domains to contact Lpd1 and the Kgd-Kgd2 core. A, Schemes of the proteins containing either the N- or the C-terminal domain of Kgd4. MPP, cleavage site of the mitochondrial processing peptidase (MPP). (B) Mitochondria containing Kgd4ΔC were lysed, and complexes containing Kgd4ΔC were purified on Ni-NTA beads. The fractions of the purifications were analyzed by Western blotting. (C) Mitochondria from wild type and Kgd4ΔN were analyzed as in 2A. T, Total of the lysate; NB, not bound fraction; E, elution fraction; * signal of Lpd1 from previous decoration.
To test this, we purified complexes of Kgd4ΔN and found that Kgd1 and Kgd2 but not Lpd1 could be copurified with this protein (Figure 7C). This demonstrates that the C-terminal domain of Kgd4 directly interacts with the assembled Kgd1-Kgd2 core because a direct interaction of Kgd4 with Kgd2 can be ruled out according to previous experiments (Figure 5A). Taken together, these data establish that Kgd4 contains two functionally separable domains. While the C-terminal domain is required to establish interactions with the Kgd1-Kgd2 core, the N-terminal domain binds to the E3 subunit Lpd1 (Figure 8A). This architecture likely enables Kgd4 to act as a molecular adaptor between the E3 subunit and the core of KGDH.
FIGURE 8:
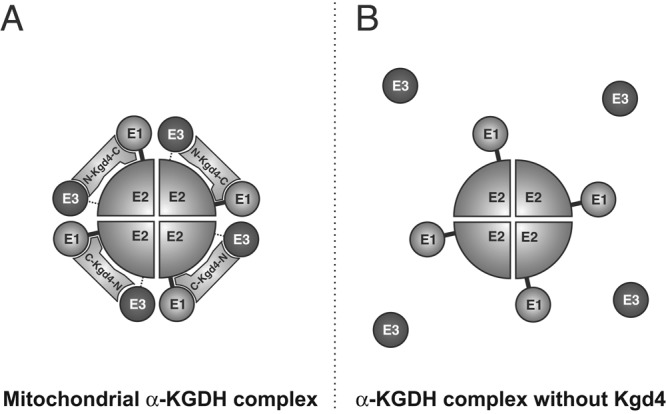
Model for the organization of KGDH and the role of Kgd4. (A) Model. The novel subunit Kgd4 recruits the E3 subunit to the core of KGDH, formed by E1 and E2 subunits. The C-terminus of Kgd4 contacts Kgd1-Kgd2 core, while the N-terminus interacts with Lpd1. (B) In the absence of Kgd4, the contact of E3 to the core is dramatically reduced.
DISCUSSION
In this paper, we have identified a novel subunit of the mitochondrial KGDH. The data presented here suggest a model for the general organization of the mitochondrial KGDH complex (Figure 8A). According to this model, the E2 subunits assemble into a homo-oligomeric core that forms independently of the E1 or E3 subunits, as shown earlier (Reed and Hackert, 1990). The E1 subunit is attracted to this E2 particle to form the E1-E2 core of the enzyme. The newly identified subunit Kgd4 then binds to this E1-E2 core, most likely to the E1 subunit, to recruit the E3 subunit into the complex. In absence of Kgd4, the contact of E3 to the core is dramatically reduced (Figure 8B).
A number of independent lines of evidence support the notion that Kgd4 is a structural subunit of the KGDH complex and not a subunit of the mitochondrial ribosome. First, Kgd4 specifically copurified with the catalytic KGDH subunits but not with ribosomal proteins when C-terminally tagged Kgd4, Kgd1, or Lpd1 was used for affinity purifications. The detailed data presented in this article are in accordance with data obtained by large-scale proteomic approaches aimed at identifying protein complexes in yeast. These approaches revealed that KGDH subunits specifically, not mitochondrial ribosomal proteins, were copurified with Kgd4 and that the protein was copurified together with Kgd1 and Lpd1 but not with the mitochondrial ribosome (Gavin et al., 2002, 2006; Krogan et al., 2006). Accordingly, general analyses of gene expression in yeast have shown that Kgd4 is coregulated with genes involved in mitochondrial energy metabolism and the citric acid cycle but not with genes encoding subunits of the mitochondrial ribosome (Hibbs et al., 2007). Moreover, we also did not observe a defect of the mitochondrial ribosome caused by the absence of Kgd4. In fact, absence of a protein subunit of the mitochondrial ribosome is typically accompanied by loss of organellar protein synthesis and a subsequent loss of the mitochondrial genome (Myers et al., 1985), and yeast mutant lacking Kgd4 or murine cells with reduced KGD4 accumulate normal amounts of the mitochondrially encoded cytochrome oxidase subunit Cox2.
The second line of evidence supporting Kgd4 as a structural subunit of KGDH is that genetic ablation of Kgd4 by deletion of KGD4 in yeast or by siRNA-mediated knockdown of KGD4 in murine cells disrupted the KGDH complex, leading to the dissociation of the complex into an E1-E2 core and free E3. Importantly, Kgd4 quantitatively comigrated with the KGDH complex and not with smaller particles. If Kgd4 were an assembly factor that acts during the biogenesis of the complex, it should not be present in the fully assembled complex, a behavior seen for the assembly factors/chaperones that mediate respiratory chain complex assembly (Mick et al., 2011; Fox, 2012). The notion that Kgd4 is a structural component is further supported by our data showing that the protein contains two biochemically separable domains in its N- or C-terminus that establish contacts to Lpd1 or the Kgd1-Kgd2 core, respectively. Clearly, high-resolution structural data will allow conclusive determination in the future of exactly how Kgd4 is organized in the KGDH complex. In addition, Kgd4 is present as two different forms of different sizes in the KGDH complex. It will be exciting for future research to identify the nature and role of these two forms.
In its function as an adaptor between the E3 subunit and the rest of the complex, Kgd4 functions analogous to protein Pdx1 of the pyruvate dehydrogenase (PDH) complex, which is necessary to stably bind the E3 subunit into PDH (Behal et al., 1989). PDH and KGDH share a similar architecture, with the difference that PDH is substantially larger (Milne et al., 2006). Interestingly, the E3 subunit in both complexes is the identical gene product. It is therefore likely that the specific adaptors Pdx1 and Kgd4 evolved to direct the E3 subunit to their respective complexes. Branched-chain amino acid dehydrogenase that is not present in yeast is the third member of the E1-E3 family of enzyme complexes that also contains the same E3 subunit as PDH or KGDH. It is tempting to speculate that this complex also contains a dedicated E3 adaptor that still awaits identification.
Mitochondrial dysfunction plays a central role in the development of human diseases as observed in neurodegeneration, cancer, and the aging process. A detailed understanding of the functions of key mitochondrial enzymes and their composition is therefore required to diagnose patients and develop strategies for treatment. The reannotation of Kgd4 from being a subunit of the mitochondrial ribosome to being a novel subunit of KGDH is therefore also an important aspect for the interpretation of high-throughput data. Interestingly, the murine KGD4 gene (formerly annotated as MRPS36) was identified as a candidate gene aimed at identifying metabolic and longevity regulators using mouse population genetics (Houtkooper et al., 2013). A detailed study of the mammalian KGD4 protein and its impact on mitochondrial KGDH could reveal how the protein and KGDH affect mitochondrial biology and aging. This would be an exciting area for future research.
MATERIALS AND METHODS
Yeast strains and growth media
All strains used in this study were isogenic to the wild-type strain YPH499 (Sikorski and Hieter, 1989). Yeast cultures were grown at 30°C in YP (1% yeast extract, 2% peptone) medium supplemented with 2% dextrose (D), 2% galactose (Gal), or 2% glycerol (G). His7- or protein A–tagged variants of Kgd1, Kgd4, and Lpd1 were generated by directed homologous recombination as previously described (Gruschke et al., 2012). KGD1, KGD2, and KGD4 were disrupted using a kanamycin resistance cassette (KanMX); Lpd1 was disrupted using an URA3 cassette. Protein A–His7–Kgd4ΔN was expressed from pYX132 in a Δkgd4 strain. Upstream of the protein A–His7–Kgd4Δ1-68, the MTS from Oxa1 (amino acids 1–48) was inserted to ensure mitochondrial import of the construct.
Native purification of Kgd1-His7, Kgd4-His7, Lpd1-His7, Kgd4Δ69-123, and Kgd4Δ1-68
Mitochondria from the wild-type or mutant strains expressing His7-tagged proteins were lysed for 30 min with lysis buffer (1% Triton X-100, 150 mM KCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× Complete Protease Inhibitor Mix [Roche Diagnostics, Bromma, Sweden], 20% glycerol, and 20 mM imidazole, pH 7.4, 20 mM HEPES, pH 7.4). After a clarifying spin for 10 min at 25.000 × g, 4°C, tagged proteins were purified at 4°C on Ni-NTA beads (Qiagen, Hilden, Germany). The beads were washed three times with lysis buffer containing 0.1% Triton X-100 and eluted with either 500 mM imidazole pH 7.4 or 2 mM EDTA.
KGDH activity assay
Isolated mitochondria were lysed in buffer containing 1% Triton X-100 and 50 mM KPi (pH 7.4) for 10 min at 4°C. After a clarifying spin for 10 min at 25.000 × g and 4°C, the lysate was mixed with 400 μl assay buffer (2.5 mM NAD, 0.2 mM thiamine pyrophosphate, 0.1 mM CoA, 0.3 mM dithiothreitol, 1 mM MgCl2, and 50 mM KPi, pH 7.4). The reaction was started by addition of 5 mM α-ketoglutarate. KGDH activity was followed by measuring the NADH production at 340 nm.
MDH activity assay
Isolated mitochondria were adjusted to a concentration of 1 mg/ml in isotonic buffer (0.6 M sorbitol, 20 mM HEPES, pH 7.4). For starting the reaction, 200 μl of this solution was added to 1 ml assay buffer (2.5 mM NAD, 37 mM dl-malate, 50 mM glycine/NaOH, pH 10.0). MDH activity was followed by measuring the NADH production at 340 nm.
Fractionation of mitochondrial protein complexes on linear sucrose gradients
Isolated mitochondria were lysed for 15 min at 4°C in lysis buffer (1% Triton X-100, 150 mM KCl, 20 mM HEPES, pH 7.4, 1 mM PMSF, and 1× Complete Protease Inhibitor Mix [Roche]). After a clarifying spin for 10 min at 25,000 × g, 4°C, the extract was layered on top of a continuous 4 ml sucrose gradient (0.3–1 M sucrose, 0.1% Triton X-100, 25 mM KCl, 20 mM HEPES, pH 7.4, 1 mM PMSF) and centrifuged for 1 h and 15 min at 484,183 × g and 4°C in an SW60 Ti rotor (Beckman Coulter, Fullerton, CA). Fractions were collected, and proteins were precipitated with 12% trichloroacetic acid.
Phylogenetic analyses
For visualization purposes, we constructed a phylogenetic tree of Kgd4 homologues. A multiple sequence alignment was generated using MUSCLE (Edgar, 2004) and the regions of poor alignment were removed using Gblocks (Talavera and Castresana, 2007; maximum nonconserved positions = 8; minimum block length = 5). The tree was constructed using PhyML 3.0 (Guindon et al., 2010; Jones–Taylor–Thornton matrix) with 100 bootstrap replicates and visualized using iTOL (http://itol.embl.de).
Knockdown of KGD4 in a murine cell line
The murine microglial BV2 cells were cultured in DMEM (GIBCO) and supplemented with 10% fetal bovine serum, 55 IU/ml penicillin and 55 μg/ml streptomycin (Sigma-Aldrich). All cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. BV2 cells were transfected with Lipofectamine 2000 transfection reagent following manufacturer's instructions, using nontargeting SMARTpool siRNA (D-001810-01-05) and SMARTpool siRNA for KGD4 (MRPS36–L-056188-01) from Thermo Scientific (Fisher Scientific, Göteborg, Sweden). Cells were analyzed 48 h after the transfection.
Miscellaneous methods
Antisera against yeast Kgd4 or murine COX2 were generated by injecting rabbits with recombinantly expressed and purified Kgd4 or COX2 C-terminus, respectively. Antibodies against OGDH, DLST, OLD, and MRPS36 were from Atlas Antibodies (Stockholm, Sweden), and the antibody against β-actin was from Sigma-Aldrich. Mitochondrial isolation, chemical cross-linking, purification of cross-linking products under denaturing conditions, and mass-spectrometric identification were done as previously described (Gruschke et al., 2010).
Acknowledgments
We thank all members of our groups and the groups of Per Ljungdal and Claes Andreasson (Stockholm University, Sweden) for stimulating discussions. The antibodies against Lpd1, Kgd1, Kgd2, and lipoic acid were a kind gift of Luke Swedza (Oklahoma Medical Research Foundation). Work in our laboratory was supported by the Swedish Research Council, the Center for Biomembrane Research at Stockholm University, the Carl Tryggers Foundation, and the Knut and Alice Wallenberg Foundation.
Abbreviations used:
- α-KGDH
α-ketoglutarate dehydrogenase
- MDH
malate dehydrogenase
- MTS
mitochondrial targeting signal
- PDH
pyruvate dehydrogenase
- PMSF
phenylmethylsulfonyl fluoride
- siRNA
small interfering RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E14-07-1178) on August 27, 2014.
REFERENCES
- Aevarsson A, Seger K, Turley S, Sokatch JR, Hol WG. Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes. Nat Struct Biol. 1999;6:785–792. doi: 10.1038/11563. [DOI] [PubMed] [Google Scholar]
- Behal RH, Browning KS, Hall TB, Reed LJ. Cloning and nucleotide sequence of the gene for protein X from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:8732–8736. doi: 10.1073/pnas.86.22.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calingasan NY, Ho DJ, Wille EJ, Campagna MV, Ruan J, Dumont M, Yang L, Shi Q, Gibson GE, Beal MF. Influence of mitochondrial enzyme deficiency on adult neurogenesis in mouse models of neurodegenerative diseases. Neuroscience. 2008;153:986–996. doi: 10.1016/j.neuroscience.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox TD. Mitochondrial protein synthesis, import, and assembly. Genetics. 2012;192:1203–1234. doi: 10.1534/genetics.112.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gibson GE, Blass JP, Beal MF, Bunik V. The alpha-ketoglutarate-dehydrogenase complex: a mediator between mitochondria and oxidative stress in neurodegeneration. Mol Neurobiol. 2005;31:43–63. doi: 10.1385/MN:31:1-3:043. [DOI] [PubMed] [Google Scholar]
- Gruschke S, Grone K, Heublein M, Holz S, Israel L, Imhof A, Herrmann JM, Ott M. Proteins at the polypeptide tunnel exit of the yeast mitochondrial ribosome. J Biol Chem. 2010;285:19022–19028. doi: 10.1074/jbc.M110.113837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S, Rompler K, Hildenbeutel M, Kehrein K, Kuhl I, Bonnefoy N, Ott M. The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc1 complex assembly in yeast mitochondria. J Cell Biol. 2012;199:137–150. doi: 10.1083/jcb.201206040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc EC, Cimen H, Kumcuoglu B, Abu N, Akpinar G, Haque ME, Spremulli LL, Koc H. Identification and characterization of CHCHD1, AURKAIP1, and CRIF1 as new members of the mammalian mitochondrial ribosome. Front Physiol. 2013;4:183. doi: 10.3389/fphys.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Matsushita Y, Kitakawa M, Isono K. Cloning and analysis of the nuclear genes for two mitochondrial ribosomal proteins in yeast. Mol Gen Genet. 1989;219:119–124. doi: 10.1007/BF00261166. [DOI] [PubMed] [Google Scholar]
- McLain AL, Szweda PA, Szweda LI. α-Ketoglutarate dehydrogenase: a mitochondrial redox sensor. Free Radic Res. 2011;45:29–36. doi: 10.3109/10715762.2010.534163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Fox TD, Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JL, Wu X, Borgnia MJ, Lengyel JS, Brooks BR, Shi D, Perham RN, Subramaniam S. Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J Biol Chem. 2006;281:4364–4370. doi: 10.1074/jbc.M504363200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Pape LK, Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985;4:2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan CL, Goncalves RL, Hey-Mogensen M, Yadava N, Bunik VI, Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J Biol Chem. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Hackert ML. Structure-function relationships in dihydrolipoamide acyltransferases. J Biol Chem. 1990;265:8971–8974. [PubMed] [Google Scholar]
- Repetto B, Tzagoloff A. Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Mol Cell Biol. 1989;9:2695–2705. doi: 10.1128/mcb.9.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto B, Tzagoloff A. Structure and regulation of KGD2, the structural gene for yeast dihydrolipoyl transsuccinylase. Mol Cell Biol. 1990;10:4221–4232. doi: 10.1128/mcb.10.8.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto B, Tzagoloff A. In vivo assembly of yeast mitochondrial alpha-ketoglutarate dehydrogenase complex. Mol Cell Biol. 1991;11:3931–3939. doi: 10.1128/mcb.11.8.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy DJ, Dawes IW. Cloning and characterization of the gene encoding lipoamide dehydrogenase in Saccharomyces cerevisiae. J Gen Microbiol. 1987;133:925–933. doi: 10.1099/00221287-133-4-925. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga T. Cancer metabolism: key players in metabolic reprogramming. Cancer Sci. 2013;104:275–281. doi: 10.1111/cas.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Trofimova LK, Araujo WL, Strokina AA, Fernie AR, Bettendorff L, Bunik VI. Consequences of the alpha-ketoglutarate dehydrogenase inhibition for neuronal metabolism and survival: implications for neurodegenerative diseases. Curr Med Chem. 2012;19:5895–5906. doi: 10.2174/092986712804143367. [DOI] [PubMed] [Google Scholar]