Derivation of induced pluripotent stem cells (iPSCs) by somatic cell reprogramming offers an alternative strategy for identifying the cellular mechanisms contributing to autism spectrum disorders (ASDs) and developing new treatment options. Normal and ASD-specific iPSCs can be differentiated toward a neural stem cell phenotype and terminally differentiated into action-potential firing neurons and glia. Some drug treatments have shown promise in reversing the neurobiological abnormalities in iPSC-based models of ASD-associated diseases.
Keywords: Induced pluripotent stem cells, Neural stem cell, Neuron, Disease modeling, Differentiation, Autism spectrum disorder
Abstract
The autism spectrum disorders (ASDs) comprise a set of neurodevelopmental disorders that are, at best, poorly understood but are the fastest growing developmental disorders in the United States. Because animal models of polygenic disorders such as the ASDs are difficult to validate, the derivation of induced pluripotent stem cells (iPSCs) by somatic cell reprogramming offers an alternative strategy for identifying the cellular mechanisms contributing to ASDs and the development of new treatment options. Access to statistically relevant numbers of ASD patient cell lines, however, is still a limiting factor for the field. We describe a new resource with more than 200 cell lines (fibroblasts, iPSC clones, neural stem cells, glia) from unaffected volunteers and patients with a wide range of clinical ASD diagnoses, including fragile X syndrome. We have shown that both normal and ASD-specific iPSCs can be differentiated toward a neural stem cell phenotype and terminally differentiated into action-potential firing neurons and glia. The ability to evaluate and compare data from a number of different cell lines will facilitate greater insight into the cause or causes and biology of the ASDs and will be extremely useful for uncovering new therapeutic and diagnostic targets. Some drug treatments have already shown promise in reversing the neurobiological abnormalities in iPSC-based models of ASD-associated diseases. The ASD Stem Cell Resource at the Children’s Hospital of Orange County will continue expanding its collection and make all lines available on request with the goal of advancing the use of ASD patient cells as disease models by the scientific community.
Introduction
Autism, or as it is now named in the recent 2013 fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM) [1], the set of autism spectrum disorders (ASDs), is a diverse group of debilitating pediatric neurodevelopmental disorders that are typically diagnosed early in childhood and can last throughout a person’s life [2]. ASDs now encompass the previous DSM-IV autistic disorder (i.e., autism), Asperger disorder, childhood disintegrative disorder, and pervasive developmental disorder not otherwise specified (PDD-NOS) [1]. People with ASD have substantial challenges in social interaction, communication and learning, and display restrictive repetitive behaviors, interests, and activities. However, not everyone with an ASD has the same challenges; a varied distribution of characteristics, phenotypes, and traits can be present, such as megalencephaly, seizures, and a high or low intelligence quotient (IQ) [3]. In 2014, the Centers for Disease Control and Prevention (http://www.cdc.gov/ncbddd/autism/data.html) estimated that, in the United States, ASDs have increased to affect 1 in 68 children, with ASD almost 5 times as common among boys (1 in 42) as among girls (1 in 189). The total societal cost of caring for children with ASD in 2011 was more than $9 billion, and the overall estimated economic burden of ASDs to the United States is $137 billion annually, with much of this expense related to adult care. A comparison of the prevalence of ASDs in countries across the globe was recently published [4].
ASDs are now more common than childhood cancer, diabetes, and pediatric AIDS combined. Despite 75 years of research since Leo Kanner’s seminal description of “Autistic Disturbances of Affective Contact” in a pediatric population [5], we still have no cure, and we still do not fully understand the neurological manifestations and causes of all the ASDs. One of the difficulties related to the study and diagnosis of the ASDs is that no definitive genetic test is available for most of the ASDs [6]. Typically, a clinical diagnosis will be determined from clinical observations, parent interviews, developmental histories, psychological testing, speech and language assessments, and the use of one or more standardized psychometric tests [7]. Variation in the diagnosis among clinicians can also be a confounding factor. The study of ASD is also complicated because children with ASD can have a specific developmental disorder that can lead to ASD, such as fragile X syndrome (FXS), Rett syndrome, Down syndrome, or tuberous sclerosis [8]. Additionally, some children will have mental health problems, such as depression or anxiety, and others could have attention problems, sensory issues, sleep problems, and digestive disorders. Studies have suggested that several hundred loci [9] might contribute to the complex genetic heterogeneity of this group of disorders, with many impinging on the neural processes related to synaptic development and function [10], axon targeting, and neuron motility [11]. Recent studies have indicated that metabolic abnormalities, environmental factors such as air pollution and pesticides [12], and maternal immune dysfunction and/or infection can also be involved [13].
Analysis of the postmortem brain has provided some of the most valuable data for advancing our understanding of ASD pathophysiology [14, 15]. A detailed examination of the proliferation and differentiation of neural stem cells (NSCs) derived from ASD brains, compared with those derived from normal brains, would likely yield important data regarding the etiology of the disease [16, 17]. However, the procurement of postmortem ASD brains has proved difficult, severely limiting the numbers of ASD NSC lines available to satisfy power analyses.
The advent of patient-derived induced pluripotent stem cells (iPSCs) now provides a unique opportunity to explore human genomic heterogeneity using in vitro “disease in a dish” models [18–20]. The development of iPSCs, which can subsequently be differentiated into NSCs and then terminally differentiated into a myriad of neural cell subtypes, now allows for successful iPSC-based modeling of the ASDs. This approach has been validated through recent studies documenting the successful modeling of human neurological disease through disorder-specific iPSC-derived neural cells [21–23]. Efficient generation of ASD NSC lines now allows for studies (a) of the detailed pathophysiology of the ASD neuron compared with normal controls; (b) of sufficient statistical power to evaluate the effects of ASD on iPSC differentiation to NSCs and, ultimately, to neuron- and glial-specific subtypes; and (c) of the influence of pharmacological agents on these processes. These studies will allow for large screens of ASD and normal neural development, facilitating diagnostic marker discovery, future ASD genetic classification and subclassification, and drug development.
We established more than 200 lines (fibroblasts, iPSC clones, NSCs, glia) derived from clinically well-defined ASD-specific patients and unaffected volunteers. Of these lines, approximately one third have a clinical diagnosis of either fragile X syndrome (full mutation) or FMR1 premutation with or without an ASD diagnosis. Using methods developed in our laboratory [24, 25], we have confirmed, using a subset of the normal and ASD-specific iPSCs, that these cells can be differentiated toward a neural stem cell phenotype and terminally differentiated into action-potential firing neurons and glia [25]. This cell line repository represents a significant resource that will facilitate the use of patient-derived cell models in ASD research; additional lines will continue to be developed as patients are identified and recruited for participation. The lines themselves are available to the scientific community through our existing National Human Neural Stem Cell Resource (NHNSCR; http://www.nhnscr.org) at Children’s Hospital of Orange County (CHOC Children’s). This will leverage our efforts for maximal benefit to patients and families affected by ASD.
Materials and Methods
Patient Consent and Inclusion Criteria
All patients recruited to donate tissue to this resource received study and patient information sheets and signed patient consent forms that were reviewed and approved by University of California (UC), Davis, and CHOC Children’s institutional review boards. In addition, the generation and distribution of the human fibroblasts and iPSCs were approved by the UC Davis human stem cell research oversight committee. Specific patients with ASD were recruited through the UC Davis MIND (Medical Investigation of Neurodevelopmental Disorders) Institute. Subgroups of patients with ASD were evaluated on a regular basis, and a detailed medical history for retrospective studies is available. When we started this resource, the patients were clinically diagnosed into several groups (Table 1), including (a) unaffected controls, (b) FXS without autism, (c) FXS with autism, (d) premutation without autism, (e) premutation with autism, (f) ASD (defined, in this context, as those not meeting the full criteria for an idiopathic autism diagnosis), and (g) idiopathic autism (no known cause). Normal unaffected volunteers served as controls. These ethnically balanced groups were chosen for inclusion because some of the patients with autism were genetically very well-defined, having FXS. Within the fragile X groups were patients with and without autism; thus, comparisons between groups could be made. In addition, all patients with autism could be similarly compared against all patients and controls without autism. Our patient inclusion and exclusion criteria were as follows. Those patients with the fragile X mutation were required to have genetic test results documenting their CGG repeat number as a full mutation [26]. All patients with idiopathic autism were required to be negative for the FMR1 mutation, to have negative high-resolution cytogenetic study results, and a medical evaluation that was negative for an identifiable cause. The patients had to be at least 8 years of age and to have a full scale IQ of 40 or higher. Most patients were male (n = 99). We recruited only 6 female patients. These patients were SC127 (Rett syndrome), SC206 (premutation), SC221, SC226, and SC234 (all idiopathic autism), and SC232 (control). Patients with an additional genetic disorder or a medical condition that affects central nervous system functioning (e.g., brain trauma, severe seizure disorder, or stroke) were excluded. Finally, those with autism who had features of known genetic disorders, other than FXS, that could be associated with autism, such as tuberous sclerosis, were excluded from the present study. Controls were recruited from healthy volunteers within the UC Davis MIND Institute who were not family members of the patients with ASD. The controls were screened negative for an ASD using the Social Communications Questionnaire and for IQ using the Wechsler Abbreviated Scale of Intelligence (WASI) test. The unaffected controls were also screened for CGG repeat size and/or FMR1 expression.
Table 1.
Overview of specific autism spectrum disorder patient groups and phenotypes contained within the collection at Children’s Hospital of Orange County

ASD Assessment
Patients underwent the Autism Diagnostic Observation Schedule–General (ADOS-G), Autism Diagnostic Interview–Revised (ADI-R), and WASI tests at the MIND Institute. The ADOS-G is a semistructured grouping of activities designed to evaluate social and communicative behaviors related to an ASD diagnosis [27]. The ADOS has four modules, each designed for different levels of verbal fluency. The ADOS can be administered to individuals with a mental age of 18 months or older. The individual’s behavior and responses were coded by the examiner according to whether they were typical for mental age and development (score of 0) or characteristic of autism (score of 3). Scores of 2 and 3 were collapsed and then summed for particularly sensitive items, providing empirically derived algorithms for both autism and ASD. The clinical evaluation teams have extensive experience with this measure in children and adults with ASD and in patients with FXS.
The ADI-R is a standardized interview conducted with a primary caregiver—the patient’s mother in our project—that elicits explicit behavioral descriptions in the domains that define autism and does so with reference to current behavior and behavior at age 4–5 years [28]. The inter-rater reliability ranges from 90% to 93% and κ from 0.62 to 0.89. Cronbach’s α ranges from 0.69 to 0.95 across the core domains. The test-retest reliability for a 3-month period with different raters yielded a mean agreement of 91%. The interview differentiates from other developmental disorders at high levels of sensitivity and specificity (more than 0.90 for both). All clinical interviewers were trained and certified in the ADI-R. The inter-rater agreement was evaluated for 15% of each diagnostic group, with 85% agreement or better required. The ADI-R was administered to all mothers of the patients with FXS and idiopathic autism. In addition, the lifetime algorithm was used to decide the appropriateness of the autism diagnosis. In this algorithm, a positive diagnosis requires scores in excess of prespecified cutoffs for each of the core domains of autism, as well as a developmental delay observed before 36 months of age.
The WASI is a reliable measure of intelligence for individuals 6 to 89 years that can be administered within 30 minutes [29]. It has been nationally standardized using 2245 cases. It provides the three traditional verbal, performance, full-scale IQ scores and is linked to the “Wechsler Intelligence Scale for Children, Third Edition,” and the “Wechsler Adult Intelligence Scale, Third Edition.” All patients and controls included in the resource were required to have an IQ of 40 or higher.
All patients were required to undergo a medical evaluation, which included a physical and neurological evaluation to assess for the causes of autism. Dysmorphic features and cutaneous stigmata, if present, were also documented.
Skin Biopsy, Blood Sampling, and Transport
All skin biopsies were procured at the MIND Institute. This was to ensure uniformity of the clinical team and skin biopsies, such that all were obtained in the same manner and from the same anatomical location. A 1.5-mm diameter skin punch biopsy was taken from the back of the shoulder of the patient after numbing the skin with lidocaine and immediately placed into 15-ml conical tubes containing skin biopsy medium (SBM; Leibovitz’s L-15 medium [SAFC Biosciences, Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com] containing 1% GlutaMAX [Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com] and 10 µM Primocin [InvivoGen, San Diego, CA, http://www.invivogen.com]). Blood sampling was performed by certified phlebotomists at the MIND Institute, and the blood was placed in heparinized (purple top) and unheparinized (amber top) tubes for isolation of plasma or peripheral blood mononuclear cells and serum, respectively. The times of the skin biopsy and blood sampling were recorded, and the skin biopsy and peripheral blood samples were shipped on ice overnight to CHOC Children’s Research Institute.
Dermal Fibroblast Derivation and Expansion
Before receipt of the skin biopsy, a T25 tissue culture flask was coated overnight at 37°C with human fibronectin (catalog no. 356006; BD Biosciences, San Diego, CA, http://www.bdbiosciences.com) at 10 µg/ml in phosphate-buffered saline PBS+/+ (catalog no. SH30264.01; HyClone, Logan, UT, http://www.hyclone.com ).The next morning, the fibronectin solution was aspirated and the flask allowed to dry in the back of the biological safety cabinet for at least 1 hour. On receipt, the skin biopsies were transferred into sterile glass Petri dishes containing 4 ml freshly prepared human dermal fibroblast medium (HDFM; Dulbecco’s modified Eagle’s medium high glucose with GlutaMAX [catalog no. 10569; Life Technologies], 10% certified fetal bovine serum [catalog no. 16000; Life Technologies], 1× nonessential amino acids [HyClone], and 10 µM Primocin [InvivoGen]). The biopsies were minced into small 1-mm pieces using 2 Bard-Parker no. 11 protected disposable stainless steel blade scalpels (catalog no. 372611; BD Biosciences). The HDFM containing biopsy pieces was taken up into a 10-ml serological pipette and transferred onto the fibronectin-precoated T25 tissue culture flask. Then, 6 ml of medium was used to wash any remaining tissue from the scalpel blades and the glass Petri plate. This was combined into the T25 to give a total volume of 10 ml. Explant fibroblast cultures were established from the skin biopsies after approximately 2–3 weeks. The explants were fed weekly with a complete medium change of HDFM. Nonadherent material was recovered by centrifugation (200g for 5 minutes), gently triturated in HDFM, and added back to the flask. As fibroblasts grew out from the explants and expanded to 50%+ confluence of the T25 surface area, they were detached from the flask using prewarmed 37°C TrypLE Express (catalog no. 12604; Gibco, Grand Island, NY, http://www.invitrogen.com) and split 1:3 to 1:4 into larger T75 culture flasks for expansion. Master stocks of the fibroblasts were prepared at passage 3 (P3) with further expansion (typically P4-P8) for banking, distribution, or use. The cells were typically frozen at a density of 1.0 × 106 cells per milliliter per cryovial (see below).
Isolation of ASD Patient Plasma, Peripheral Blood Mononuclear Cells and Serum
Detailed protocols for isolation of ASD patient plasma, peripheral blood mononuclear cells, and serum are provided in the supplemental online data.
Nomenclature
In order to accurately define and successfully keep track of the cell phenotypes and genotypes derived from our ASD patients’ fibroblasts, we developed a robust nomenclature system that was chronological, adaptable, easy to follow, and provided a brief history of each cell line and/or labeled cryovial [25]. This was critical, because multiple clones were often derived from any given patient, and these were subsequently transduced, passaged, and cultured using a number of different methods.
In our ASD resource, each cell line starts with the SC indicator (National Human Neural Stem Cell Resource) and a unique patient identifier number and disease category number, followed by a listing of the tissue from which the biopsy was taken (S = skin) and the particular cells grown from that biopsy (F = fibroblasts). Each single dash divides the cellular phenotypes through which a particular line has transitioned or progressed (e.g., fibroblasts [F], iPSCs [I], NSCs [N], to neurons [Nn] and/or astrocytes [A]). Within these divisions, the specific culture history was also documented (culture condition, passage number, length of differentiation in days). Using this system, any unique manipulation of the cells, such as green fluorescent protein transduction, is indicated using parentheses at the phenotype at which it took place. Finally, because subculture has a propensity to result in a line with slightly different properties [30, 31], we designated this occurrence by the introduction of a double dash in that part of the sequence at which the subculture occurred.
iPSC Generation and Characterization
Fibroblasts were reprogrammed to iPSCs using both traditional integrative (Lentivirus; 1IX) [32] and nonintegrative (Sendai virus; 2IX) methods [25].
FMR1 CGG Repeat Length
Genomic DNA was isolated from patient fibroblasts using standard methods. The CGG repeat length was determined using Southern blot analysis and polymerase chain reaction amplification of genomic DNA, as previously described [26, 33]. The methylation status was determined by densitometry, as previously described [34]. The FMR1 mRNA expression levels were measured as detailed previously [35].
Karyotyping and Mycoplasma Detection
Karyotype analysis and mycoplasma detection were performed by Cell Line Genetics (Madison, WI, http://www.clgenetics.com).
Cryopreservation of ASD Fibroblast and iPSC Lines
Detailed protocols for the cryopreservation of the ASD fibroblast and iPSC lines are provided in the supplemental online data.
Generation of ASD-Specific NSCs
ASD iPSCs cultured using traditional mouse embryonic fibroblasts (MEFs) were transitioned for long-term, feeder-free culture using Accutase passaging and defined StemPro medium (Life Technologies), as previously described [24, 25]. Differentiation of iPSCs to NSCs was performed using DMH1 and SB431454, as previously described [25]. Immunocytochemistry characterization of pluripotency and neural differentiation was performed as previously described [25, 36].
Lentivirus Generation and Fluorescent Labeling of NSCs and Neurons
Detailed protocols of Lentivirus generation and fluorescent labeling of NSCs and neurons are provided in the supplemental online data.
Terminal Differentiation and Electrophysiology of ASD-Specific Neurons
Terminal differentiation and electrophysiology of ASD-specific neurons was performed as previously described [25], with the following modification. The NSCs were plated onto mouse cortical astrocytes on coverslips at a density of 1 × 105 cells per centimeter squared. Glial differentiation was also performed as previously described [25].
Results
ASD Fibroblast and iPSC Resource at CHOC Children’s
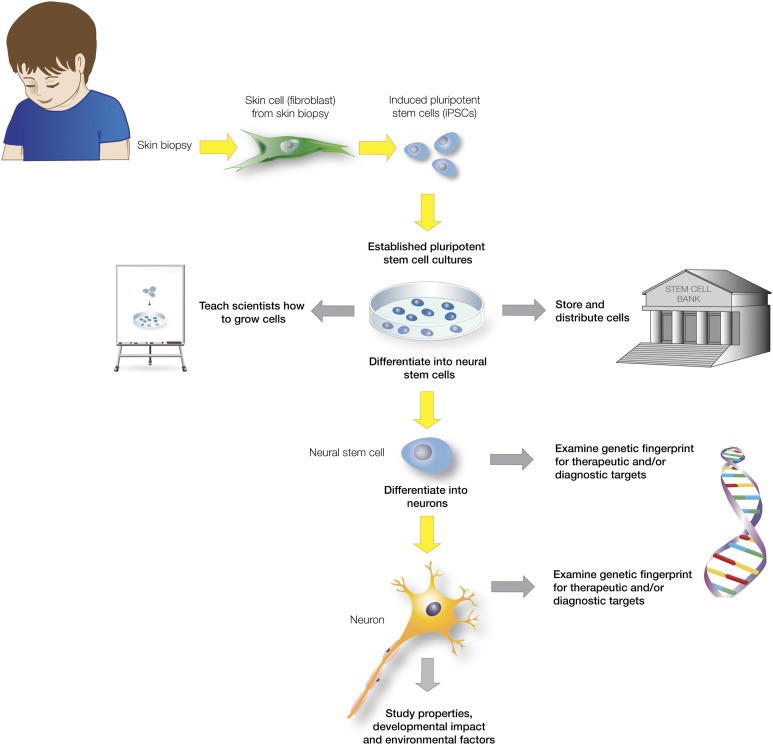
The overall purpose of this undertaking was to overcome a critical barrier in the field of ASD research (i.e., accessibility to statistically relevant numbers of patient-derived neural tissues for analysis). Our overall strategy (Fig. 1) was to obtain fibroblasts from specific ASD, FXS, and unaffected control patients, to derive iPSCs from the fibroblasts, and then to differentiate NSCs from the iPSCs, thus facilitating characterization and comparison of neural derivatives using a variety of next generation sequencing and bioinformatics approaches. We have successfully generated a resource bank of more than 200 different patient-specific lines. Plasma, serum, and peripheral blood mononuclear cells were also banked from each of the patients for follow-up study, when and if required. The patient lines were given a unique indicator and identifier that was not derived from information about the donor, denoted SCXXX (SC for National Human Neural Stem Cell Resource) and starting at SC101 [25]. Most ASD patient samples and unaffected controls were male (as described in Materials and Methods).
Figure 1.
Creating brain cells from skin cells to study the impact of autism on the living brain. Shown is an overview of the autism spectrum disorders (ASDs) fibroblast and induced pluripotent stem cell (iPSC) resource at the Children’s Hospital of Orange County. Skin biopsies from clinically well-defined patients with ASD were obtained, explant fibroblast cultures were established, expanded, and banked, and iPSCs were generated and characterized. After differentiation to the neural lineage, the ASD-specific cells can be compared with normal unaffected controls, allowing a myriad of biochemical, next-generation sequencing, and bioinformatics approaches.
Many of the patients had a clinical diagnosis of mutations that are linked with ASD, such as FXS and premutation involvement, and were assigned to specific patient groups (.1–.7) (Table 1). Also included were patients with ASD who had been diagnosed with autistic disorder (autism; idiopathic), PDD-NOS, Asperger syndrome, or Williams syndrome. The unaffected control lines were taken from neurologically normal volunteers who were not family members of the patients with ASD. All the clinical diagnoses were performed at one center, the UC Davis MIND Institute, by one group of clinical specialists, providing uniformity and confidence in the assigned ASD diagnoses of the patient cell lines. The detailed patient histories included race and ethnicity, age at biopsy, occipital-frontal circumference (OFC) percentile, seizure history (supplemental online Table 1), and the psychometric test scores for the Autism Diagnostic Observation Schedule–General (supplemental online Table 1), Autism Diagnostic Interview–Revised, and Wechsler Abbreviated Scale of Intelligence (supplemental online Table 1). The patient samples were also screened for CGG repeat size length, FMR1 expression, and methylation status (supplemental online Table 1).
The distribution of subtypes in our patient population represents another of our assets. As can be seen in Table 1, the collection contains various numbers of ASD phenotypes, with idiopathic autism (autistic disorder) predominating (n = 43), and also a significant cohort (n = 21) of unaffected controls that will also be useful for general iPSC disease model studies. The collection also contains several phenotypes known to be important in the ASDs. For example, the collection contains autism patient samples with and without megalencephaly, with and without seizures, and with high or low functioning (determined by IQ) (supplemental online Table 1). Furthermore, information on the presence or absence of seizures and the specific type of seizure previously presented (convulsions, petit mal, partial motor, temporal lobe, febrile or vasovagal, staring spells) were also documented (supplemental online Table 1) and available for study. For all these phenotypes, a variety of ages and race and ethnic backgrounds exist (supplemental online Table 1) within individual groups, allowing one to screen for and eliminate differences between groups that could arise simply from the age or demographics of the patient.
One of the advantages that this collection of ASD and control cell lines housed within the NHNSCR has to offer is uniformity. All skin biopsies were obtained in the same manner and from the same anatomic location (back of the shoulder) at UC Davis. In order to ensure uniformity among all the fibroblast lines, each of the fibroblast cell lines was derived by one group at CHOC Children’s using documented standard operating procedures for media preparation (SBM, HDFM), skin biopsy processing, expansion, and cryopreservation. Additionally, all iPSCs were generated, expanded, and characterized according to published protocols by the same group of individuals [25, 32]. Multiple iPSC clones were characterized by pluripotent stem cell (PSC) morphology, uniformity of staining for classic PSC markers such as Tra-1-60, Nanog, Sox-2, and Oct-4 and normal karyotype (seven NSCs were also karyotyped and were shown to be normal similar to the iPSC from which they were derived; thus, this was not routinely done). All equipment, media, and reagents associated with the generation and storage of the ASD fibroblasts, iPSCs, NSCs, and neural derivatives (tissue culture CO2 incubators, 4°C refrigerators, −20°C and −80°C freezers, and automatic-fill liquid nitrogen cryogenic freezers) at CHOC Children’s are continually monitored by an automated Centron Monitoring Alarm System (Rees Scientific, Trenton, NJ, http://www.reesscientific.com); thus, critical environmental parameters such as temperature and carbon dioxide levels remained constant and within range.
Furthermore, to provide quality control over the identity and history of the lines, we developed a standardized nomenclature to accurately define the multiple cell phenotypes and genotypes that could possibly be derived [25]. This robust nomenclature reflects that fact that we were deriving several different cellular phenotypes, as well as different clones, from any given patient with any given ASD and manipulating these cells (passage, transduction, culture, differentiation) in any number of different ways. This nomenclature gives a reasonable, but not exhaustive, picture of the cell culture history of any given line; thus, it would be relatively straightforward to determine whether one was actually working with two equivalent cell lines or, if they were from the same patient, that they were either the same or different in some definable way.
Access to these ASD fibroblast and iPSC lines is open to the scientific community, and they are available to all researchers (either nonprofit or commercial) worldwide for use in basic research. This collection will be continually expanded and will be a valuable resource for research into the basic neurobiology of the ASDs. An up-to-date list of cell lines available for request from the National Human Neural Stem Cell Resource at CHOC Children’s is available from http://www.nhnscr.org. Users wishing to request cells are asked to provide their curriculum vitae and complete both a statement of research intent and a CHOC Children’s materials transfer agreement. Researchers wishing to use the ASD-specific iPSCs and ASD-specific neural derivatives for study, but lacking sufficient expertise, can also request to receive hands-on training through the NHNSCR.
Functional Derivation of ASD-Specific Neurons and Glia
Reprogramming of somatic cells into iPSCs provides a unique opportunity to gain insight into the molecular and cellular basis of disease. Specifically, development of disease in dish models using previously inaccessible cell types affected in a tissue such as brain holds immense interest for the study and treatment of neurological disease such as the ASDs. At the pluripotent stage, cells multiply indefinitely; thus, large numbers of cells can be generated for studies designed to link unique aspects of patients’ disease manifestations and drug responses with their complex genetic causes.
However, efficient neuronal differentiation of functional neuronal subtypes from iPSCs must be achieved to delineate differences between unaffected controls and ASD states and facilitate the promise of iPSC technologies for effective screening of drug candidates to help restore normal phenotype and function.
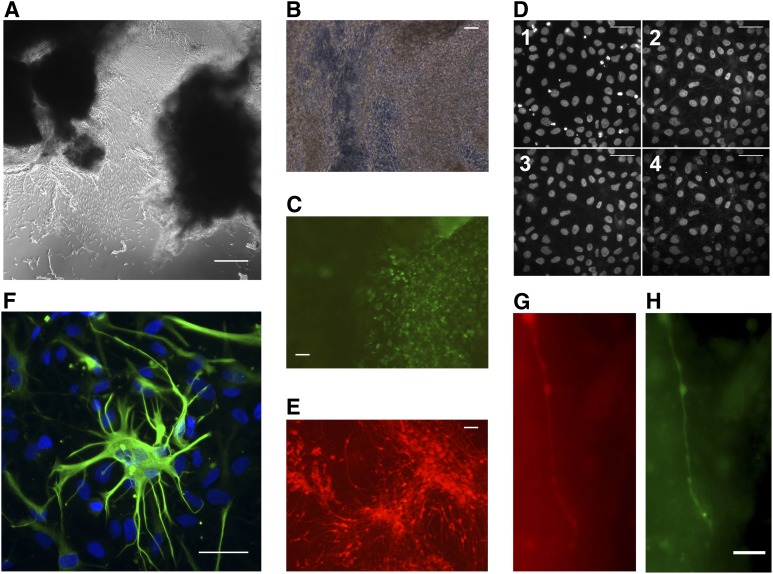
We developed a process-based system to ensure uniformity across all fibroblasts, iPSCs, and NSCs produced. Explant fibroblast cultures (Fig. 2A) developed usually within 2–3 weeks after dicing of the skin biopsy and plating on fibronectin-coated flasks. Fibroblasts were expanded, banked, and transduced to produce iPSCs (Fig. 2B, 2C). Live cell staining of iPSCs with Alexa Fluor Tra-1-60-labeled antibody (BD Biosciences) proved useful in distinguishing bona fide iPSC colonies for picking and expansion (Fig. 2C) [37]. Previously, we have found that iPSCs grown under traditional culture conditions using MEFs can be easily transitioned to single-cell passaging conditions using Accutase passaging and defined StemPro medium (Life Technologies) [24]. StemPro-adapted cultures show uniformity of staining for classic PSC markers such as Nanog and Sox-2 (Fig. 2D) while maintaining the normal karyotype [25].
Figure 2.
Process-based expansion of human autism spectrum disorder (ASD) explant fibroblast cultures to neural stem cells (NSCs) and terminal differentiation to glia and synaptic neurons. (A): Phase contrast image of SC234.7-SF0 explant fibroblast culture formation on a fibronectin-coated T25 flask. (B): Phase contrast of SC157.7-SF4-2I4.M5 induced pluripotent stem cell (iPSC) colonies. (C): Live Tra-1-60 surface immunofluorescence staining of the same field in (B), illustrating discrimination of “true” iPSC colonies. (D): Immunofluorescence staining of StemPro-adapted SC105.9-SF5-1I6.M8S7 showing nuclear counterstaining by 4′6-diamidino-2-phenylindole (DAPI) (D1) and maintenance of the pluripotency markers Nanog (D2), Oct4 (D3), and Sox-2 (D4). (E): Fluorescent-labeled NSCs [SC105.9-SF5-1I6/M8S7—S2-N2G8 (Tdt)]. (F): Terminal differentiation to glia [SC176.1-SF6-2I2.M5S3—S2-N2G5-A127]—composite image showing expression of the glial-specific marker glial fibrillary acidic protein (green) with nuclear counterstaining by DAPI. (G, H): Labeling of synaptic neurons [SC176.1-SF6-2I2-M5S3—S3S2N2G5(Tdt)-4Nn85 (green fluorescent protein)-20]. Tdt-labeled NSCs [SC176.1-SF6-2I2-M5S3—S3S2N2G5(Tdt)] were plated on mouse glia and terminally differentiated to neurons for 85 days before transduction with pHIV7/Syn-EGFP lentivirus. The cells were stained and observed for colabeling of red (neurons derived from iPSC-derived NSCs) and synapsin expressing neurons (green) at 20 days after transduction. Scale bars = 100 µm (A–F) (×100), 20 µm (G, H).
Critically, this feeder-free and defined culture system allowed for the efficient, highly reproducible scale up of ASD-specific iPSC cultures for NSC differentiation and more robust, cleaner, downstream evaluation. This is critical for high throughput iPSC or NSC screens for marker discovery and/or drug screening. Using a completely defined chemical neural induction protocol (SB431452 and DMH1), we were able to effectively differentiate adherent feeder-free ASD iPSCs en masse into Pax-6-, Sox-1-, neural cell adhesion molecule-expressing NSCs. These NSCs can be effectively labeled for cell tracking purposes (Fig. 2E), and terminal differentiation of these ASD-specific iPSC-derived NSCs into glia (Fig. 2F) and bona fide action-potential-firing neurons has also been achieved [25]. These ASD neurons not only fire trains of action potentials on depolarization [25] but are now found to also form functional synaptic connections and fire spontaneous bursts of action potentials (Fig. 3). Blocking K+ and Ca2+ currents, using an internal solution with Cs+ instead of K+ and an external solution with Co2+ instead of Ca2+, with the addition of 2.5 mM tetraethylammonium chloride and 1 mM 4-aminopyridine, reveals isolated sodium currents with a large transient (INaT) component and a small persistent component (INaP) typical of mammalian neurons (Fig. 3B).
Figure 3.
Electrical activity in autism spectrum disorder (ASD) neurons differentiated from induced pluripotent stem cell (iPSC)-derived neural stem cells (NSCs). Whole-cell voltage clamp recording of a neuron derived from SC105.7-SF4-1I1M20S13-N2G2-4Nn21 revealing spontaneous synaptic currents (A); spontaneous action potential bursts (B); voltage steps under K+ and Ca2+ blockade (C); resulting Na+ currents (D); voltage steps under Na+ and Ca2+ blockade (E); and resulting K+ currents (F).
Thus, one can traverse from a skin biopsy ASD fibroblast cell line (Fig. 2A) to a defined NSC population that immunologically stains positive for NSC markers such as Pax-6, nestin, and Sox-1 [25], morphologically resembles brain derived NSCs [16] to ultimately ASD-specific neurons and glia. Our overall strategy using iPSC technology to model the functional ASD brain could well be very powerful and effective because terminal differentiation of these cells has consistently produced glia and has resulted in the generation of bona fide action-potential-firing, synaptic (synapsin+, MAP2ab+, PSD-95+ staining) neurons [25]. Furthermore, colabeling of tandem dimer tomato ASD-specific neurons after infection with a lentivirus (pHIV7/Syn-EGFP) encoding enhanced green fluorescent protein under control of the neuron-specific synapsin promoter is also possible (Fig. 2G, 2H), adding additional confirmation to their functional synaptic identity [38, 39].
Discussion
The Autism Spectrum Disorders Fibroblast and iPSC Resource at CHOC Children’s was developed with funding from the NIH to meet the scientific need for a comprehensive collection of ASD-specific cell lines for study. Most of the ASDs are still rather poorly understood at the molecular, cellular, and genetic levels, and they remain a substantially unmet medical need. Successful iPSC-based modeling of genetically complex diseases such as the ASDs requires a population-based approach to create a comprehensive collection of cell lines for statistically valid investigation. We have generated an ASD fibroblast and iPSC resource that we believe has significant potential to affect our understanding of ASD disease mechanisms and improve treatment options through ASD-specific disease modeling, target discovery, and drug discovery and development.
The NHNSCR has currently received more than 100 patient skin biopsy samples that have been expanded and banked for distribution. Many have also been reprogrammed to iPSCs, characterized for quality and banked, and are available for distribution to interested investigators. Critically, the ASD donor-specific demographic, medical, and complete ASD diagnostic information (ADOS-G, ADR-I, WASI) associated with each ASD patient cell line and information on head circumference (OFC) percentiles and seizure status are also available, allowing for additional phenotypic comparative studies.
Successful disease modeling, in particular, for the ASDs, will rely on the development of reliable, reproducible, cell-based assays and models that effectively reflects patient phenotypes [40]. The establishment of iPSC-based models is a complex process with multiple steps, including tissue biopsy collection, establishment of explant fibroblast cultures, clonal iPSC derivations and culture, and expansion and differentiation into relevant cell types. Because each of these steps is prone to introduce experimental variation that can skew and confound disease phenotype analyses, we have chosen to create a bank of ASD fibroblasts and iPSCs in-house using one clinical institute’s ASD patient diagnoses, one site of skin biopsy, and one laboratory site of fibroblast and iPSC derivation. We sought to establish rigorous quality control protocols for the establishment of the ASD cell lines using written standard operating procedures, the development of a robust system of cellular nomenclature that describes an accurate genotype and phenotype of the cells at any point, and automated environmental monitoring of all equipment used to culture and bank the cells for distribution. This overall strategy should minimize variation in both clinical ASD diagnoses among centers and fibroblast and iPSC generation between different laboratories and reduce the noise in downstream analysis, which can be critical in modeling the ASDs owing to their phenotypic variability.
It is currently thought that ASDs result from disruptions in early brain development involving, variably or concomitantly, changes in cortical growth or patterning or changes in synaptic signaling, perhaps in the context of sexual dimorphism [41, 42]. We initially focused the patient recruitment on those with idiopathic autism and those with FXS, the most-common known, single-gene associated with ASD, which is caused by the absence of the FMR1 gene encoded protein, FMRP. FMRP is an RNA binding protein that controls the translation of several other genes that regulate synaptic development and plasticity, glutamate and γ aminobutyric acid neurotransmission, and mammalian target of rapamycin and phosphatase and tensin homolog pathways (reviewed in [43]). ASD occurs in approximately 60% of cases [44]. Premutation repeat expansions (55–200 CGG repeats) can also result in ASDs, through a molecular mechanism that involves a direct toxic effect of the expanded CGG-repeat FMR1 mRNA [45].
We envisage that using the shared and unshared neurobiology of ASD and FXS [8] as a comparative framework for studies that focus on genetics, brain mechanisms, and pharmacological and biological interactions will be revolutionary for the understanding of the ASDs and the development of novel therapeutic agents. This approach, however, requires sufficient numbers of patient samples that satisfy power analysis considerations of large multivariable studies in next-generation sequencing bioinformatics approaches. Thus, we have created this ASD-specific cell resource to satisfy this need for access to ASD-specific lines within the ASD research community. This ASD resource and analysis thereof can also draw on existing ASD and FXS clinical, basic science, and Gene Expression Omnibus databases.
The conversion of ASD fibroblasts to ASD-specific NSCs and ASD-specific neural derivatives is a major breakthrough in research aimed at delineating the differences between those with ASDs and the unaffected controls. It is now well accepted that iPSC-derived disease-in-a-dish models faithfully reproduce the phenotype [20]. Disease-specific phenotypes in iPSC cell-derived neurons generated from patients with spinal muscular atrophy [46], familial dysautonomia [47], Rett and Timothy syndromes [48], schizophrenia [49], and Parkinson disease [50], among others, have been reported.
A challenge in using iPSC disease models is to efficiently produce relevant differentiated and functional cell types for analysis. We have developed a reliable method for long-term, single-cell passaging of iPSCs using a feeder-free, defined culture system that facilitates scale up and expansion of iPSCs and allows for the efficient, highly reproducible, fully defined, en masse neural differentiation of confluent, adherent iPSC cultures. For downstream analysis, this eliminates traditional coculture and inefficient embryoid body neural differentiation methods. We have applied this process-based approach to several of the ASD patient samples in our collection. Terminal differentiation of ASD, iPSC-derived, NSCs produced large numbers of β-III-tubulin-positive immature neuronal cells that can be matured into bona fide repetitive action-potential-firing neurons. Thus, we are confident that the iPSCs and neural cells so produced for analysis are functionally relevant and will be extremely useful and facilitate the induction of a multitude of ASD neuronal cell types for study. This is important for large-scale comparative studies of iPSC-derived models, because previous studies, using NSCs derived from fmr1-mouse brain or from the brain of a fragile X fetus, have suggested that FMRP-deficient cells result in a higher proportion of β-III-tubulin-positive cells (immature neurons) than unaffected cell populations and that these cells had fewer and shorter neurites [51–53].These cells also had alterations in intracellular Ca2+ after neurotransmitter exposure [54]. Furthermore, evidence of a larger population of early progenitors in the neurogenic zone suggested changes in early maturation. Remarkably, similar changes were seen in the fragile X permutation, in which, in addition to the presence of intranuclear inclusions in both neurons and glia, alterations in cell migration and differentiation were seen, along with less complex dendritic arborization and a higher frequency of spike bursting [55–58]. The iPSC-based neural model platform we have developed allows for additional confirmation and exploration of these earlier seminal findings.
Terminal differentiation of these ASD-specific iPSCs to cells of the glial lineage has also been possible using recently devised methods, and these resulted in the production of a large number of S-100 β-positive glial cells [25]. The latter are particularly important to consider [59], because it has recently been shown in two disease states (Parkinson disease and amyotrophic lateral sclerosis) that these diseases, long thought to be cell autonomous diseases of neurons, might be, in fact, cell nonautonomous glial diseases [60]. Separate examination of glia versus neurons will address the hypothesis that neuronal dysfunction is cell autonomous. Furthermore, each cellular phenotype can also be differentiated on human glia of contrasting ASD and control phenotypes to elucidate the participation of glia in the neuronal phenotypes seen. Alterations in the pathways involved in proliferation, migration, differentiation, and maintenance of synaptic homeostasis can serve to distinguish certain ASD patient genotypes and phenotypes. Collectively, these results validate our overall approach in that ASD-specific patient skin fibroblasts contained in our collection can be effectively reprogrammed to iPSCs that can be expanded in large numbers to satisfy power analyses and produce functionally active neurons and glia for downstream analysis and study. Similarly, the ASD iPSCs and NSCs can be differentiated into a myriad of neuronal subtypes [61], such as interneurons and excitatory and inhibitory neurons, using the most up-to-date induction protocols available and compared with unaffected controls.
Because ASD appears to be a primary brain disorder, it seems clear that cellular neural derivatives are most likely to yield mechanistic insights and possible therapeutic targets (the in vitro representation of the in vivo brain). Critically, all the cellular iPSC derivatives are produced from every patient; thus, genetic matching is not only possible, but this also greatly enhances the statistical analytical power. iPSC models could have many advantages compared with the traditional transcriptome analysis of ASD postmortem brain-derived samples [62, 63]. The brain samples, although clearly neural, are not, however, genetic matches to any of the iPSC-derived cellular derivatives currently available for study. Moreover, the brain parenchyma is predominately glial, not neuronal; thus, the data derived from it will necessarily be a complex mixture (not so with the cellular derivatives) of cell types. It serves the useful purpose, however, of providing an in vivo correlate that is the only one we presently have. Moreover, postmortem RNA, although subject to degradation during the postmortem interval, has proved useful and has been shown to provide data that are reasonably consistent with our current theories of the underpinnings of the ASDs (e.g., synaptic dysfunction).
Analysis of the non-neural ASD cellular derivatives (fibroblasts and peripheral blood mononuclear cells) present in the collection could also serve an entirely different purpose—they might provide diagnostic targets in easily obtainable tissues (blood and skin). If one can find commonalities between the ASD neural and ASD non-neural derivatives, which are unaffected in controls, one might be able to provide the rationale for genetic and/or biochemical tests that can help in the early diagnosis of patients with ASD, who currently are diagnosed by what are necessarily subjective behavioral tests.
Importantly, recent next-generation sequencing studies [64–66] suggest the importance of de nova mutations in the etiology of some cases of ASDs. In this respect, independent of the well-known RNAs that encode for genes, noncoding RNAs (e.g., microRNAs [miRNAs]) have recently also begun to emerge as key players that alter the expression of genes [67]. miRNAs exist in the tissues and as circulating miRNAs in several body fluids, including plasma or serum, cerebrospinal fluid, urine, and saliva. Because studies have indicated significant differences between the circulating miRNA expression profiles of healthy and affected individuals in a variety of neurological diseases [68, 69], circulating miRNAs could also be a novel class of noninvasive and sensitive biomarkers to test. Because early behavioral intervention mechanisms help improve the outcomes of children affected with ASDs [70], the development of methods for early screening, detection, and diagnosis of ASD will be of immense importance.
Collectively, the patient-derived ASD fibroblasts, iPSCs, NSCs, and neural derivatives contained within our collection represent a valuable resource to the ASD community and provide a powerful, surrogate, cell-based platform that will enable unprecedented insights into ASD mechanisms, genetics, and contributing factors. Critically, this should facilitate the identification and development of new ASD-specific biomarkers, diagnostics, and/or novel therapeutic agents. We have already distributed many of the ASD and control lines to investigators worldwide, and it is our hope that the lines will become extensively used within the research community. The unaffected control iPSC lines (n = 21) will also be useful to anyone outside the ASD field interested in well-characterized unaffected controls for a general iPSC-based disease model and drug discovery studies. Together, we hope that the analysis of these lines will help unravel the “autisms” that are contained within the ASDs. This will lead to novel interventions developed through high-throughput screening of ASD patient-specific iPSC-derived cells and has the potential to rapidly and significantly affect a large and growing ASD patient population and their families currently coping with an ASD diagnosis.
Conclusion
A critical barrier to ASD research is the limited availability of statistically relevant numbers of patient-derived tissue samples, especially those of neural origin. We describe a newly established ASD-specific fibroblast and stem cell resource. We currently have more than 200 lines, available for request, derived from clinically well-defined ASD-specific patients and unaffected volunteers. We have confirmed, using a subset of the normal and ASD-specific iPSCs, that these cells can be differentiated toward a neural stem cell phenotype and terminally differentiated into spontaneous action-potential firing neurons and glia. This represents a significant resource that will advance the use of ASD-specific patient stem cells as disease models by the scientific community.
Supplementary Material
Acknowledgments
We thank the patients with ASDs and their families for participation in this study. We also thank the unaffected controls at UC Davis, who generously volunteered to be a part of the study. We are extremely grateful to Dr. Lily Ngotran, Andrew Ligsay, and Jonathan Polussa for the coordination of patient recruitment and procurement of patient samples at the Medical Investigation of Neurodevelopmental Disorders Institute. We thank Dr. Jane Pickett of the Autism Tissue Program of Autism Speaks and Dr. Clara Lajonchere of the Autism Genetic Resource Exchange of Autism Speaks for their enthusiasm for the development of this ASD-specific resource. We thank Dr. Atsushi Miyanohara, UC San Diego, for the kind gift of the pHIV7/Syn-EGFP transfer vector. We thank Dr. Omar Khalid, Siranush Herculian, Daniel C. Mendez, Dean R. Perusse, and Matthew Kunicki for tissue culture assistance. This work was funded in part by NIH Grant R01-HD059967 and Autism Speaks Trailblazer Grant 7379 to P.H.S., Eunice Kennedy Shriver National Institute of Child Health and Development Grants HD036071 and HD02274 to R.J.H. and F.T., NIH Grant R01-NS083009 to D.K.O., and CHOC Children’s continued support of the National Human Neural Stem Cell Resource. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, Autism Speaks, the University of California, Irvine, the University of California, Davis, or CHOC Children’s. The National Cancer Institute Preclinical Repository generously supplied basic fibroblast growth factor-2.
Author Contributions
D.J.B.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; R.J.H.: patient recruitment and biopsy collection, conception and design, collection and assembly of data, data analysis and interpretation; F.T. and D.K.O.: conception and design, collection and assembly of data, data analysis and interpretation; H.E.N., S.M., M.G.B., A.E.S., and S.S.S.: assembly of data, data analysis and interpretation; D.R.H. and M.O.: data collection, assembly and interpretation; P.H.S.: conception and design, data analysis and interpretation, financial support, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
R.J.H. has compensated consulting, research funding, and travel reimbursement. D.R.H. is a compensated consultant and speaker. F.T. has uncompensated employment.
References
- 1.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. 5th ed. Arlington, VA: American Psychiatric Publishing; 2013. [Google Scholar]
- 2.Belmonte MK, Cook EH, Jr, Anderson GM, et al. Autism as a disorder of neural information processing: Directions for research and targets for therapy. Mol Psychiatry. 2004;9:646–663. doi: 10.1038/sj.mp.4001499. [DOI] [PubMed] [Google Scholar]
- 3.Grzadzinski R, Huerta M, Lord C. DSM-5 and autism spectrum disorders (ASDs): An opportunity for identifying ASD subtypes. Mol Autism. 2013;4:12. doi: 10.1186/2040-2392-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atladottir HO, Gyllenberg D, Langridge A, et al. The increasing prevalence of reported diagnoses of childhood psychiatric disorders: A descriptive multinational comparison. Eur Child Adolesc Psychiatry. 2014 doi: 10.1007/s00787-014-0553-8. [DOI] [PubMed] [Google Scholar]
- 5.Kanner L. Autistic disturbances of affective contact. Nervous Child. 1943;2:217–250. [PubMed] [Google Scholar]
- 6.Skafidas E, Testa R, Zantomio D, et al. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol Psychiatry. 2012;19:504–510. doi: 10.1038/mp.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lord C. Methods and measures of behavior in the diagnosis of autism and related disorders. Psychiatr Clin North Am. 1991;14:69–80. [PubMed] [Google Scholar]
- 8.Hagerman R, Hoem G, Hagerman P. Fragile X and autism: Intertwined at the molecular level leading to targeted treatments. Mol Autism. 2010;1:12. doi: 10.1186/2040-2392-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo R, Sanders SJ, Tian Y, et al. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am J Hum Genet. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santini E, Huynh TN, MacAskill AF, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14:161–176. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- 12.Shelton JF, Hertz-Picciotto I, Pessah IN. Tipping the balance of autism risk: Potential mechanisms linking pesticides and autism. Environ Health Perspect. 2012;120:944–951. doi: 10.1289/ehp.1104553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiao EY, McBride SW, Chow J, et al. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc Natl Acad Sci USA. 2012;109:12776–12781. doi: 10.1073/pnas.1202556109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickett J, London E. The neuropathology of autism: A review. J Neuropathol Exp Neurol. 2005;64:925–935. doi: 10.1097/01.jnen.0000186921.42592.6c. [DOI] [PubMed] [Google Scholar]
- 15.Haroutunian V, Pickett J. Autism brain tissue banking. Brain Pathol. 2007;17:412–421. doi: 10.1111/j.1750-3639.2007.00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz PH, Bryant PJ, Fuja TJ, et al. Isolation and characterization of neural progenitor cells from post-mortem human cortex. J Neurosci Res. 2003;74:838–851. doi: 10.1002/jnr.10854. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz PH, Tassone F, Greco CM, et al. Neural progenitor cells from an adult patient with fragile X syndrome. BMC Med Genet. 2005;6:2. doi: 10.1186/1471-2350-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park IH, Lerou PH, Zhao R, et al. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- 19.Lewitzky M, Yamanaka S. Reprogramming somatic cells towards pluripotency by defined factors. Curr Opin Biotechnol. 2007;18:467–473. doi: 10.1016/j.copbio.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Saporta MA, Grskovic M, Dimos JT. Induced pluripotent stem cells in the study of neurological diseases. Stem Cell Res Ther. 2011;2:37. doi: 10.1186/scrt78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peitz M, Jungverdorben J, Brüstle O. Disease-specific iPS cell models in neuroscience. Curr Mol Med. 2013;13:832–841. doi: 10.2174/1566524011313050014. [DOI] [PubMed] [Google Scholar]
- 22.Bellin M, Marchetto MC, Gage FH, et al. Induced pluripotent stem cells: The new patient? Nat Rev Mol Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- 23.Kondo T, Asai M, Tsukita K, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Stover AE, Schwartz PH. Adaptation of human pluripotent stem cells to feeder-free conditions in chemically defined medium with enzymatic single-cell passaging. Methods Mol Biol. 2011;767:137–146. doi: 10.1007/978-1-61779-201-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover AE, Brick DJ, Nethercott HE, et al. Process-based expansion and neural differentiation of human pluripotent stem cells for transplantation and disease modeling. J Neurosci Res. 2013;91:1247–1262. doi: 10.1002/jnr.23245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filipovic-Sadic S, Sah S, Chen L, et al. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin Chem. 2010;56:399–408. doi: 10.1373/clinchem.2009.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord C, Risi S, Lambrecht L, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- 28.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 29.Wechsler D. Wechsler Abbreviated Scale of Intelligence (WASI) San Antonio: Harcourt Assessment, Inc.; 1999. [Google Scholar]
- 30.Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23:699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- 31.Maitra A, Arking DE, Shivapurkar N, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 32.Nethercott HE, Brick DJ, Schwartz PH. Derivation of induced pluripotent stem cells by lentiviral transduction. Methods Mol Biol. 2011;767:67–85. doi: 10.1007/978-1-61779-201-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tassone F, Pan R, Amiri K, et al. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 2008;10:43–49. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tassone F, Longshore J, Zunich J, et al. Tissue-specific methylation differences in a fragile X premutation carrier. Clin Genet. 1999;55:346–351. doi: 10.1034/j.1399-0004.1999.550508.x. [DOI] [PubMed] [Google Scholar]
- 35.Tassone F, Hagerman RJ, Taylor AK, et al. Elevated levels of FMR1 mRNA in carrier males: A new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nethercott HE, Brick DJ, Schwartz PH. Immunocytochemical analysis of human pluripotent stem cells. Methods Mol Biol. 2011;767:201–220. doi: 10.1007/978-1-61779-201-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan EM, Ratanasirintrawoot S, Park IH, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 38.Kucharova K, Hefferan MP, Patel P, et al. Transplantation of rat synapsin-EGFP-labeled embryonic neurons into the intact and ischemic CA1 hippocampal region: distribution, phenotype, and axodendritic sprouting. Cell Transplant. 2011;20:1163–1178. doi: 10.3727/096368910X564544. [DOI] [PubMed] [Google Scholar]
- 39.Pruszak J, Sonntag KC, Aung MH, et al. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells. 2007;25:2257–2268. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Walsh CA, Morrow EM, Rubenstein JL. Autism and brain development. Cell. 2008;135:396–400. doi: 10.1016/j.cell.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bagni C, Tassone F, Neri G, et al. Fragile X syndrome: Causes, diagnosis, mechanisms, and therapeutics. J Clin Invest. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris SW, Hessl D, Goodlin-Jones B, et al. Autism profiles of males with fragile X syndrome. Am J Ment Retard. 2008;113:427–438. doi: 10.1352/2008.113:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagerman R, Hagerman P. Advances in clinical and molecular understanding of the FMR1 premutation and fragile X-associated tremor/ataxia syndrome. Lancet Neurol. 2013;12:786–798. doi: 10.1016/S1474-4422(13)70125-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebert AD, Yu J, Rose FF, Jr, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee G, Papapetrou EP, Kim H, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krey JF, Paşca SP, Shcheglovitov A, et al. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16:201–209. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brennand KJ, Simone A, Jou J, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung CY, Khurana V, Auluck PK, et al. Identification and rescue of α-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342:983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tervonen TA, Louhivuori V, Sun X, et al. Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiol Dis. 2009;33:250–259. doi: 10.1016/j.nbd.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 52.Bhattacharyya A, McMillan E, Wallace K, et al. Normal neurogenesis but abnormal gene expression in human fragile X cortical progenitor cells. Stem Cells Dev. 2008;17:107–117. doi: 10.1089/scd.2007.0073. [DOI] [PubMed] [Google Scholar]
- 53.Comery TA, Harris JB, Willems PJ, et al. Abnormal dendritic spines in fragile X knockout mice: Maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao Z, Hulsizer S, Cui Y, et al. Enhanced asynchronous Ca(2+) oscillations associated with impaired glutamate transport in cortical astrocytes expressing Fmr1 gene premutation expansion. J Biol Chem. 2013;288:13831–13841. doi: 10.1074/jbc.M112.441055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greco CM, Hagerman RJ, Tassone F, et al. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 56.Greco CM, Berman RF, Martin RM, et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 57.Jacquemont S, Hagerman RJ, Leehey M, et al. Fragile X premutation tremor/ataxia syndrome: Molecular, clinical, and neuroimaging correlates. Am J Hum Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tassone F, Hagerman RJ, Garcia-Arocena D, et al. Intranuclear inclusions in neural cells with premutation alleles in fragile X associated tremor/ataxia syndrome. J Med Genet. 2004;41:e43. doi: 10.1136/jmg.2003.012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacobs S, Doering LC. Astrocytes prevent abnormal neuronal development in the fragile X mouse. J Neurosci. 2010;30:4508–4514. doi: 10.1523/JNEUROSCI.5027-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchetto MC, Muotri AR, Mu Y, et al. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz PH, Brick DJ, Stover AE, et al. Differentiation of neural lineage cells from human pluripotent stem cells. Methods. 2008;45:142–158. doi: 10.1016/j.ymeth.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garbett K, Ebert PJ, Mitchell A, et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helsmoortel C, Vulto-van Silfhout AT, Coe BP, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31:20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin XF, Wu N, Wang L, et al. Circulating microRNAs: A novel class of potential biomarkers for diagnosing and prognosing central nervous system diseases. Cell Mol Neurobiol. 2013;33:601–613. doi: 10.1007/s10571-013-9940-9. [DOI] [PubMed] [Google Scholar]
- 69.Rege SD, Geetha T, Pondugula SR, et al. Noncoding RNAs in neurodegenerative diseases. ISRN Neurol. 2013;2013:375852. doi: 10.1155/2013/375852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harris SL, Handleman JS. Age and IQ at intake as predictors of placement for young children with autism: A four- to six-year follow-up. J Autism Dev Disord. 2000;30:137–142. doi: 10.1023/a:1005459606120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.