Human embryonic stem cell lines produce a number of clusters of pigmented retinal pigment epithelium (RPE) cells when allowed to spontaneously differentiate, but the timing and efficiency of differentiation is highly variable. Neural induction using dorsomorphin significantly enhanced the RPE yield in one cell line but significantly reduced it in another, underlining the necessity of tailoring differentiation protocols to suit the innate properties of different cell lines.
Keywords: Retinal pigment epithelium, Cell differentiation, Human embryonic stem cells, Cell therapy, Bioprocess engineering
Abstract
Human embryonic stem cells (hESCs) are a promising source of retinal pigment epithelium (RPE) cells: cells that can be used for the treatment of common and incurable forms of blindness, such as age-related macular degeneration. Although most hESC lines will produce a number of clusters of pigmented RPE cells within 30–50 days when allowed to spontaneously differentiate, the timing and efficiency of differentiation is highly variable. This could prove problematic in the design of robust processes for the large scale production of RPE cells for cell therapy. In this study we sought to identify, quantify, and reduce the sources of variability in hESC-RPE differentiation. By monitoring the emergence of pigmented cells over time, we show how the cell line, passaging method, passage number, and seeding density have a significant and reproducible effect on the RPE yield. To counter this variability, we describe the production of RPE cells from two cell lines in feeder-free, density controlled conditions using single cell dissociation and seeding that is more amenable to scaled up production. The efficacy of small molecules in directing differentiation toward the RPE lineage was tested in two hESC lines with divergent RPE differentiation capacities. Neural induction by treatment with a bone morphogenetic protein inhibitor, dorsomorphin, significantly enhanced the RPE yield in one cell line but significantly reduce it in another, generating instead a Chx10 positive neural progenitor phenotype. This result underlines the necessity to tailor differentiation protocols to suit the innate properties of different cell lines.
Introduction
The retinal pigment epithelium (RPE) is a monolayer of pigmented cells that lie adjacent to the photoreceptors in the retina. The RPE is critical to the function and survival of the photoreceptors, and a number of degenerative eye disorders begin with dysfunction and death of RPE cells [1]. Transplantation of healthy adult or fetal RPE cells into the subretinal space has proved beneficial for the treatment of a number of degenerative retinal diseases in animal models and humans [1, 2]. The scarcity of such material, however, has made this an impractical approach to treat common retinal disorders such as age-related macular degeneration, which is the most common cause of blindness in people older than 50 years [3, 4]. Human embryonic stem cells (hESCs) are pluripotent and have proved capable of differentiating into RPE and a number of early neural retinal cell types [5, 6]. In contrast to other sources of cells, hESCs can be expanded almost indefinitely in culture, making them a potentially unlimited resource for transplantation therapies. Clusters of pigmented epithelial cells appearing in confluent hESC cultures were first described in 2001 by Odorico et al. in the H9 cell line [7]. Subsequent studies in which the pigmented cells were enriched and characterized confirmed their RPE-like properties, including cobblestone morphology, RPE protein and RNA expression profiles, morphological features such as apical microvilli, polarized secretion of growth factors, and phagocytosis of rod and cone photoreceptor outer segments [8–12]. The RPE cells tend to appear in differentiating cultures as heavily pigmented clusters that are easily identifiable by eye and can be manually dissected away from the nonpigmented cells using a fine needle or microcresent blade [9].
The relative lack of intervention required for the hESC-RPE differentiation process, coupled with the simplicity with which the target cell type can be identified and isolated, has no doubt contributed to the speed with which this potential therapy has been brought to the clinic. Nevertheless, the time required to generate a pure population of RPE cells ready for transplantation, the cost of the materials, and the labor intensive nature of current hESC culture practices mean that the efficiency of the differentiation process will be critical to the economic feasibility of any hESC-RPE therapy. RPE cells have been successfully generated from a large number of hESC lines and a variety of protocols have been used [13], with some progress made in identifying small molecules and growth factors that increase hESC to retinal and/or RPE differentiation These have included the addition of factors to induce neuroectoderm/forebrain formation (Nicotinamide, dickkopf-related protein 1, Noggin, LeftyA, SB431542) and factors thought to act later in the developmental time line (insulin-like growth factor 1, sonic hedgehog, fibroblast growth factor, activin) [5, 13–16]. The efficacy of these differentiation protocols has proven highly variable between cell lines [14, 17, 18]. Reaching a consensus regarding the most efficient protocol has not been helped by the variety of methods used to quantify the yield and/or efficiency of RPE differentiation and the timing at which these properties are measured [13].
In this study, we have developed a simple, robust method for quantifying the relative hESC-RPE conversion efficiency. A novel, feeder-free differentiation protocol that uses single cell dissociation and density-optimized seeding was used. This method enhances reproducibility in hESC-RPE differentiation and is more amenable to scaled up and/or automated manufacturing processes than traditional methods. Using this method, we demonstrate and quantify a cell line-dependent effect of time, culture adaptation, seeding density, and passaging method on RPE differentiation efficiency. These will be critical variables to optimize and control during the scaled up manufacturing of RPE cells for clinical use. Using this protocol, we demonstrate the divergent effects of small molecules on RPE differentiation in different cell lines, suggesting that the optimal RPE manufacturing strategy is likely to be highly cell line specific and a function of a particular cell line’s innate differentiation preferences.
Materials and Methods
Cell Culture
Undifferentiated Shef6 and Shef3 hESC lines were obtained from the UK Stem Cell Bank. The cells were maintained at 37°C and 5% vol/vol CO2 in a humidified Sanyo IncuSafe incubator (Sanyo, Leicestershire, U.K., http://www.sanyo-biomedical.co.uk). The cells were seeded onto 0.1% porcine gelatin (Sigma-Aldrich, Gillingham, U.K., http://www.sigmaaldrich.com) coated T25 flasks on mitomycin-C inactivated mouse embryo fibroblasts (MEFs) in knockout Dulbecco’s modified Eagle’s medium supplemented with 20% (vol/vol) knockout serum replacement, 2 mM GlutaMAX 1% (vol/vol) nonessential amino acids, 4 ng/ml basic fibroblast growth factor (FGF2) (Invitrogen, Paisley, U.K., http://www.invitrogen.com), and 0.1 mM β-mercaptoethanol (Sigma-Aldrich). hESC colonies were passaged every 3–4 days when most colonies were large enough to fill a 10× field of view, and the cells were treated with 0.125 mg/ml collagenase IV (Invitrogen) for 4 minutes at 37°C. The colonies with undifferentiated hESC morphology were then dissected into smaller clumps under a dissecting microscope using a fine tip Pasteur pipette. The detached clumps were seeded onto fresh MEF feeders in hESC medium at a split ratio of one flask into two.
hESC-RPE Differentiation
Shef6 and Shef3 were mechanically passaged after 4 minutes of collagenase IV treatment at 37°C (Invitrogen). Unless otherwise stated, the flasks were split at a ratio of 1:2. If the experiments tested more than 2 conditions, multiple flasks were pooled and split at a 1:2 ratio. The cells were seeded onto mitomycin inactivated MEFs (9200 MEFs/cm2). The same batch of MEFs was used for differentiation experiments throughout the study. The day of seeding was designated day 0; the cells were left to attach to the flask for 48 hours and then fed daily with hESC medium plus bFGF (6 ml). On the 10th day, the medium was changed to hESC medium without bFGF. Thereafter, the hESC differentiation medium without bFGF was exchanged every 2 days (5 ml) until days 50–80.
Feeder-Free Differentiation and Single Cell Dissociation
For feeder-free differentiation, the hESCs were passaged as clumps or dissociated into single cells. For single cell seeding, the flasks of hESC were first observed under a dissecting microscope and differentiating colonies removed with a Pasteur pipette. The cells were then incubated with TryPLE select enzyme (Invitrogen) for 10 minutes. Detached single cells were counted and seeded at 35,000 cells per cm2 onto growth factor reduced Matrigel (1:30×, BD Biosciences, Oxford, U.K., http://www.bdbiosciences.com). hESCs were fed daily with mTeSR1 (Stem Cell Technologies, Grenoble, France, http://www.stemcell.com) until confluence, after which the medium was changed to hESC without bFGF differentiation medium for long-term differentiation.
Quantifying RPE Foci Yield
Pigmented foci emerging among other nonpigmented cell types are easily distinguishable by eye. In order to rapidly and reproducibly acquire images of the flasks, we designed a mounting device that attaches to an Epson V7500 flatbed scanner (Epson, Suwa-gun, Japan, http://www.global.epson.com). The mounting device contains a hole fitted to a T25 into which flasks were placed before closing the lid. The lid was designed to eliminate background lighting and capture images with sufficient illumination to distinguish pigmented RPE foci. The flasks and plates were wiped with a lint free tissue before acquiring images at a resolution of 480 dots per inch at multiple points during the differentiation and were processed in ImageJ (NIH, Bethesda, MD, http://www.imagej.nih.gov/ij/). The images were converted to gray scale before applying the “find edges” function to detect and highlight the sharp changes in intensity at which the pigmented foci appeared. The “analyze particles” function was then used on the processed image to detect and count the areas highlighted more than 18 pixels in size. These parameters were validated by comparison with manual counts performed at high magnification on flasks with various levels of pigmentation. The average number of foci per cm2 was determined by dividing the number of foci by the total area.
hESC-RPE Enrichment and Maintenance
Pigmented foci were mechanically isolated from nonpigmented tissue after treating with 1 mg/ml collagenase IV (Invitrogen) for 4 hours at 37°C. Microcresent blades were used to cut nonpigmented tissue away from pigmented foci under a dissection microscope. The foci were then washed in fresh differentiation medium before seeding onto Matrigel-coated plates (BD Biosciences). The foci were left for 2–3 days to adhere, before exchanging medium every 2–3 days (hESC differentiation medium without bFGF).
Immunocytochemistry
The cells were fixed with 4% (wt/vol) paraformaldehyde (Sigma-Aldrich) at room temperature for 20 minutes. The cells were then simultaneously permeabilized and blocked in 0.3% vol/vol Triton-X100, 5% vol/vol goat serum in Dulbecco’s phosphate-buffered saline (DPBS) (all Sigma-Aldrich) for 1 hour at room temperature. The primary antibodies were diluted to the specified concentrations in 1% goat serum, 0.3% Triton-X100, DPBS solution (OTX2; 1:1,000; Millipore, Watford, U.K., http://www.millipore.com), ZO-1 (1:200; Life Technologies), CRALBP (1:300; Invitrogen), MitF (1:50; Santa Cruz Biotechnology, Inc, Santa Cruz, CA, http://www.scbt.com), bestrophin (1:300; Millipore), Chx10 (1:300; Sigma-Aldrich), Pax6 (1:300; Millipore), Oct4 (1:300; Santa Cruz Biotechnology, Inc). The primary antibodies were incubated at room temperature overnight. The next day, the cells were washed 3× in DPBS before incubating for 2 hours with the appropriate Alexa Fluor conjugated secondary antibody (Invitrogen) diluted 1:200 in 2% goat serum in DPBS. For each experiment, control wells containing isotype control antibodies (mouse/rabbit IgG/IgM, 1:200, Abcam, Cambridge, U.K., http://www.abcam.com) or no primary antibody were used to detect the absence of nonspecific staining. The cells were counterstained with 4′,6-diamidino-2-phenylindole (1:1,000; Invitrogen). Fluorescence and bright field images were acquired using a Nikon, Eclipse TE2000-U microscope and analyzed with NIS-element software (Nikon UK Ltd., Surrey, U.K., http://www.europe-nikon.com).
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from cells using the RNeasy Mini Kit and treated with RNase-free DNase I (both from Qiagen, Hilden, Germany, http://www.qiagen.com). The cDNA synthesis reaction was performed using Ambion RETROscript Kit First Strand Synthesis Kit (Ambion, Warrington, U.K., http://www.lifetechnologies.com) from 1 µg of RNA according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (PCR) was performed using an Applied Biosystems 7900HT Fast Real-Time PCR system using the SYBR Green method. Master mixes were made using Power SYBR (Applied Biosystems, Paisley, U.K., http://www.appliedbiosystems.com). Each sample was analyzed in triplicate in a 25-µL reaction performed in a 96-well plate. The data were analyzed in SDS, version 2.2.2 (Applied Biosystems) and exported to DART-PCR (version 1.0) to calculate the theoretical starting fluorescence value, Ro [19], which were then normalized to the geometric mean of the expression of the housekeeping genes POLR2A, MAN1B1, and GNB2L1 (chosen for their stability across the sample groups using the GeNorm algorithm [20]). The quantitative PCR data are expressed as the fold changes relative to normalized expression in control samples from the same differentiation run.
Statistical Analysis
The values for the number of pigmented foci per cm2, fold changes in pigmentation or gene expression are expressed as the mean of three or more biological repeats ± SEM. Significance was assessed by one- or two-way analysis of variance and Student’s t tests. Probabilities less than .05 were considered significant.
Results
Spontaneous Differentiation Efficiency Is Cell Line Dependent
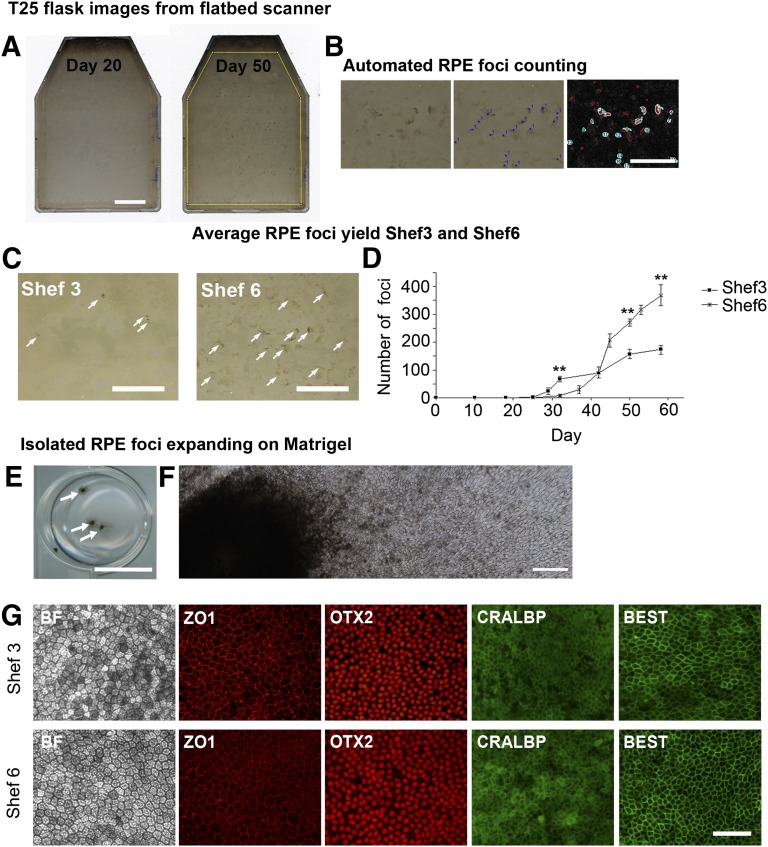
Our laboratory has previously performed RPE differentiation in several lines derived in Sheffield [21] and noted cell line-specific differences in the RPE differentiation capacity. We selected two cell lines, one female, Shef6, which could generate high RPE yields, and one male, Shef3, which produced poor RPE yields after spontaneous differentiation. In order to quantify the relative yields, Shef6 and Shef3 from three separate passages were grown to confluence for 10 days before initiating spontaneous differentiation. After 4–5 weeks of differentiation, pigmented foci were observed against a background of nonpigmented cells. When large enough, these pigmented foci were manually dissected and replated to produce a pure population of RPE cells. In order to determine the rate of differentiation, whole flasks of differentiating Shef6 and Shef3 were placed in our modified scanning device and scanned at fixed points from days 25 to 60 after seeding. Images of the flasks were imported into ImageJ (NIH) and thresholded by pigment intensity and particle size before automated counting (Fig. 1B). Pigmented foci were typically detected in Shef3 by day 28, whereas in Shef6, they were not detected until day 32. The total foci number, and the average size of the foci increased with time in both cell lines. Despite some variability between passages, the rate of foci accumulation per cm2 from days 30–50 was consistently significantly higher in Shef6 (0.5 foci per cm2 per day vs. 0.3 foci per cm2 per day; p < .001; Fig. 1C, 1D). In order to confirm the RPE identity of these pigmented areas, the foci were routinely dissected out of the differentiating flasks and replated onto Matrigel. The pigmented cells proliferated away from the attached foci and formed monolayers of cobblestone epithelium that were immunopositive for the RPE markers Zo1, OTX2, Cralbp, and bestrophin (Fig. 1E, 1F).
Figure 1.
Pigmented foci of RPE begin to appear between days 25 and 32 after human embryonic stem cell seeding. (A): Images of T25 flasks acquired on a flatbed scanner showing the emergence of pigmented foci at day 50. Scale bar = 10 mm. (B): Pigmented areas are manually counted to determine the number of foci (center), with automated highlighting and counting of pigmented areas using a size and pigment intensity threshold (right). Scale bar = 5 mm. (C): Pigmented foci in typical flasks of Shef3 and Shef6 at day 50. Consistently more RPE per cm2 were present in Shef6. Scale bar = 10 mm. (D): The accumulation of individual RPE foci over time in T25 flasks of Shef3 (n = 3) and Shef6 (n = 3) was measured using a flatbed scanner and automated foci counting in ImageJ (NIH). Error bars = SEM of biological replicates. A significant difference was seen in total foci numbers over time and between the two cell lines (p < .001). (E): Manually isolated RPE expanding on a Matrigel-coated 12-well plate. Arrows highlight individual foci. Scale bar = 10 mm. (F): Stitched photomicrographs at 20× magnification depicting RPE cells expanding on Matrigel from a single, manually isolated foci. Scale bar = 500 µm. (G): Immunostaining of pigmented cells derived from Shef3 (upper panel) and Shef6 (lower panel). RPE have typical cobblestone morphology and express RPE markers ZO1, OTX2, CRALBP, and BEST. Scale bar = 100 µm. Abbreviations: BEST, bestrophin; RPE, retinal pigment epithelium.
RPE Yield Is Compromised at High Passage Numbers and at Low Seeding Density
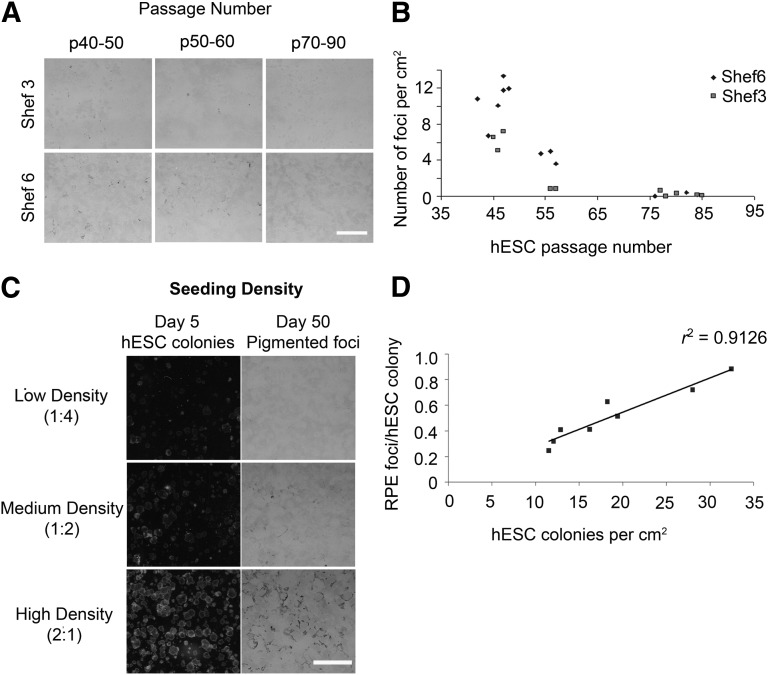
hESCs have been heralded for their capacity to expand indefinitely in culture, making them a theoretically unlimited source of cells for cell therapy. In order to determine whether the age of hESC cultures has a significant effect on their RPE differentiation capacity, Shef6 and Shef3 were cultured up to passage 85. Both cell lines retained good morphology, hESC marker expression, and normal karyology up to this time (data not shown). The cells were differentiated for 50 days, and the number of foci per cm2 was determined from the scanned images. A significant decrease in foci numbers per cm2 was observed with increasing passage number in both Shef6 and Shef3 (p < .01; Fig. 2A, 2B). The cells from the earliest passage tested (p40–50) produced the highest RPE yields in both cell lines, suggesting that low passage cells are a requirement for efficient large scale RPE production.
Figure 2.
Passage number and hESC seeding density are critical factors in ensuring efficient RPE differentiation. (A): Pigmented foci in Shef3 and Shef6 flasks at day 50 for a range of passages showing the decline in average RPE foci numbers with increasing passage number. Scale bar = 5 mm. (B): Scatter graph showing foci number per cm2 versus passage number. The passage number had a significant effect on the pigmented foci number per cm2, with lower passage cells producing significantly greater RPE yields in both cell lines (p < .05). (C): RPE yield is highly dependent on the seeding density. (Left): hESC colony density at day 5. Flask split ratios shown in brackets. (Right): RPE foci day at 50. (D): Scatter plot depicting the number of RPE foci derived per hESC colony versus the total number of hESC colonies seeded, demonstrating improved differentiation efficiency with increasing hESC seeding density (r2 = 0.912). Abbreviations: hESC, human embryonic stem cell; RPE, retinal pigment epithelium.
The seeding density is another major source of variability in hESC culture and is rarely controlled for when cells are manually passaged into clumps of undefined size. In order to determine the relationship between seeding density and RPE differentiation efficiency, pluripotent Shef6 hESC colonies within a small range of passage numbers (passages 44–47) were mechanically passaged as clumps and seeded at different split ratios. At day 5, the numbers of expanding hESC colonies were counted from the scanned images. At day 10, the medium was changed to hESC differentiation medium without bFGF, and the cells were left to differentiate until day 50 (Fig. 2C). The numbers of RPE foci produced per flask were then counted from the scanned images and used to determine the average number of RPE foci per hESC colony seeded (Fig. 2D). A close correlation between the initial seeding density and the RPE foci per colony was observed in Shef6, with the efficiency of differentiation improving with increasing seeding density. The highest density cultures exhibited the highest conversion rates, with a maximum of 0.88 foci produced per hESC colony. The cultures were all fed with the same volume of medium throughout the differentiation; thus, high density culturing is more efficient in terms of both hESC and medium usage.
Single Cell Dissociation and Feeder-Free Expansion Enhances Differentiation Yield
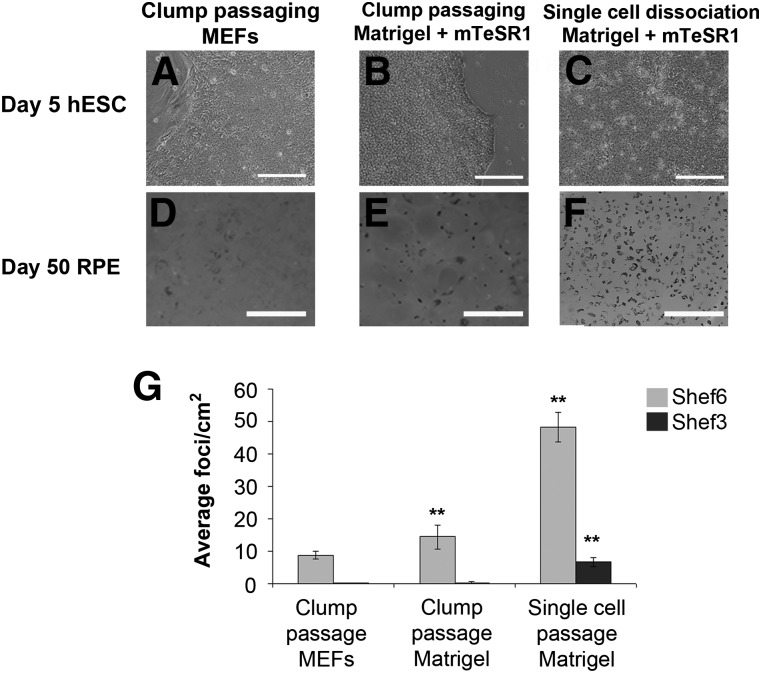
Despite the importance of hESC density on RPE differentiation, it is challenging to routinely quantify and control the number and size of mechanically passaged cell clumps. Recent reports have shown that RPE cells can be generated in feeder-free conditions from clonally propagated hESCs [22]. Such conditions are more suitable for automation and large scale RPE production. We sought to determine the relative differentiation efficiency in enzymatically dissociated hESCs seeded as single cells. Shef6 were passaged by traditional clump passaging methods and differentiated on either MEFs on Matrigel (BD Biosciences) in mTeSR1 or on Matrigel in mTeSR1 after dissociation into single cells (Fig. 3A–C). Dissociated hESCs seeded at a range of relatively high densities of 17,000–100,000 cells per cm2 formed confluent monolayers of pluripotent cells within 5 days in the absence of Rho-associated protein kinase (ROCK) inhibitor. At densities less than 10,000 cells per cm2, however, cell survival was poor, and the cells did not reach confluence before differentiating. In high-seeding density wells, once confluent, mTeSR1 medium was replaced with hESC differentiation medium without bFGF, and pigmented foci emerged within the normal period observed in clump-passaged cells (Fig. 3D–3F). The average RPE yield from single cell dissociated Shef6-hESC was significantly higher at 48.3 ± 4.6 foci per cm2 (n = 6, p < .01; Fig. 3F, 3G) compared with traditionally clump-passaged cells. A similar effect was observed in high passage Shef3 cells, although the RPE yields were significantly lower than Shef6 in all conditions (Fig. 3G). These results suggest that single cell dissociation is conducive to more efficient RPE differentiation, irrespective of innate RPE differentiation capacity.
Figure 3.
Single cell dissociation and feeder free expansion improves differentiation yield. Photomicrographs showing typical hESC colony morphology when cell colonies are passaged as clumps onto MEFs (A), as clumps onto Matrigel (B), and after single cell dissociation onto Matrigel (C). Scale bar = 200 µm. (D–F): Cultures at day 50 after differentiation in the three conditions, illustrating the increase in pigmented foci yield after single cell dissociation of hESC before seeding. Scale bar = 1 cm. (G): Quantification of average RPE foci numbers per cm2 after clump passaging onto MEFs (Shef6, n = 9, Shef3, n = 7), clump passaging onto Matrigel (Shef6 n = 4, Shef3 n = 3), and single cell passaging onto Matrigel (Shef6 n = 6, Shef3 n = 6). Error bars = SEM of the indicated number of biological repeats (∗∗, p < .01). Abbreviations: hESC, human embryonic stem cell; MEFs, mouse embryo fibroblasts; RPE, retinal pigment epithelium.
Seeding Density Optimization Produces a Fivefold Increase in RPE Yield
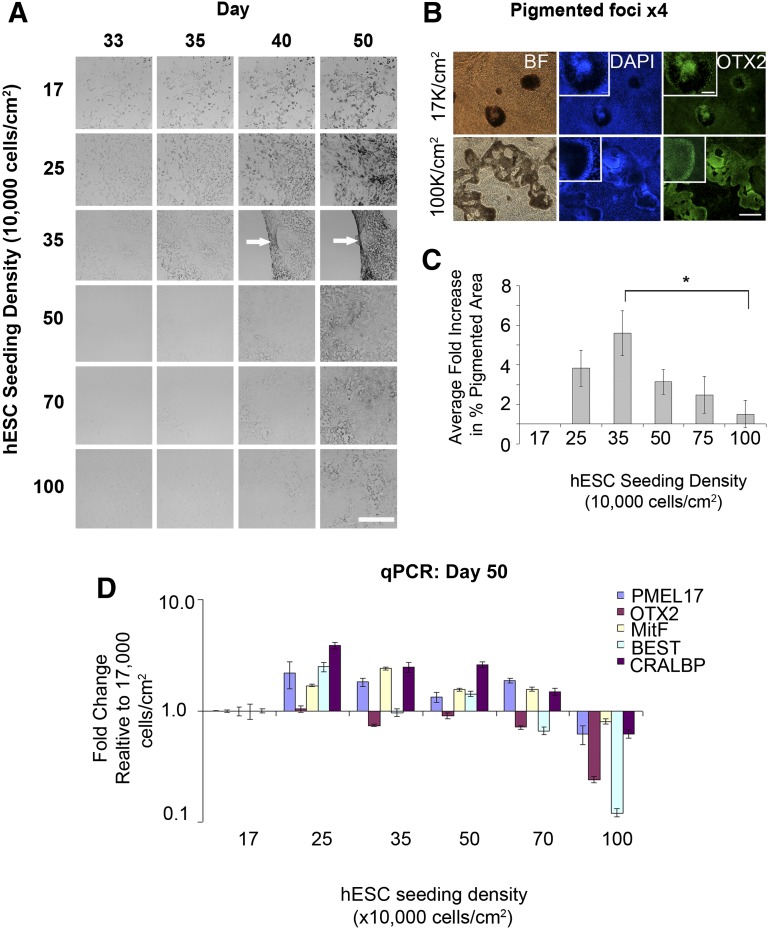
Having established that cells were able to generate higher RPE yields after single cell dissociation, we sought to determine the optimal density. The cells were seeded at densities from 17,000 to 100,000 per cm2, and the resultant pigmented area per well was quantified from days 33 to 50 in 4–6 separate experimental runs in Shef6. In Shef3, the RPE yields were too low to detect a density mediated effect. In high-RPE yielding Shef6, however, the pigmented surface area at day 50 was highly dependent on initial seeding density (p < .05; Fig. 4A). The morphology of emerging OTX2 positive pigmented areas also varied with the density, with discrete, tightly packed foci emerging at low density and continuous, less densely pigmented monolayers emerging at high densities (Fig. 4B). On average, a fivefold increase in the pigmented surface area was observed at 35,000 relative to 17,000 cells per cm2 (Fig. 4C). This was accompanied by a slight increase in expression of RPE genes CRALBP, bestrophin, MitF, and PMEL17 at 25,000–35,000 cells per cm2 (Fig. 4D). At seeding densities greater than 35,000 cells per cm2, RPE differentiation was inhibited or delayed, with only lightly pigmented areas visible at day 50 and a reduced relative expression of RPE genes. These results suggest that the mechanisms of in vitro RPE differentiation is highly dependent on cell density and that optimization of this variable can further enhance the process efficiency in high yielding cell lines.
Figure 4.
Optimal seeding density for retinal pigment epithelium (RPE) differentiation. (A): Images of differentiating wells during a course of 33–50 days, demonstrating differences in the timing of pigmentation at different seeding densities. The greatest yield was typically achieved at a seeding density of 35,000 cells per cm2. (B): Immunocytochemistry for OTX2 in wells seeded at 17,000 hESCs per cm2 and 100,000 cells per cm2. In the very high density wells, the OTX2 positive pigmented areas are spread out. In contrast, low density wells appear as discrete OTX2 positive pigmented foci. Scale bars = 500 µm and 100 µm, inset. (C): Bar graph showing the average fold change in the percentage of pigmented surface area at day 50 across a range of hESC seeding densities over several passages (n = 4–6; ∗, p < .05). (D): qPCR showing the normalized expression of RPE markers at day 50 in the differentiating wells shown in (A). Data show the average normalized expression in triplicate reactions relative to the lowest seeding density (17,000 cells per cm2). Abbreviations: BEST, bestrophin; BF, bright field; DAPI, 4′,6-diamidino-2-phenylindole; hESCs, human embryonic stem cells; qPCR, quantitative polymerase chain reaction.
Low RPE Yields Can Be Rescued by Early Neuroectoderm Induction With Small Molecule Bone Morphogenetic Protein Inhibitor, Dorsomorphin
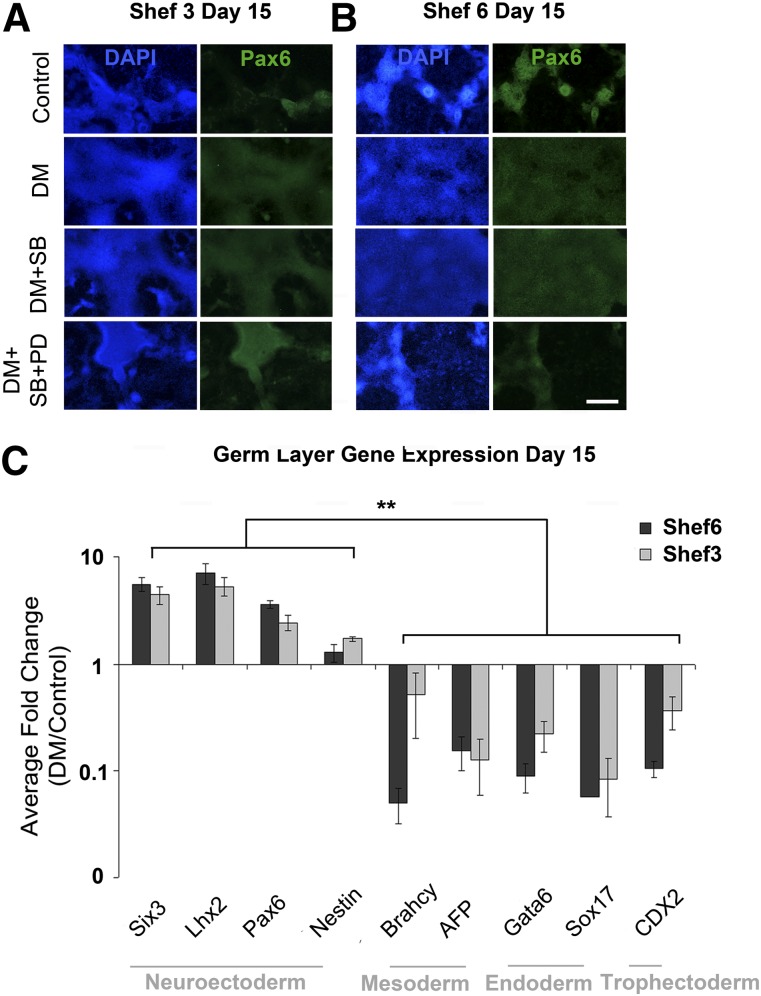
Spontaneous differentiation is likely to yield a range of cell types, the relative ratios of which could depend on a cell line’s innate propensity for differentiation. The Shef3 cell line generates a particularly low RPE yield, perhaps owing to early differentiation lineage preferences. We sought to improve the RPE yield in Shef3 by directing differentiation with small molecules. Three combinations of small molecules known to interfere with bone morphogenetic protein, activin/nodal, and FGF signaling in hESCs were added to medium for the first 10 days of differentiation. Dorsomorphin has been shown to induce neuroectodermal differentiation in a number of hESC and induced pluripotent stem cell lines via a mechanism of dual SMAD inhibition [23]. Addition of dorsomorphin generated a contiguous monolayer of Pax6 positive cells in Shef6 and Shef3 (Fig. 5A, 5B). In contrast, the dimethyl sulfoxide control wells contained only isolated clumps of Pax6 positive cells at the same point. The addition of activin/nodal inhibitor SB431542 produced a similar result but the addition of FGF/ERK inhibitor PD0325901 reduced the total area of Pax6 positive cells. The expression of genes involved in specification of the various germ layers was measured after dorsomorphin treatment, over three passages in both cell lines (Fig. 5C). Dorsomorphin treatment consistently upregulated the neuroectodermal genes Pax6, Six3, Lhx2, and Nestin and significantly downregulated the genes involved in mesoderm, endoderm, and trophectoderm differentiation in both Shef6 and Shef3 (n = 4, Fig. 5C). Thus, we deduced that 10 days of dorsomorphin treatment was sufficient to direct neuroectoderm differentiation and inhibit differentiation into other germ layers.
Figure 5.
Early treatment with small molecules selectively enhance neuroectoderm differentiation. Immunocytochemistry for Pax6 in Shef3 (A) and Shef6 (B) after 10 days of differentiation in the presence of small molecules. Scale bar = 500 µm. (C): qPCR showing the normalized average relative fold change in germ layer gene expression after 10 days of differentiation in the presence of dorsomorphin relative to the same cells in control conditions in four separate passages. Neuroectoderm genes are consistently upregulated after DM treatment in both Shef6 and Shef3. In contrast, markers of other germ layers are significantly downregulated. ∗∗, p ≤ .01 (n = 4). Error bars = SEM. Abbreviations: Control, dimethyl sulfoxide carrier only; DAPI, 4′,6-diamidino-2-phenylindole; DM, bone morphogenetic protein/activin inhibitor dorsomorphin; DM+SB, dorsomorphin plus activin/nodal inhibitor SB431542; DM+SB+PD, dorsomorphin plus SB431542 plus FGF/ERK inhibitor PD0325901.
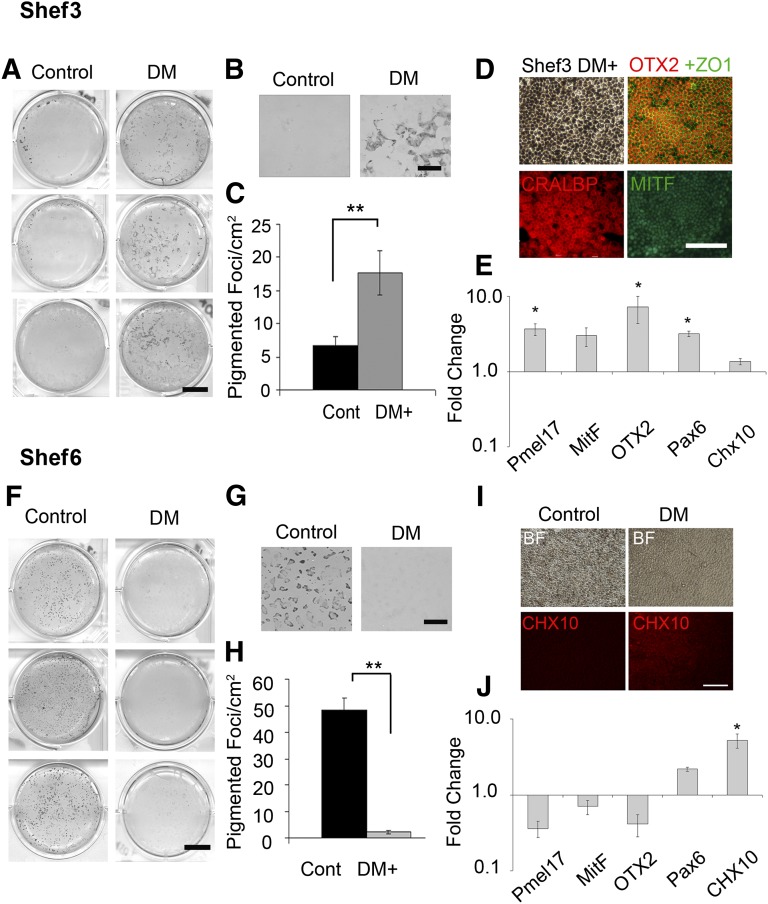
Shef6 and Shef3 cells were differentiated for an additional 35 days after dorsomorphin treatment, and the relative RPE yield was compared with untreated controls. In control conditions, Shef3 produced few pigmented foci, with pigmentation appearing at the edges of the wells only (Fig. 6A); in the dorsomorphin treated wells, however, pigmented foci were abundant (Fig. 6A, 6B). On average, a threefold increase was seen in the numbers of pigmented foci per cm2 over a range of passages (p < .01, n = 6; Fig. 6C), which was accompanied by a significant increase in RPE gene expression (n = 3, Fig. 6E). Pigmented foci isolated from dorsomorphin treated Shef3 expanded to form pigmented, cobblestone, epithelial monolayers that were immunopositive for RPE markers (Fig. 6D).
Figure 6.
Early dorsomorphin treatment augments retinal pigment epithelium (RPE) differentiation in Shef3 but suppresses it in Shef6 in favor of nonpigmented Chx10 positive neural retinal progenitors. (A): Scanned images of wells of Shef3 at day 50 showing the significant increase in pigmented cell yield in response to dorsomorphin treatment across a range of passages. Scale bar = 10 mm. (B): Magnification ×5. Scale bar = 2 mm. (C): The average number of pigmented foci per cm2 was significantly higher in dorsomorphin treated wells (17.7 ± 3.3 foci per cm2) than in control wells (6.7 ± 1.3 foci per cm2; ∗∗, p < .01, n = 6). Error bars = SEM. (D): Foci isolated from dorsomorphin treated wells have typical RPE morphology and are immunopositive for RPE markers OTX2, ZO1, CRALBP, and MitF. (E): Quantitative polymerase chain reaction (qPCR) at day 50 in four separate experiments shows the average upregulation in RPE associated genes pmel17, mitF, otx2 in dorsomorphin treated wells relative to controls (n = 4). ∗, p < .05). Neural retina progenitor marker Chx10 was not significantly upregulated in Shef3. (F–H): Pigmentation is abundant in Shef6 control wells but sparse or absent in the dorsomorphin treated group (n = 6). ∗∗, p < .01. (I): At day 50, RPE morphology was observed in control wells but not in dorsomorphin treated wells, in which a Chx10 positive nonpigmented cell type is abundant. Scale bar = 100 µm. (J): qPCR at day 50 over four passages revealed a consistent downregulation in RPE genes pmel17, otx2, mitF in dorsomorphin treated wells and an upregulation in early neural retina markers chx10 and pax6. One-way analysis of variance revealed a significant difference in the dorsomorphin induced expression of RPE genes between cell lines (n = 4, p < .01). Abbreviations: BF, bright field; Cont, control; DM, dorsomorphin.
In feeder-free, density optimized conditions, Shef6 produced high RPE yields by day 50 (Fig. 6F, 6G); on average, 48 ± 4 foci per cm2 across a range of passages (n = 6). The effect of dorsomorphin pretreatment was tested on Shef6 at the same seeding density as that for Shef3. In contrast to Shef3, Shef6 cells pretreated with dorsomorphin had a highly significant reduction in pigmentation at day 50 (p < .01, n = 6; Fig. 6H), with emergent cells adopting a neural progenitor morphology in place of the pigmented cells seen in the controls (Fig. 6G, 6I). Quantitative PCR confirmed a corresponding downregulation in RPE genes (Fig. 6J). During development, eye field cells adopt either a neural retina or RPE fate, according to the divergent expression of chx10 and mitF in response to signals emanating from adjacent tissues. In dorsomorphin treated wells, emergent cells were immunopositive for Chx10 (Fig. 6I), suggesting that the dorsomorphin induction step pushed cells toward an early neural retina lineage instead of toward RPE. Activin A has been shown to pattern multipotent eye field cells toward an RPE lineage [12, 13, 24]. The addition of 100 ng/ml activin A from days 20 to 40, as described previously [12, 13], did not restore pigmentation in the dorsomorphin induced cells (n = 3, data not shown), suggesting that additional chemical cues or more prolonged treatment is required to induce the RPE lineage in favor of Chx10 neural progenitor cells after dorsomorphin treatment.
Discussion
In this study, hESC-RPE differentiation was performed in two hESC lines over a wide range of passage numbers in the hope of developing a robust and efficient hESC-RPE differentiation protocol. RPE cells have been generated from many hESC lines, since the discovery that hESCs undergo spontaneous differentiation toward the anterior neuroectoderm and eye field lineages. A range of directed and nondirected differentiation methods have been tested [13], and, despite the many successes in generating functionally mature RPE cells, the process of converting cells from pluripotent hESCs to mature and functional RPE cells is lengthy, unpredictable, and inefficient. Some improvements in differentiation efficiency by the addition of small molecules and growth factors have been demonstrated in recent years, but many of these have proved limited in their efficacy across cell lines and among laboratories with diverse cell culturing practices [11, 14, 17, 18].
For this study, a simple method for nondestructive, semiautomated measurement of the hESC-RPE differentiation rates was created using a flatbed scanner and ImageJ (NIH). The combination of the flatbed scanner and ImageJ (NIH) is an inexpensive and flexible method of obtaining a macro analysis of large cell culture areas. Similar methods have been used previously in the determination of mammalian cell colony numbers [25–27]. The scanner quickly imaged the culture vessels, allowing quantification of the emerging pigmented RPE clusters. This is important because the reduction in temperature resulting from the prolonged period out of the incubator (such as is required for manual counting methods) has been shown to cause a decrease in medium pH, affecting both cell proliferation and differentiation [28]. We observed a significant difference between the two cell lines in the rate of RPE foci emergence and endpoint yield. For scaled up hESC-RPE manufacture, it will be important to determine the rate of differentiation for individual cell lines, such that the optimal time for harvesting RPE cells can be established.
Subtle changes in parameters such as the growth rate and differentiation capacity after prolonged culture have been reported in hESC cultures [29, 30], most likely reflecting the selective pressure applied as a result of serial passaging, in which the “best” hESC colonies are selected for propagation. The effect of progressive culture adaptation on differentiation capacity will be an important consideration for large-scale hESC-RPE manufacturing. We tested the effect of prolonged culture by keeping the differentiation conditions constant and measuring the RPE yield at day 50 using standardized image-capturing methods. A highly significant reduction in RPE differentiation efficiency over time in culture was observed and quantified in our experiments. Although the passage number is not a robust method for quantifying the numbers of population doublings, and the rate of culture adaptation is likely to depend on the individual cell culturing techniques used, these results, replicated in two cell lines, have clearly demonstrated the potent effect of the time in culture on the differentiation efficiency. For research purposes, this result highlights the importance of controlling for the passage number when comparing cell lines or protocols. For industrial purposes, this result demonstrates the need to use low passage hESCs for RPE production and the importance of carefully monitoring the process of culture adaptation over the time scale of manufacturing. In this study, we were not able to obtain cells at lower passage numbers in sufficient quantities to test; thus, whether these would generate higher or lower yields remains an open question.
In order to design a cost-effective RPE manufacturing process, it will be necessary to define the number of RPE cells that can be generated per quantity of hESC, per volume of raw material within a given time [31]. By comparing the hESC colony numbers per flask to the final RPE foci numbers per flask, we were able to determine typical conversion efficiencies. An optimal hESC-RPE manufacturing process should generate the largest number of RPE foci, using the smallest amount of laboratory space, time, manual labor, and raw materials. Our data showed that in a relatively high density environment (∼30 colonies per cm2), hESCs generate more pigmented foci per HESC colony seeded within a given time, using the same volume of medium and incubator space.
Passaging pluripotent hESCs has traditionally involved the constant observation and selection of colonies with appropriate size and morphology. The “good” colonies are then microdissected into smaller pieces before seeding at an undefined density. This technique is widely used but is not practical for scaled-up or automated manufacturing [32]. In this study, we found that survival, proliferation, and differentiation of the two hESC lines into RPE cells after single cell dissociation is possible after a gentle enzymatic treatment. RPE differentiation after single cell dissociation has recently been described by others [22]; in this study the cells were seeded at clonal densities (1,000 cells per cm2) and required the addition of blebbistatin to enable cell survival. We observed that at a density of 17,000 cells per cm2, the cells survived and proliferated to form pluripotent, confluent monolayers without the addition of chemical inhibitors. In addition to the effect on processing efficiency, evidence has suggested that seeding density has an effect on cell fate decisions during early differentiation [33–35]. We observed a trend for an increasing RPE yield up to a peak at 35,000 hESCs per cm2, after which the yields were reduced. Although variability was present in the extent of pigmentation between experimental runs, the relative fold change between wells and the optimal seeding density was consistent. In earlier experiments, we did not observe significant effects of seeding density on early (day 15) Pax6 expression, suggesting that the density-mediated effects on RPE differentiation are important later in the differentiation process. These results demonstrate the significant room for enhancing the target cell yield without the addition of exogenous factors.
Having established a feeder-free, density-controlled differentiation protocol, we sought to test the efficacy of small molecules in further directing the spontaneous differentiation toward the eye field and RPE lineages. We hypothesized that the reduced yields observed in the Shef3 cell line could be improved by prompting early neuroectodermal conversion with small molecules. Small molecules, which interfere with key signaling pathways, are powerful tools in stem cell research. Compared with growth factors, they are less vulnerable to degradation in culture, are significantly less expensive, and have less batch to batch variability; properties that make them excellent tools for industrial application [36]. Several groups have reported enhanced neuroectoderm conversion of pluripotent cell lines using dorsomorphin [23, 37, 38]. In Shef6 and Shef3, dorsomorphin selectively upregulated neuroectoderm gene expression and simultaneously suppressed mesoderm, endoderm, and trophectoderm expression. However, diverse effects were found on RPE yield, with Shef6 failing to pigment and generating, instead, a Chx10-positive, early neural retinal phenotype. These two cell types emerge from the same common progenitor during human development [39], and Chx10-positive cells, emerging alongside pigmented RPE cells, have been observed in several studies of hESC differentiation [6, 13, 14, 40]. During human development, multipotent cells in the optic vesicle differentiate into RPE or neural retina, dependent on their relative exposure to the growth factors emanating from the extraocular mesenchyme or surface ectoderm, respectively [24, 39]. We hypothesize that the relative ratios of early Pax6-positive neuroectodermal cells to other cell types is key to the RPE versus neural retina decision. It is clear from this study that different cell lines will have diverse responses to the same small molecule, making it unlikely that one strategy could be applied to all cell lines in a cell therapy manufacturing context. The preferential differentiation of some hESC lines into particular germ layers could restrict their cell therapy applications. Just as seen in this study, small molecules can be used to direct otherwise recalcitrant lines to differentiate into the desired lineage; it would be preferable, from a cell therapy manufacturing perspective, if all cell lines exhibited similar behaviors. Recent developments in hESC culture technology have enabled the conversion of epiblast-like hESCs to a more stable “ground state” [41]. Directed or spontaneous differentiation from ground state hESCs might have more reliable and predictable results across cell lines and be less vulnerable to the differentiation inhibiting effects of culture adaptation observed in this study. Because of the time required to perform these differentiation experiments, it would be beneficial to develop an early stage assay that could predict the eventual RPE yield such that the most appropriate cell line-specific long-term differentiation strategy could be applied.
Conclusion
An understanding of the key variables that govern the efficiency of hESC-RPE differentiation will be critical to the clinical and commercial success of hESC-RPE based therapies. We have demonstrated how a small number of process parameters, including cell line, passage number, and initial cell seeding density, have a significant effect on the RPE yield. Using single cell dissociation and feeder-free differentiation, we were able to optimize the RPE yields after spontaneous differentiation, leading to a more than fourfold increase in differentiation efficiency. Using small molecules to direct early differentiation, we demonstrated a robust increase in RPE yield in an otherwise low yielding cell line but caution that the efficacy of such directed differentiation strategies are likely to be cell line specific. These findings should facilitate the development of more robust hESC-RPE manufacturing processes and highlight areas in which additional optimization is likely to be fruitful.
Acknowledgments
This work was supported by the Engineering and Physical Sciences Research Council and the Rosetrees Trust. We thank Martin Town in the UCL Biochemical Engineering Workshop for his help in designing and making the scanning device.
Author Contributions
A.L.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; L.R.P.: collection and/or assembly of data; L.R. and C.M.: conception and design; K.F. and M.S.: data analysis and interpretation; A.C.: data analysis and interpretation, final approval of manuscript; P.C.: conception and design, data analysis and interpretation, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Ramsden CM, Powner MB, Carr A-JF, et al. Stem cells in retinal regeneration: Past, present and future. Development. 2013;140:2576–2585. doi: 10.1242/dev.092270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.da Cruz L, Chen FK, Ahmado A, et al. RPE transplantation and its role in retinal disease. Prog Retin Eye Res. 2007;26:598–635. doi: 10.1016/j.preteyeres.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Owen CG, Jarrar Z, Wormald R, et al. The estimated prevalence and incidence of late stage age related macular degeneration in the UK. Br J Ophthalmol. 2012;96:752–756. doi: 10.1136/bjophthalmol-2011-301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman DS, O’Colmain BJ, Muñoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 5.Osakada F, Ikeda H, Mandai M, et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26:215–224. doi: 10.1038/nbt1384. [DOI] [PubMed] [Google Scholar]
- 6.Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 8.Klimanskaya I, Hipp J, Rezai KA, et al. Derivation and comparative assessment of retinal stem cells from human embryonic stem cells using transcriptomics. Cloning Stem Cells. 2004;6:217–245. doi: 10.1089/clo.2004.6.217. [DOI] [PubMed] [Google Scholar]
- 9.Vugler A, Carr A-J, Lawrence J, et al. Elucidating the phenomenon of HESC-derived RPE: Anatomy of cell genesis, expansion and retinal transplantation. Exp Neurol. 2008;214:347–361. doi: 10.1016/j.expneurol.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Carr AJ, Vugler A, Lawrence J, et al. Molecular characterization and functional analysis of phagocytosis by human embryonic stem cell-derived RPE cells using a novel human retinal assay. Mol Vis. 2009;15:283–295. [PMC free article] [PubMed] [Google Scholar]
- 11.Idelson M, Alper R, Obolensky A, et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Liao JL, Yu J, Huang K, et al. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum Mol Genet. 2010;19:4229–4238. doi: 10.1093/hmg/ddq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowland TJ, Buchholz DE, Clegg DO. Pluripotent human stem cells for the treatment of retinal disease. J Cell Physiol. 2012;227:457–466. doi: 10.1002/jcp.22814. [DOI] [PubMed] [Google Scholar]
- 14.Meyer JS, Howden SE, Wallace KA, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahabi A, Shahbazi E, Ahmadieh H, et al. A new efficient protocol for directed differentiation of retinal pigmented epithelial cells from normal and retinal disease induced pluripotent stem cells. Stem Cells Dev. 2012;21:2262–2272. doi: 10.1089/scd.2011.0599. [DOI] [PubMed] [Google Scholar]
- 16.Buchholz DE, Pennington BO, Croze RH, et al. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Translational Medicine. 2013;2:384–393. doi: 10.5966/sctm.2012-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Carido M, Meinhardt A, et al. Three-dimensional neuroepithelial culture from human embryonic stem cells and its use for quantitative conversion to retinal pigment epithelium. PLoS One. 2013;8:e54552. doi: 10.1371/journal.pone.0054552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowland TJ, Blaschke AJ, Buchholz DE, et al. Differentiation of human pluripotent stem cells to retinal pigmented epithelium in defined conditions using purified extracellular matrix proteins. J Tissue Eng Regen Med. 2013;7:642–645. doi: 10.1002/term.1458. [DOI] [PubMed] [Google Scholar]
- 19.Peirson SN, Butler JN, Foster RG. Nucleic Acid Res. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. . 2003;31:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:34.1–34.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aflatoonian B, Ruban L, Shamsuddin S, et al. Generation of Sheffield (Shef) human embryonic stem cell lines using a microdrop culture system. In Vitro Cell Dev Biol Anim. 2010;46:236–241. doi: 10.1007/s11626-010-9294-2. [DOI] [PubMed] [Google Scholar]
- 22.Maruotti J, Wahlin K, Gorrell D, et al. A simple and scalable process for the differentiation of retinal pigment epithelium from human pluripotent stem cells. Stem Cells Translational Medicine. 2013;2:341–354. doi: 10.5966/sctm.2012-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Su P, Li D, et al. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor-beta superfamily receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuhrmann S, Levine EM, Reh TA. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development. 2000;127:4599–4609. doi: 10.1242/dev.127.21.4599. [DOI] [PubMed] [Google Scholar]
- 25.Cai Z, Chattopadhyay N, Liu WJ, et al. Optimized digital counting colonies of clonogenic assays using ImageJ software and customized macros: Comparison with manual counting. Int J Radiat Biol. 2011;87:1135–1146. doi: 10.3109/09553002.2011.622033. [DOI] [PubMed] [Google Scholar]
- 26.Dahle J, Kakar M, Steen HB, et al. Automated counting of mammalian cell colonies by means of a flat bed scanner and image processing. Cytometry A. 2004;60:182–188. doi: 10.1002/cyto.a.20038. [DOI] [PubMed] [Google Scholar]
- 27.Guzmán C, Bagga M, Kaur A, et al. ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014;9:e92444. doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veraitch FS, Scott R, Wong J-W, et al. The impact of manual processing on the expansion and directed differentiation of embryonic stem cells. Biotechnol Bioeng. 2008;99:1216–1229. doi: 10.1002/bit.21673. [DOI] [PubMed] [Google Scholar]
- 29.Baker DEC, Harrison NJ, Maltby E, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman LM, Carpenter MK. Human embryonic stem cell stability. Stem Cell Rev. 2005;1:139–144. doi: 10.1385/SCR:1:2:139. [DOI] [PubMed] [Google Scholar]
- 31.Kirouac DC, Zandstra PW. The systematic production of cells for cell therapies. Cell Stem Cell. 2008;3:369–381. doi: 10.1016/j.stem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Ellerström C, Strehl R, Noaksson K, et al. Facilitated expansion of human embryonic stem cells by single-cell enzymatic dissociation. Stem Cells. 2007;25:1690–1696. doi: 10.1634/stemcells.2006-0607. [DOI] [PubMed] [Google Scholar]
- 33.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauwens CL, Peerani R, Niebruegge S, et al. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 35.Heng BC, Liu H, Rufaihah AJ, et al. Human embryonic stem cell (hES) colonies display a higher degree of spontaneous differentiation when passaged at lower densities. In Vitro Cell Dev Biol Anim. 2006;42:54–57. doi: 10.1290/0510071.1. [DOI] [PubMed] [Google Scholar]
- 36.Allsopp TE, Bunnage ME, Fish PV. Small molecule modulation of stem cells in regenerative medicine: Recent applications and future direction. MedChemComm. 2010;1:16–29. [Google Scholar]
- 37.Kim D-S, Lee JS, Leem JW, et al. Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev. 2010;6:270–281. doi: 10.1007/s12015-010-9138-1. [DOI] [PubMed] [Google Scholar]
- 38.Boulting GL, Kiskinis E, Croft GF, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Morales JR, Rodrigo I, Bovolenta P. Eye development: A view from the retina pigmented epithelium. BioEssays. 2004;26:766–777. doi: 10.1002/bies.20064. [DOI] [PubMed] [Google Scholar]
- 40.Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]