Summary
Protein machines are multi-subunit protein complexes that orchestrate highly regulated biochemical tasks. An example is the Anaphase-Promoting Complex/Cyclosome (APC/C), a thirteen-subunit ubiquitin ligase that initiates the metaphase-anaphase transition and mitotic exit by targeting proteins such as securin and cyclin B1 for ubiquitin-dependent destruction by the proteasome1,2. Because blocking mitotic exit is an effective approach for inducing tumor cell death3,4, the APC/C represents a potential novel target for cancer therapy. APC/C activation in mitosis requires binding of Cdc205, which forms a co-receptor with the APC/C to recognize substrates containing a Destruction box (D-box)6-14. Here we demonstrate that we can synergistically inhibit APC/C-dependent proteolysis and mitotic exit by simultaneously disrupting two protein-protein interactions within the APC/C-Cdc20-substrate ternary complex. We identified a small molecule, called apcin (APC inhibitor), which binds to Cdc20 and competitively inhibits the ubiquitylation of D-box-containing substrates. Analysis of the crystal structure of the apcin-Cdc20 complex suggests that apcin occupies the D-box-binding pocket on the side face of the WD40-domain. The ability of apcin to block mitotic exit is synergistically amplified by co-addition of tosyl-L-arginine methyl ester (TAME), a small molecule that blocks the APC/C-Cdc20 interaction15,16. This work suggests that simultaneous disruption of multiple, weak protein-protein interactions is an effective approach for inactivating a protein machine.
We identified apcin (Fig. 1a) in an earlier study as an inhibitor of cyclin proteolysis in mitotic Xenopus egg extract17, but its mechanism of action remained unknown. Analysis of the structure-activity relationship (Fig. 1b and Extended Data Fig. 1a) revealed that elimination of one nitrogen in the pyrimidine ring of apcin (apcin-P) reduced activity slightly, but replacement with a morpholino group (apcin-M) eliminated activity. In contrast, elimination of the nitro-imidazole moiety (apcin-A) had little effect. To identify the target of apcin, we coupled apcin-A to beads via its amino group, incubated the beads with mitotic Xenopus extract, and then removed the beads. We found that Cdc20 was depleted from the extract (Fig. 1c), resulting in stabilization of a cyclin B-luciferase reporter protein (Fig. 1d). Cyclin degradation was rescued by adding in vitro-translated Cdc20 (Fig. 1d), implicating Cdc20 as the target of apcin. Cdc20 interacted with apcin-A beads in a manner that could be competed by free apcin (Fig. 1e), but not by the inactive analog apcin-M or the APC/C inhibitor TAME15,16 (Extended Data Fig. 1b). A cyclin B1 N-terminal fragment (cycB1-NT) also competed for Cdc20 binding to the apcin-A resin, but the same fragment with a mutated D-box did not (Fig. 1f). Among a panel of WD40-containing proteins, Cdc20 binding to apcin-A beads was most robust, followed by Cdh1, with much less binding of other WD40-containing proteins observed (Extended Data Fig. 1c). Although we have not tested the ability of apcin to inhibit Cdh1-dependent ubiquitylation, apcin inhibited Cdh1-dependent proteolysis in interphase Xenopus extract less efficiently than Cdc20-dependent proteolysis in mitotic Xenopus extract (Extended Data Fig. 1d). Apcin bound to endogenous Cdc20 in Xenopus extract in a dose-dependent manner (Extended Data Fig. 1e) that correlated with its ability to inhibit formation of high-molecular weight ubiquitin conjugates of cycB1-NT (Fig. 1g) or full-length cyclin B1 (Extended Data Fig. 2a). Kinetic analysis of a reconstituted APC/C-dependent ubiquitylation reaction16 showed that apcin caused a significant increase (P=0.0039) in Km (Ki=23 μM) but no reduction in kcat (Extended Data Fig. 2b). Together these results suggest that apcin competitively inhibits APC/C-dependent ubiquitylation by binding to Cdc20 and preventing substrate recognition.
Figure 1.
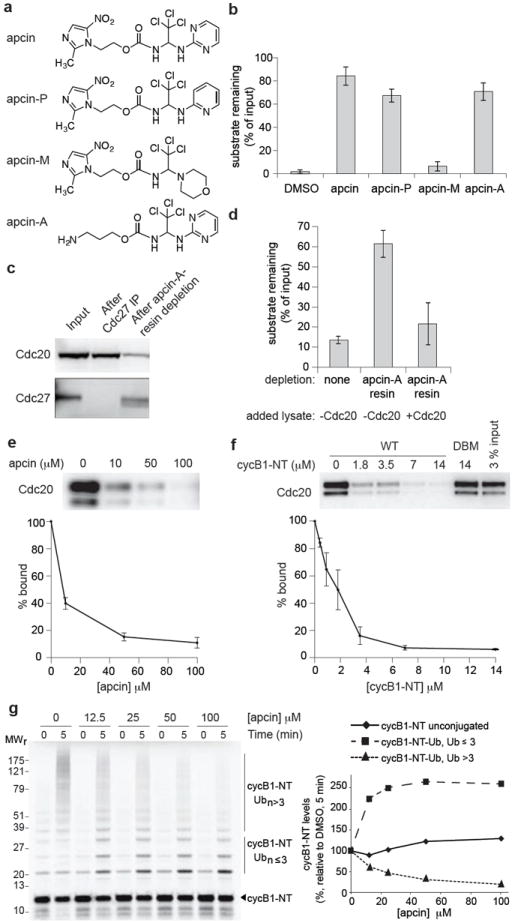
Apcin binds to Cdc20 and inhibits APC/C-dependent ubiquitylation. a, Structures of apcin and derivatives. b, Effects of apcin and derivatives (200 μM) on proteolysis of an N-terminal fragment of cyclin B1 (cycB1-NT) in mitotic Xenopus egg extract. Substrate levels were measured at 40 minutes. N=3. c, Apcin-A resin depletes Cdc20 from mitotic Xenopus egg extract. Cdc20 and Cdc27 levels were measured by western blotting. d, Depletion with Apcin-A resin stabilizes a cyclin-luciferase reporter protein and degradation can be rescued by addition of in vitro-translated Cdc20. Substrate levels were measured at 60 minutes. N=3. e, Cdc20 expressed in reticulocyte lysate binds to apcin-A resin and can be competed by free apcin. Cdc20 was detected by western blotting. N=3. f, Wild-type (WT) CycB1-NT, but not a D-box mutant (DBM, mutation of R42A, L45A), competes with Cdc20 binding to apcin-A resin. Cdc20 was detected by western blotting. N=4. g, Apcin inhibits formation of high-molecular weight ubiquitin conjugates of 35S-labeled cycB1-NT in mitotic Xenopus extract. Bars represent mean +/- standard error of the mean (S.E.M.).
To identify the site on Cdc20 that binds apcin, we soaked apcin into Cdc20 protein crystals and determined the structure of the Cdc20-apcin complex to 2.1Å resolution (Extended Data Table 1). We found that apcin bound a small pocket on the side of the WD40 domain that has been implicated in binding the D-box (Fig. 2a,b and Extended Data Fig. 3a,b)14,18,19. The binding mode of acpin is consistent with the structure-activity relationship, as the pyrimidine ring and aminal nitrogens make hydrogen bonds with backbone atoms from D177. The hydrophobic trichloromethyl group is buried in the pocket occupied by leucine of the D-box (Extended Data Fig. 3b). The nitro-imidazole moiety is positioned facing solvent, explaining why apcin-A retains activity and can be used to isolate Cdc20 when coupled to beads.
Figure 2.
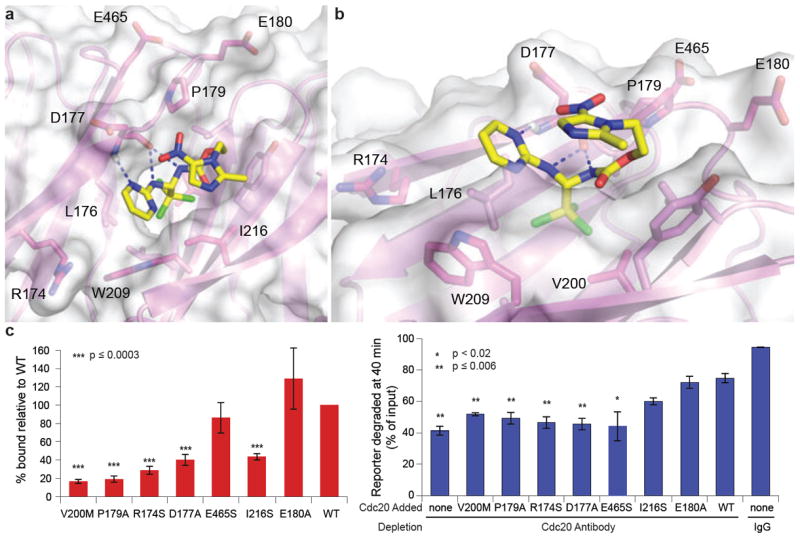
Apcin binds to the D-box binding site of Cdc20. a, Crystal structure of the apcin-Cdc20 complex. Apcin atoms are labeled in yellow (carbon), blue (nitrogen), red (oxygen), and green (chlorine). Cdc20 is shown in magenta. Dotted blue lines indicate hydrogen bonds. b, View is rotated to show the position of V200 at the base of hydrophobic binding pocket. c, Mutation of residues in the binding pocket reduces Cdc20 binding to apcin-A resin (red bars) and the capacity of in vitro-translated Cdc20 protein to rescue cyclin-luciferase degradation in mitotic Xenopus egg extract immunodepleted of Cdc20 (blue bars). Bars represent mean of three experiments +/- S.E.M. P values were calculated by an unpaired t-test in comparison to WT.
We tested whether mutations in the presumptive binding site affect the binding of Cdc20 to apcin-A beads (Fig. 2c and Extended Data Fig. 3c,d). V200 is located at the base of the hydrophobic pocket and mutation to methionine blocked binding of Cdc20 and inactivated the ability of Cdc20 to rescue cyclin degradation in a Cdc20-depleted extract. Mutation of D177, P179 or I216, which line the binding pocket, also reduced binding to apcin and function of Cdc20. Mutation of R174, which lies near the pyrimidine ring of apcin, also reduced apcin binding and blocked Cdc20 rescue activity, consistent with a role of this residue in interacting with negatively charged amino acids in the D-box14. In contrast, mutation of E465, which interacts with the conserved arginine of the D-box14 but lies distant from the apcin-binding pocket, decreased the ability of Cdc20 to rescue degradation, but had little effect on apcin binding. E180 lies further away and mutation to alanine had no effect on apcin binding or rescue activity. Overall, we observed a strong correlation between effects of mutations on apcin binding and their effects on Cdc20 function for residues that line the apcin-binding pocket.
Cdc20 is recruited to the APC/C through multiple weak interactions6,8,20-23. For example, substrates can promote cooperative Cdc20 binding to the APC/C through a co-receptor interaction in which the substrate is simultaneously recognized by Cdc20 and the APC/C6-14. Consistent with this idea, we found that addition of substrate increased Cdc20 loading onto the APC/C in a concentration- and D-box-dependent manner in Xenopus extract (Fig. 3a and Extended Data Fig. 4a,b). Substrate-induced loading of Cdc20 could be blocked by addition of apcin (Fig. 3a and Extended Data Fig. 4a), indicating that the leucine-binding pocket of Cdc20 is critical for co-receptor function. Because the binding of Cdc20 to the APC/C was variable in the absence of added substrate, we were not able to assess the effects of apcin under this condition. Thus we cannot exclude the possibility that apcin might also decrease Cdc20 binding to the APC/C in the absence of substrate. The small molecule TAME, which antagonizes the IR-tail interaction between Cdc20 and the APC/C15,16, also antagonized Cdc20 loading. At high concentrations of substrate, the combined use of apcin and TAME was more effective at blocking Cdc20 binding to the APC/C than either compound used alone, suggesting that simultaneous disruption of multiple interactions between substrate, Cdc20, and APC/C may be an effective strategy for inhibiting APC/C.
Figure 3.
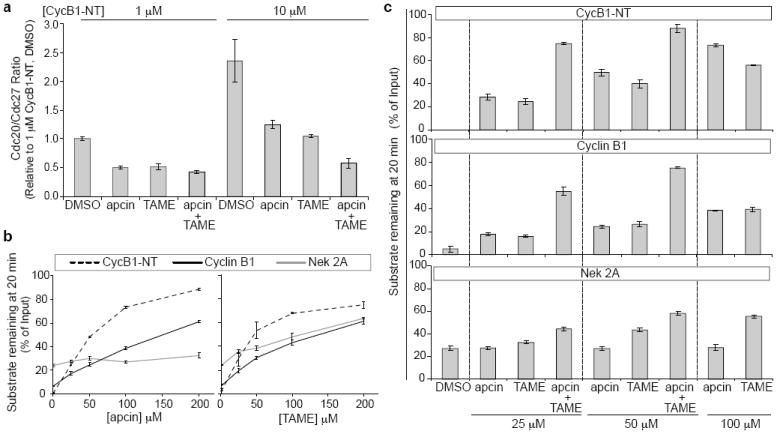
Effects of apcin on Cdc20 binding to APC/C and stability of APC/C substrates in mitotic Xenopus extract. a, Apcin blocks co-receptor-dependent binding of Cdc20 to APC/C. Substrate (cycB1-NT, 1 μM or 10 μM), apcin and/or TAME (50 μM each) were added, and APC/C was isolated with anti-Cdc27 antibody. Levels of Cdc20 were assessed by western blotting and normalized to the levels of Cdc27. b, Effect of apcin and TAME on APC/C substrate stability. Levels of 35S-labeled substrates were assessed by gel electrophoresis and phosphorimaging. c, As in b except that combinations of apcin and TAME were examined. Values and error bars in a, b, c represent mean of three experiments +/- S.E.M.
Because substrates can be recruited to the APC/C through both D-box-dependent and - independent mechanisms, we compared the ability of apcin to stabilize different APC/C substrates in mitotic Xenopus extract. Apcin stabilized cycB1-NT and securin most effectively (Fig. 3b and Extended Data Figs. 4c and 5b), with somewhat weaker effects against full-length cyclin B1 (Fig. 3b and Extended Data Fig. 4c). Interestingly, even high concentrations of apcin failed to stabilize cyclin A2 or Nek2A (Fig. 3b and Extended Data Fig. 4c), suggesting that apcin is highly specific for the D-box binding site. In contrast, TAME inhibited the degradation of all APC/C substrates tested (Fig. 3b and Extended Data Fig. 4c and 5a,b), consistent with TAME’s ability to directly block recruitment of Cdc20 to the APC/C15,16. The ability of apcin to inhibit formation of high-molecular weight substrate-ubiquitin conjugates closely correlated with its ability to stabilize each substrate (Extended Data Fig. 6). These results suggest that the leucine-binding pocket of Cdc20 plays an essential role in recruiting the D-boxes of securin and the cycB1-NT, and that apcin can effectively compete with these interactions. The ability of full-length cyclin B1 and cyclin A2 to bind Cks1 via Cdk1 may facilitate their D-box-independent recruitment to the APC/C24,25, helping these substrates partially overcome the effects of apcin. Furthermore, the N-terminal region of cyclin A2 appears to bind Cdc20 with higher affinity than cyclin B126, which may further reduce the effectiveness of apcin. In contrast to these substrates whose degradation is D-box-dependent, Nek2A is recruited directly to the APC/C via a Cdc20-independent mechanism that requires its MR-tail rather than a D-box27-29, explaining why apcin fails to inhibit ubiquitylation or degradation of this protein.
Because apcin and TAME inhibit APC/C-dependent proteolysis by distinct mechanisms, we tested the effect of combining the inhibitors on the proteolysis of APC/C substrates in Xenopus extract. The combination of apcin and TAME led to synergistic stabilization of cyclin B1, cycB1-NT, securin, and cyclin A2, with a much weaker effect for Nek2A (Fig. 3c and Extended Data Figs. 4d and 5a,b). For example, combining TAME and apcin at 25 μM each was more effective at stabilizing cyclin B1 than using either compound alone at 100 μM. Apcin slightly enhanced the ability of TAME to stabilize Nek2A (Fig. 3c), suggesting that the leucine-binding pocket of Cdc20 may bind Nek2A, even though this interaction is not essential for proteolysis if the APC/C is not otherwise perturbed. We next examined the effect of apcin, proTAME15 (a cell-permeable TAME prodrug), and the combination, on mitotic exit in four different human cell lines. Apcin and proTAME synergized to increase the mitotic fraction in all cell lines examined (Fig. 4a and Extended Data Fig. 7a,b). Apcin-M was inactive whereas apcin-P retained activity (Extended Data Fig. 7c), consistent with effects on Cdc20 binding and APC/C-dependent proteolysis in Xenopus extract. In live-cell imaging experiments in RPE1 cells (Fig. 4b, Extended Data Figs. 8a-d and Extended Data Videos 1 and 2), 25 μM apcin had no detectable effect on mitotic duration (P= 0.279) unless mitosis was artificially shortened by depletion of the spindle assembly checkpoint (SAC) protein Mad2 (P= 0.0001). In contrast, in the presence of proTAME, the addition of apcin dramatically slowed the rate of mitotic exit in a synergistic manner (Fig. 4b and Extended Data Fig. 8a-d): the rate of mitotic exit was 63% of that predicted by a multiplicative combination of the single compound effects (P= 0.016). Significant synergy was also observed in U2OS cells (P=2.0×10-8; Extended Data Fig. 8e). Addition of the inactive derivative apcin-M had no effect on the rate of mitotic exit in the presence of proTAME (P=0.68; Fig. 4b and Extended Data Fig. 8a-e). The response of cells to proTAME alone was biphasic, because prolongation of metaphase can cause cohesion fatigue30, which reactivates the SAC to block mitotic exit in a subpopulation of cells. Notably, the addition of apcin eliminated the biphasic response. Furthermore, the combined effect of apcin and proTAME was largely preserved when the SAC was inactivated by Mad2 depletion. When modeled quantitatively, the degree of synergy between apcin and proTAME was in fact enhanced in the absence of Mad2, as the rate of mitotic exit was reduced to 38% of the rate predicted by a multiplicative combination of the single compound effects (P= 8.92×10-5; Extended Data Fig. 8d). Together these findings suggest that synergistic inhibition of mitotic exit does not rely on the SAC, but instead likely arises from direct pharmacologic APC/C inhibition.
Figure 4.
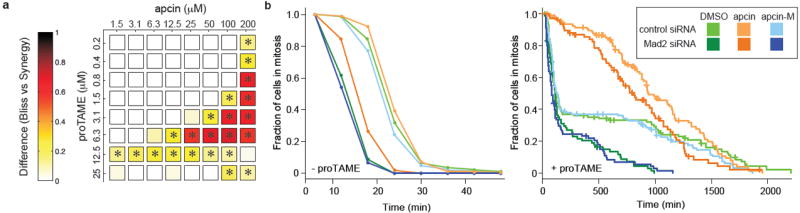
Apcin synergizes with proTAME to prolong mitotic duration. a, RPE1 cells were treated with indicated concentrations of apcin and proTAME for 18 hours, fixed, and mitotic index determined by automated high-throughput imaging. The panel shows the difference between the mitotic indices calculated by a Bliss-independence model compared to a synergy model; any positive value indicates synergy. * P<0.05 based on analysis of four technical replicates. b, Asynchronous RPE1 cells expressing H2B-GFP were transfected with siRNA and twenty-four hours later treated with apcin or apcin-M (25 μM) and/or proTAME (6 μM). Cells were then imaged every 6 minutes for 45 hours. Mitotic duration and cell fate (Extended Data Fig. 8b) were determined by manual inspection of the videos and plotted as inverse cumulative frequency (-proTAME) or Kaplan-Meier curves (+proTAME). The hatch marks on the Kaplan-Meier curves indicate censored cells that did not exit mitosis or die in mitosis before the end of the movie or before they migrated out of the field of view. Graphs include the combined results of two independent experiments.
The discovery of apcin and the elucidation of its mechanism validate the importance of the leucine-binding pocket in Cdc20 as a D-box co-receptor. Apcin preferentially stabilizes APC/C substrates whose degradation is D-box dependent. However, on its own, low concentrations of apcin do not effectively block mitotic exit, either because substrates can outcompete apcin binding to the leucine-binding pocket, or because substrates can be recruited to the APC/C through other mechanisms. The effectiveness of apcin can be dramatically enhanced by the addition of TAME, which blocks Cdc20 loading through a distinct mechanism, highlighting the importance of multiple weak protein-protein interactions in promoting activator binding and efficient substrate ubiquitylation (Extended Data Fig. 9). Our work highlights the possibility of disrupting the function of a protein machine by simultaneously inhibiting multiple protein-protein interactions. Because dynamic protein complexes regulate virtually all aspects of cell biology, simultaneous targeting of multiple weak interactions may represent a new opportunity for therapeutic targeting of protein complexes that may otherwise be difficult to inhibit with a single compound.
Methods
Reagents
Commercial antibodies used for western analysis were as follows: anti-Cdc27 (610455, BD Transduction Laboratories), anti-Cdc20 (BA8; sc-93399, Santa Cruz Biotechnology and NB 100-2646, Novus Biologicals) to recognize Xenopus Cdc20, anti-Cdc20 (H-175; sc-8358, Santa Cruz Biotechnology) to recognize human Cdc20, and anti-HA-peroxidase (3F10; 12013819001, Roche). Secondary antibodies used included anti-rabbit IgG-HRP (NA934; GE Healthcare), and anti-mouse IgG-HRP (NA931; GE Healthcare). For APC/C immunopurification from Xenopus extract, anti-Cdc27 (AF3.1; sc-9972, Santa Cruz Biotechnology) was used. For immunodepletion of Xenopus Cdc20, a rabbit polyclonal antibody was generated by Yenzym (South San Francisco, CA) by immunization with an N-terminal fragment of Cdc20 (residues 1-170; tagged at C-terminus). Chemicals used were cycloheximide (239764, Calbiochem), calcium ionophore A23187, free acid form (100105, Calbiochem), tosyl-L-arginine methyl ester (T4626, Sigma), proTAME (I-440, Boston Biochem), MG262 (I-120, Boston Biochem), apcin (T0506-3874, Enamine), apcin-P (Amb2237944, Ambinter), apcin-M (Amb1395012, Ambinter). Apcin and apcin-A were also synthesized by Sundia Meditech (Shanghai, China) according to the methods described in the Supplementary Information.
Assessment of substrate degradation in Xenopus egg extract
Use of female Xenopus laevis for production of cytoplasmic egg extracts was approved by the Harvard Medical School standing committee on animals (Protocol Number 03231). Interphase Xenopus egg extract was prepared from eggs laid overnight according to the protocol of Murray31 with the exception that eggs were activated with 2 μg/ml calcium ionophore (A23187) for 30 minutes prior to the crushing spin. Extract was frozen in liquid nitrogen and stored at -80 °C. Interphase extract was induced to enter mitosis by addition of nondegradable cyclin B (MBP-Δ90) at 20 μg/ml and incubated at 22-24 °C for 30-60 minutes. MBP-Δ90 consists of a fusion of the maltose-binding protein (MBP) to Xenopus cyclin B1 lacking its N-terminal 90 amino acids32 and was expressed in E. coli by inducing cultures at an OD600 of 0.6 with 300 μM isopropylthiogalactoside (IPTG) for 5 h at room temperature (RT). Purification was carried out following New England BioLabs (NEB) protocol. To promote degradation in interphase extract, Cdh1 protein, expressed in baculovirus, was added to extract at a final concentration of 50 nM in the presence of 75 μM roscovitine. Roscovitine addition is necessary to suppress inhibitory phosphorylation of Cdh1 by Cdk1. Extract was then pretreated with drug (DMSO, apcin, and/or TAME) for 15 minutes at 22 °C before addition of substrates. Substrates consisted of human full length cyclin B1, cyclin A2, securin, Nek2A, or an N-terminal fragment (residues 1-88) of human cyclin B1 (cyc B1-NT). Each substrate was amplified with primers by PCR to allow T7-dependent transcription of the PCR product. Substrates were expressed and labeled with 35S-methionine (Perkin Elmer NEG709A500UC) using the TNT system (Promega). To measure degradation of substrates, extract was pretreated with DMSO or test compounds for 10 minutes in the presence of 100 μg/ml cycloheximide to prevent re-incorporation of free labeled amino acid. The in vitro-translation reaction was then added to the Xenopus extract at 10% final volume. Extract was then incubated at 24 °C, with shaking at 1250 rpm, with samples taken at indicated times. Reactions were quenched with sodium dodecyl sulfate (SDS) sample buffer and processed for SDS-poly acrylamide gel electrophoresis (PAGE) and phosphor imaging (Bio-Rad PMI) and quantification was performed using Quantity One software (Bio-Rad).
Assessment of substrate ubiquitylation in mitotic Xenopus egg extract
Interphase Xenopus egg extract was supplemented with MBP-Δ90 to promote entry into mitosis. Mitotic extract was then treated for 30 minutes at 24 °C with 20 μM ubiquitinvinyl sulfone (UbVS; U-202, Boston Biochem) to suppress deubiquitylation and with 100 μg/ml cycloheximide to prevent re-incorporation of free labeled amino acid. Subsequently, mixtures containing apcin or DMSO, as indicated, proteasome inhibitor MG262 (150 μM) and wild-type ubiquitin (44 μM) were added to UbVS-treated extract and incubated for additional 10 minutes, at 24 °C with agitation. In vitro-translation reactions expressing human full length cyclin B1, cyclin A2, securin, Nek2A, or cycB1- NT with 35S-methionine labeling were also pre-treated with UbVS at 20 μM and added to pre-treated extract at 10% final volume. Extract was then incubated at 24 °C, with shaking at 1250 r.p.m., and substrate ubiquitylation monitored by taking samples at indicated times. Reactions were quenched with SDS sample buffer and processed for SDS-PAGE and phosphor imaging (Bio-Rad PMI), and quantification was performed using Quantity One software (Bio-Rad).
Measurement of the apcin-Cdc20 interaction in Xenopus egg extract using cellular thermal shift assay (CETSA)
The CETSA method33 was adapted to examine apcin engagement of endogenous Cdc20 in Xenopus egg extract. Interphase extract was diluted ten-fold and incubated with various concentrations of apcin dissolved in DMSO in a total volume of 200 μl, with a final DMSO concentration of 1%. After 10-minute incubation at RT, 50 μl of the lysate was transferred into PCR tubes (20170-012, VWR) and heated in a PCR machine (Mastercycler gradient, Eppendorf) for 3 minutes at 46 °C, followed by cooling for 3 minutes at RT. These conditions were established in preliminary experiments as the temperature that yielded the greatest degree of Cdc20 precipitation that could be rescued by apcin treatment (data not shown). The heated lysates were centrifuged at 14,000 r.p.m. (20,000 × g) for 20 minutes at 4 °C in order to separate the soluble fractions from precipitates. Twenty μl of the supernatants were mixed with SDS sample buffer and the fraction of soluble Cdc20 was analyzed by SDS-PAGE and anti-Cdc20 western blot. Quantification of soluble Cdc20 was done using Fuji Imager LAS3000 and Image J software. Soluble Cdc20 levels were normalized to soluble Cdc20 in samples treated with the highest concentration of apcin.
Measurement of Cdc20 binding to APC/C in Xenopus egg extract
To examine levels of Cdc20 associated with APC/C, the APC/C was immunopurified from mitotic Xenopus egg extract. For 100 μl extract, 2 μg of anti-Cdc27 antibody (AF3.1, Santa Cruz Biotechnology) was cross-linked to 5 μl of Affiprep Protein A beads (156-0006, Bio-Rad) and incubated for 1 h at 4 °C. Apcin, TAME or DMSO was mixed with extract upon addition to anti-Cdc27-Affiprep Protein A beads in the presence or absence of cycB1-NT containing a HA-tag at N-terminus and his tag at C-terminus, as previously described34. Following incubation with extract, beads were washed quickly three times with 20-fold volume of XB (10 mM potassium HEPES, pH 7.7, 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2) and combined with SDS sample buffer. For analysis of Cdc20 binding to APC/C, samples were processed for SDS-PAGE and immunoblotting against Cdc20 and APC/C subunit Cdc27. Chemiluminescence was imaged on a Fuji LAS 3000 with Image Reader LAS-3000 software. Levels of Cdc20 were quantified using Image J and data was normalized to respective Cdc27 levels.
Coupling of apcin-A to affigel-10 resin
Affigel-10 resin (153-6099, Bio-Rad) was washed twice with DMSO and dried. The resin was then mixed with 5 mM or 15 mM apcin-A dissolved in DMSO (2x volume of dry resin). N,N-diisopropylethylamine was diluted 50-fold into the solution. The resin was rotated at RT for 2 h and the reaction was quenched with 1/5 resin volume of ethanolamine. The resin was then washed sequentially with isopropanol, water and XB + 0.05% Tween. The resin was stored at 4 °C as a 50% slurry in XB + 0.05% Tween.
Cdc20 depletion by apcin-A resin
For a round of depletion of Cdc20 from mitotic Xenopus extract, apcin-A resin (15 mM coupling) was incubated with extract at 4 °C rotating for 30 minutes. The volume of resin used was 40% of the extract volume. Three rounds of depletion were performed. To rescue degradation in the depleted extract, reticulocyte lysate containing in vitro-translated human Cdc20 or control reticulocyte lysate, treated as for a translation reaction but with no DNA template, was added to the extract at 1/10th extract volume.
Assay for Cdc20 binding to apcin-A resin
Human Cdc20 in pCS2 vector was mutated at the various residues described with the Quikchange Site-Directed Mutagenesis Kit (Agilent Technologies) and custom primers for each sequence. All mutations were confirmed by sequencing. For a single pull down assay, 5 μl of apcin-A resin (5 mM coupling) was incubated with 30 μl diluted in vitro-translated human Cdc20 or other WD40 proteins (5 μl reticulocyte lysate diluted to 30 μl with XB + 0.05% Tween) at 24 °C with shaking (1500 RPM) for 30 minutes. Competitors were pre incubated with Cdc20 in reticulocyte lysate for 2 minutes at RT before adding to apcin-A resin. Bound and input Cdc20 were detected by western blotting (Fig. 1e,f). Cdc20, other WD40 proteins, ODC, and Cdc20 mutants in Extended Data Fig. 1b,c, Fig. 2c and Extended Data Fig. 3d were labeled with 35S methionine and detected by phosphorimaging.
Assessment of kinetics of APC/C-dependent ubiquitylation in a reconstituted system
Measurements of kinetics of ubiquitylation were performed using cycB1-NT and APC/C isolated from Xenopus extract, exactly as previously described16, in the presence or absence of 50 μM apcin. The Ki was calculated based on the assumption of a competitive inhibition model according to the equation: Kmapcin=Kmuntreated (1+[I]/Ki), where [I]=50 μM; Kmapcin (773 nM) and Kmuntreated (245 nM) were the average values from three independent experiments. P value was calculated by an unpaired t-test.
Antibody-based depletion of Cdc20 from Xenopus extract
Cdc20 antibody, covalently coupled to Affiprep protein-A beads as described15, was incubated with mitotic extract at 4 °C with rotation for 30 minutes. The volume of antibody beads used was 20% of the reaction volume. Three rounds of depletion were performed, with separation of extract from beads after each round by centrifugation in spin columns (89868, Thermo-Pierce).
Luciferase assay
A fusion of the N-terminal domain of cyclin B1 to luciferase17 was added to mitotic extract at 4 μg/ml (Fig. 1d) or to interphase extract at 250 μg/ml for 10 minutes at RT then diluted to a final concentration of 4 μg/ml final in mitotic extract (Fig. 2c). The extract was incubated at RT and 3 μL samples were taken at 0, 20, 40 and 60 minutes. The samples were mixed quickly with 30 μL luciferin assay buffer (270 μM coenzyme A, 20 mM tricine, 3.67 mM MgSO4, 0.1 mM ethylediaminetetraacetic acid (EDTA), 33.3 mM dithiothreitol (DTT), 530 μM ATP and 470 μM luciferin, pH 7.8), and the level of luminescence was measured on Wallac 1420 multilabel counter.
Protein purification and crystallization
The coding region of human Cdc20 containing residues 161-477 (Cdc20-WD40) was amplified by PCR and cloned into the modified pFastBac vector. Recombinant baculovirus encoding the N-terminal His6-tagged Cdc20 protein was constructed using the Bac-to-Bac system (Invitrogen) according to manufacturer’s protocols. A tobacco etch virus (TEV) protease cleavage site was introduced into the N-terminus of Cdc20. Sf9 insect cells were infected with the Cdc20 baculovirus and harvested at about 60 hours post-infection. His6-tagged Cdc20-WD40 was purified with Ni2+-NTA agarose resin (Qiagen) and cleaved with TEV protease to remove the His6-tag. The Cdc20-WD40 protein was further purified by anion exchange chromatography with a Mono-Q column followed by size exclusion chromatography with a Superdex 200 column (GE Healthcare). Purified Cdc20-WD40 was concentrated to 4-5 mg/ml in the Superdex 200 column buffer containing 25 mM Tris (pH 8.5), 150 mM NaCl, 1 mM MgCl2, 5% glycerol and 5 mM tris(2-carboxyethyl)phosphine.
The Cdc20-WD40 protein was crystallized at 20°C using the sitting-drop vapor-diffusion method with a reservoir solution containing 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES, pH 6.5), 15% (w/v) polyethylene glycol (PEG) 6000, and 5% MPD. The Cdc20-WD40 crystals were transferred to a new 2μl sitting drop with a reservoir solution containing 0.1 M MES (pH 6.5) and 20% (w/v) PEG 6000 and soaked for 3-5 hours to get rid of bound MPD. Apcin compound was dissolved into DMSO and added to the same drop at the final concentration of 5 mM. After overnight soaking, the crystals were cryo-protected in a solution containing 0.1 M MES (pH 6.5), 20% (w/v) PEG 6000, 10% glycerol and 5 mM apcin, and then flash-cooled in liquid nitrogen. Crystals diffracted to a minimum Bragg spacing (dmin) of about 2.1 Å and exhibited the symmetry of space group P21 with cell dimensions of a = 41 Å, b = 87 Å, c = 48 Å, and b = 113° and contained one Cdc20 molecule per asymmetric unit.
Data collection and structure determination
Diffraction data were collected at beamline 19-ID (SBC-CAT) at the Advanced Photon Source (Argonne National Laboratory, Argonne, Illinois, USA) and processed with HKL300035. Phases were obtained by molecular replacement with Phaser using the crystal structure of human Cdc20-WD40 (PDB code: 4GGC) as search model36. Iterative model building and refinements were carried out with COOT and Phenix, respectively37,38. The final model for Cdc20-WD40-apcin (Rwork = 16.5%, Rfree = 21.3%) contains 313 residues, 82 water molecules and one apcin molecule. MolProbity was used for structure validation to show that all models have good geometry, except for one surface residue that is an outlier in a Ramachandran plot39. Data collection and structure refinement statistics are summarized in Extended Data Table 1.
High-throughput image-based assay to measure mitotic fraction
Parental A549, U2OS, and hTERT-RPE1 cells were purchased from ATCC. DLD-1 cells were purchased from Sigma. For hTERT-RPE1, A549 and U20S cells, stable cell lines expressing H2B-GFP were derived using described methods40 and used in the experiments. DLD-1 cells were used without further modification. Cell lines were tested for mycoplasma contamination (Lonza kit LT07-218) after they were derived and were found negative. For each cell line, asynchronous cells were resuspended to a density of 3.75 × 104 cells/ml. A WellMate dispenser (Thermo Scientific) was used to distribute 40 μL of suspension to each well of a black, clear-bottom 384-well plate (3712, Corning). Plates were sealed with breathable white rayon sealing tape (241205, Nunc) during plating and subsequent incubation. After 24 hours incubation, the cells were treated with indicated concentrations of apcin and proTAME dissolved in DMSO, in four technical replicates. After 18 hours, cells were fixed and stained directly without wash steps to avoid loss of mitotic cells, by adding 10 μl of 6x concentrated fixing/staining reagent (60% Formalin, 0.6% Triton X-100, and 1.5 μg/ml Hoechst 33342 in DPBS). The plates were sealed with aluminum sealing tape (276014, Nunc) and incubated at RT for 40 minutes before imaging. Plates were then imaged at 4 positions per well using an ImageXpress Micro (Molecular Devices) high-throughput microscope, using a 10x objective, yielding a total of 16 images per condition (four images × four replicates). Cell images were processed automatically in ImageJ to identify the nuclei and count the number of nuclei and determine maximum fluorescence intensity of each nucleus in each image. The output files from ImageJ for each treatment were pooled and the cumulative frequency curves of maximum intensity for the cell population in each treatment was computed using Matlab. An intensity threshold was set based on the mitotic fraction in the wells treated with DMSO to separate mitotic cells from interphase cells. The interphase fraction for each treatment was indicated by the fraction below the threshold on the cumulative frequency plot. Methods for the statistical analysis of the data are presented in Supplementary Information.
Fluorescence live cell imaging
For experiments with RPE1-H2B-GFP cells, asynchronous RPE1-H2B-GFP cells were plated in 24-well glass-bottom plates (Greiner BioOne, 662892) 18–24 hours prior to siRNA transfection using RNAiMax (Invitrogen). Cells were transfected with Mad 2 siRNA (GGAACAACUGAAAGAUUGGdTdT, synthesized by Dharmacon) or non-targeting control siRNA (D-001210-01-20, Dharmacon) at a final concentration of 20 nM. Twenty-four hours following transfection, cells were treated with compounds and imaging was initiated following compound treatment. For U2OS-H2B-GFP cells, the cells were first synchronized by double thymidine block (18 hour first block, 8 hour release, 17 hour second block, thymidine concentration 2 mM). Compounds were added at 7 hours after release from the second block and imaging was initiated following compound treatment. To measure efficiency of Mad2 knockdown by siRNA, Western blot samples were each prepared from a single well of the 24-well glass-bottom plates. Twenty-four hours after transfection, cells were collected by trypsinization, pelleting, and resuspension in 2x NuPAGE sample buffer (Invitrogen) + 50 mM DTT.
For imaging, plates were inserted into a covered chamber supplied with humidified 5% CO2 and mounted onto a motorized microscope stage (Prior Scientific). Differential interference contrast and fluorescence images were captured at 6 minute intervals for 45 hours using a Nikon Ti inverted fluorescence microscope fitted with a 37°C enclosed incubation chamber and using a 20X Plan Apo 0.75 NA objective lens. A Hamamatsu ORCA cooled CCD camera collected the images with 2×2 binning using Nikon Elements software (version 3.0). Videos were manually analyzed using Nikon Elements software or ImageJ. Mitotic duration was defined as the time from nuclear envelope breakdown until anaphase, in the case of normal mitosis, or until exit from prolonged mitosis as indicated by cytoplasmic blebbing accompanied by changes in chromatin as detected by H2B-GFP. Cell fate was scored as “division” if two daughter cells were produced by mitosis of any duration, and “abnormal exit” if a single cell of interphase appearance resulted after that cell was in a mitotic state of any duration. The “death” fate describes cells that entered mitosis and died while in mitosis. Methods for the statistical analysis of live cell imaging data are presented in Supplementary Information.
Extended Data
Extended Data Figure 1. Apcin binds Cdc20 and inhibits APC/C-dependent proteolysis in Xenopus extract.
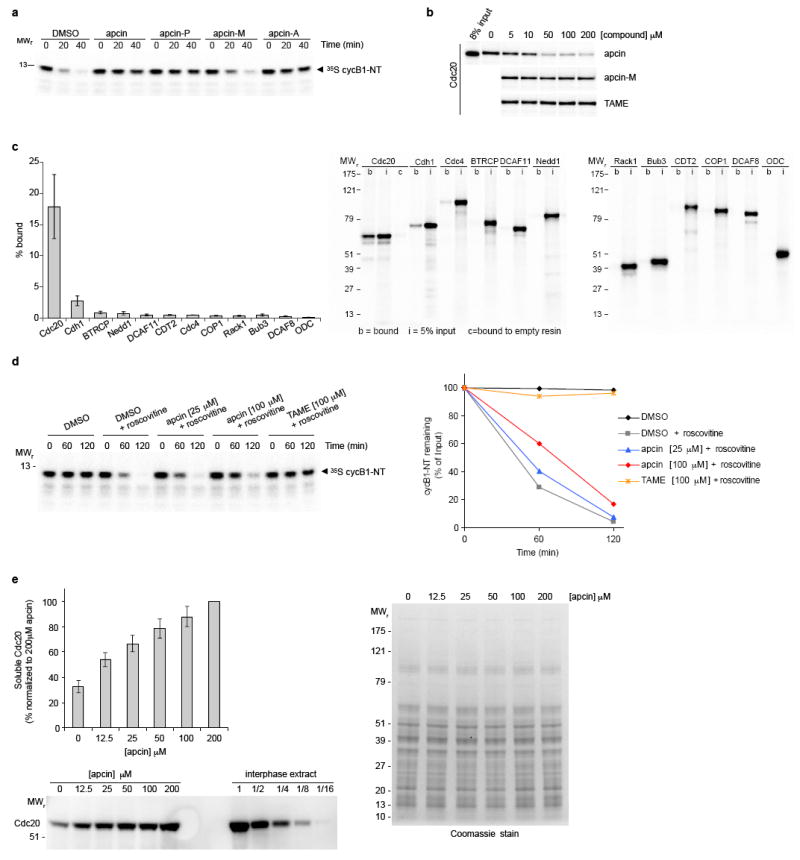
a, Effects of apcin and derivatives (200 μM) on degradation of a cyclin B1 N-terminal fragment (cycB1-NT) in mitotic Xenopus egg extracts. Substrate was expressed in reticulocyte lysate and labeled with 35S-methionine. Samples were analyzed by SDS-gel electrophoresis and phosphorimaging. Quantitation of the 40-minute time point from three independent experiments is shown in Fig. 1b. b, Apcin-M and TAME do not inhibit binding of Cdc20 to apcin-A resin. Experiment was performed as shown in Fig. 1e except that the inactive apcin derivative apcin-M or the Cdc20-IR tail antagonist TAME were also tested and 35S-labeled Cdc20 was detected by autoradiography. c, Apcin-A interacts strongly with Cdc20, weakly with Cdh1, and shows little interaction with other WD40-domain proteins. Proteins were expressed in reticulocyte lysate and labeled with 35S-methionine. Left panel, mean value for percent bound based on three experiments (error bars represent S.E.M.). Right panel, representative autoradiograph of one of three experiments. b= bound, i=5% input, c=bound to empty resin (control). d, Apcin inhibits degradation of cycB1-NT in Cdh1- treated interphase extract. The addition of roscovitine (75 μM) is required to inhibit Cdk1 activity that suppresses Cdh1-dependent proteolysis. Note that 100 μM apcin is less effective at stabilizing the substrate in Cdh1-activated interphase extract than in mitotic Xenopus extract (Fig. 3b). e, Apcin binds to endogenous Cdc20 in Xenopus extract as measured by a thermal shift assay33. Extract was incubated with varying concentrations of apcin, heated to 46 °C for three minutes, and precipitated proteins removed by centrifugation. The soluble fraction was analyzed by SDS-PAGE and western blotting for Cdc20. The left panel shows the percentage of soluble Cdc20 (mean +/- S.E.M. from three independent experiments). Western blot from one of three experiments is shown below. For comparison, total Cdc20 from interphase extract (and various dilutions) is shown. Right panel, coomassie-stained gel of soluble proteins, indicating that there is not an observable nonspecific stabilization of proteins induced by apcin addition.
Extended Data Figure 2. Apcin acts as a competitive inhibitor of APC/C-dependent ubiquitylation.

a, Apcin inhibits formation of high molecular weight ubiquitin conjugates of full-length cyclin B1. Mitotic Xenopus extract was pretreated with the deubiquitinating enzyme inhibitor ubiquitin vinyl sulfone (Ub-VS, 20 μM) and proteasome inhibitor MG262 (150 μM) to stabilize ubiquitin conjugates. 35S-labeled cyclin B1 was added together with ubiquitin (44 μM) and samples analyzed by SDSPAGE and phosphorimaging. Right panel shows quantitation of the experiment. b, Apcin acts as a competitive inhibitor of APC/C-dependent ubiquitylation. APC/C was purified from mitotic Xenopus extracts and the initial rates of ubiquitylation of HA-tagged cycB1- NT were measured in the presence of methylated ubiquitin to prevent ubiquitin chain elongation. The reaction was stopped at 45 seconds and the products were detected by anti-HA blot. The left panel shows the anti-HA blot from one experiment; substrate concentrations were 62.5, 125, 250, 500 and 1000 nM (left to right). Asterisk indicates an SDS-resistant aggregated form of substrate. Quantitation of three independent experiments, and a summary of kinetic parameters, is shown. Note that the effects of apcin in this reconstituted assay performed under initial rate conditions appear distinct from those obtained in crude Xenopus extract. See Supplementary Discussion for a more detailed discussion of these differences.
Extended Data Figure 3. Structure of the apcin-Cdc20 complex.
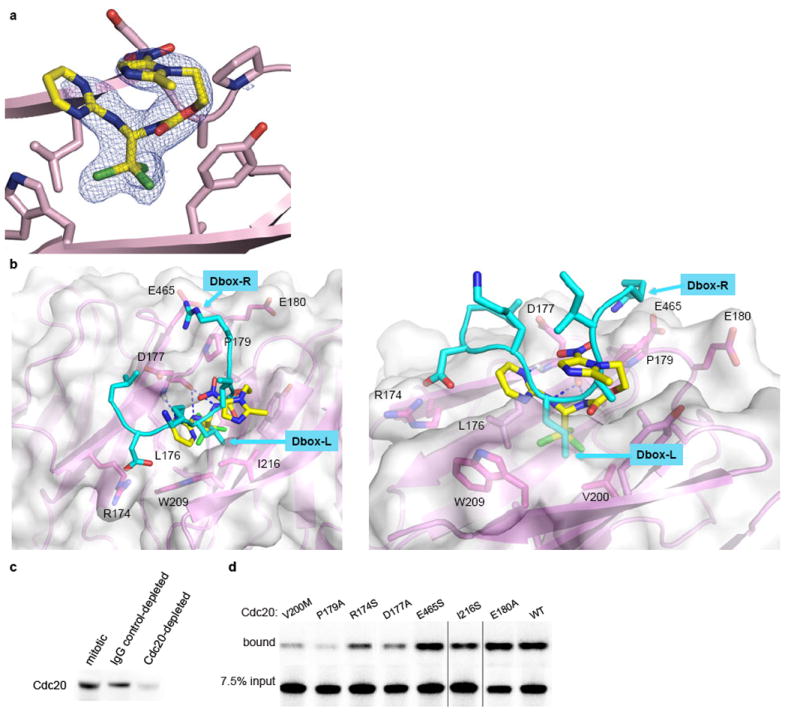
a, The Fo-Fc omit electron density map at the contour level of 3σ is shown. The density for the bound apcin conformation is unambiguous and is consistent with the structure-activity analysis and mutagenesis experiments. b, Overlay of the structure of the Cdc20-apcin structure with the structure of a D-box-containing protein bound to Cdh114. The trichloromethyl group of apcin projects into a hydrophobic pocket that is occupied by the leucine of the RXXL motif of the D-box. The position of the arginine from the RxxL motif suggests a role for E465 of Cdc20 in a charge-based interaction with the D-box, consistent with our data that the E465S mutation disrupts the ability of Cdc20 to promote substrate degradation more than it perturbs apcin binding. c, Depletion with anti-Cdc20 antibody covalently coupled to protein A beads depletes endogenous Xenopus Cdc20 from mitotic extract. d, Example autoradiogram of data shown in Fig. 2b.
Extended Data Figure 4. Effects of apcin on Cdc20 binding to APC/C and stability of APC/C substrates in mitotic Xenopus extract.
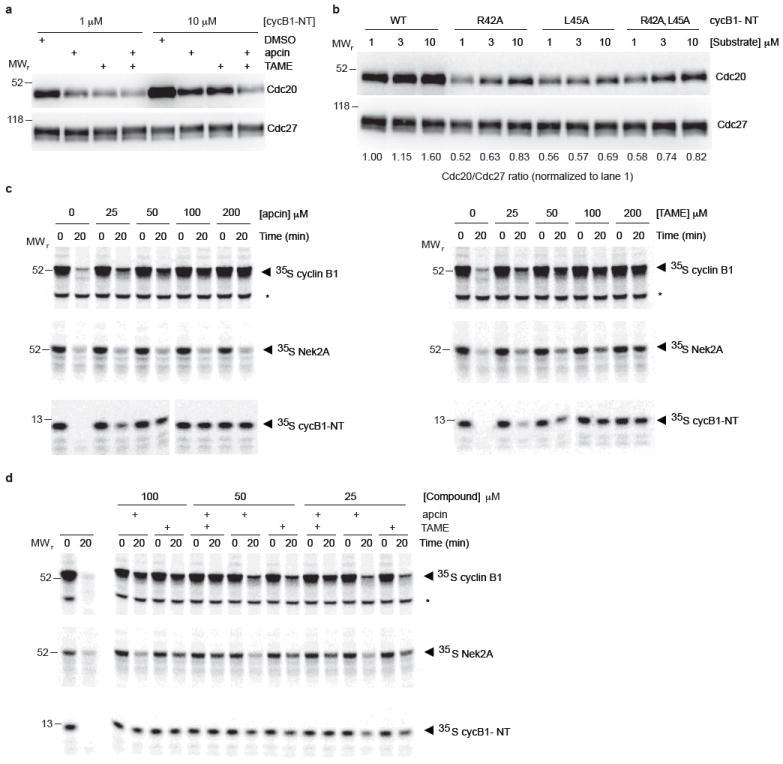
a, Example western blot for data shown in Fig. 3a. b, Substrate-mediated recruitment of Cdc20 to APC/C in Xenopus extract is dependent on the D-box motif. Increasing concentrations of wild-type or different D-box mutants of cycB1-NT were introduced into mitotically-arrested Xenopus extract and the APC/C was isolated with anti-Cdc27 antibodies by immunoprecipitation for 1 hour at 4°C. The immunoprecipitate was separated by SDS-PAGE and analyzed by western blotting against Cdc20 and Cdc27. Levels of Cdc20 were quantitated using Image J and normalized to APC/C subunit Cdc27. c, Analysis of effects of apcin and TAME on APC/C substrate degradation in mitotic Xenopus extract. Levels of 35S-labeled substrates were assessed by SDS-PAGE and phosphorimaging. Asterisk represents a non specific band. Images show one of three experiments quantitated in Fig. 3b. d, Experiment performed as in c, but examining combined effects of apcin and TAME. Image shows one of three experiments quantitated in Fig. 3c.
Extended Data Figure 5. Effects of apcin and TAME on stability of APC/C substrates in mitotic Xenopus extract.
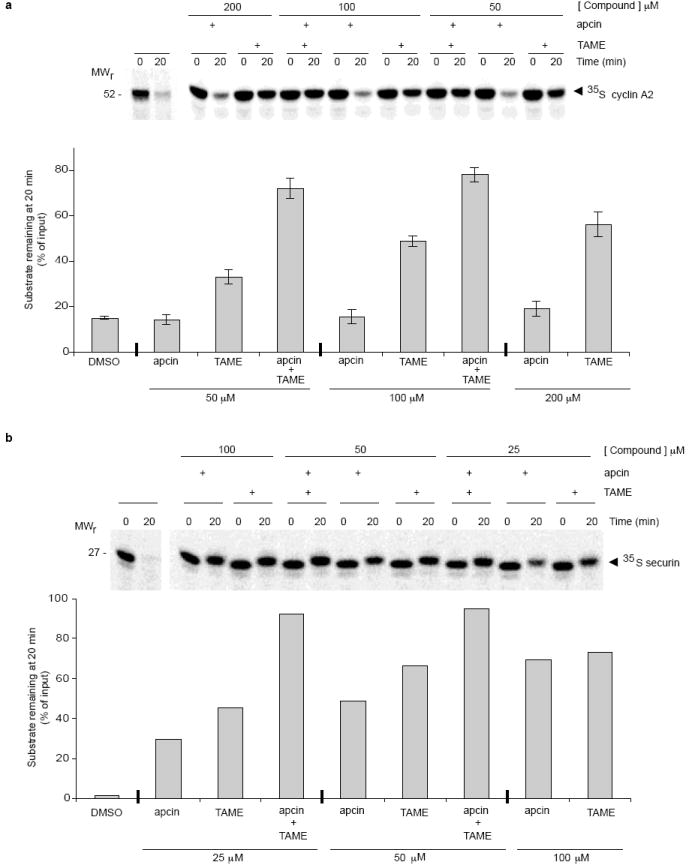
Apcin and TAME synergize in stabilizing cyclin A2 (a) and securin (b) in mitotic Xenopus extract. Levels of 35S-labeled substrates were assessed by SDS-PAGE and phosphorimaging. The change in mobility of securin between 0 and 20 minutes is likely a result of mitotic phosphorylation. Error bars in a represent mean and S.E.M. of three independent experiments. Data in b are representative of two independent experiments.
Extended Data Figure 6. Apcin has differential effects on substrate ubiquitylation in mitotic Xenopus extract that correlate with effects on proteolysis.
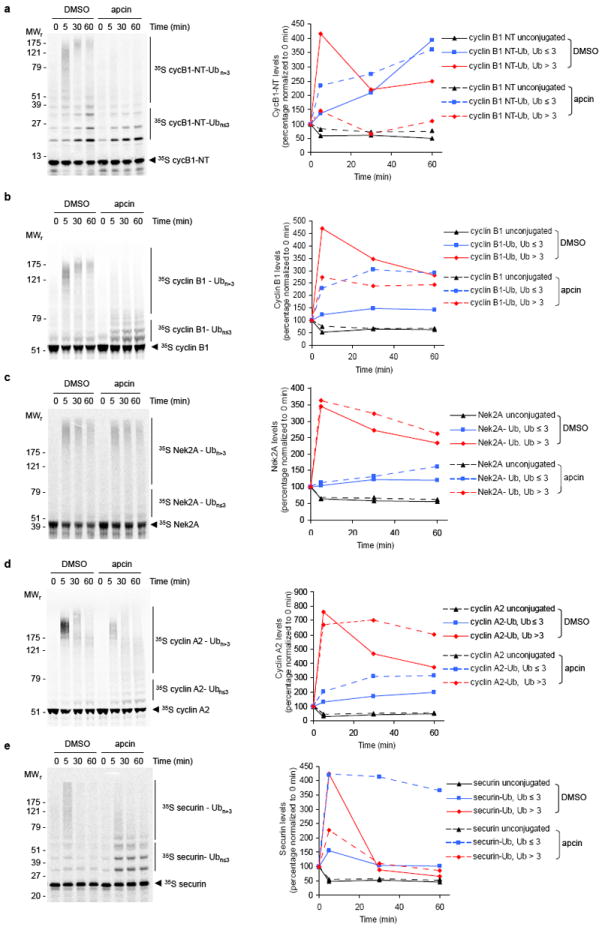
To examine the profile of ubiquitylated species generated in Xenopus extract, deubiquitylation and proteasome-mediated degradation were inhibited by pre-treatment with the general deubiquitinating-enzyme inhibitor ubiquitin-vinyl sulfone (UbVS; 20 μM), as established previously34, and the proteasome-inhibitor MG262 (150 μM). Next, wild-type ubiquitin (Ub; 44 μM) and apcin (100 μM) or DMSO were added. 35S-labeled APC/C substrates expressed in reticulocyte lysate were introduced into treated extract and their ubiquitylation at indicated times assessed by SDS-PAGE and phosphor imaging. Levels of substrates that were not modified with Ub (unconjugated), modified with 1-3 Ubs (substrate-Ub, Ub≤3) or modified with more than 3 Ub moieties (substrate-Ub, Ub>3) were quantitated by phosphorimaging and plotted relative to radiolabeled protein in the respective region of the gel at 0 minutes in the apcin or DMSO sample. a, cycB1-NT; b, cyclin B1; c Nek2A; d, cyclin A2 ; e, securin. Note that for cyclin A2, apcin reduces the amount of very high molecular weight Ub conjugates (as indicated by inspection of the gel image) but does not reduce the fraction of conjugates modified with more than three ubiquitins (as indicated by the quantitation). These results are consistent with the inability of apcin to stabilize cyclin A2 in proteolysis assays, given that the proteasome typically requires at least 4 ubiquitin molecules to be attached to a substrate for efficient proteolysis.
Extended Data Figure 7. Apcin and proTAME synergize to block mitotic exit in human cells, as measured in a fixed cell assay.
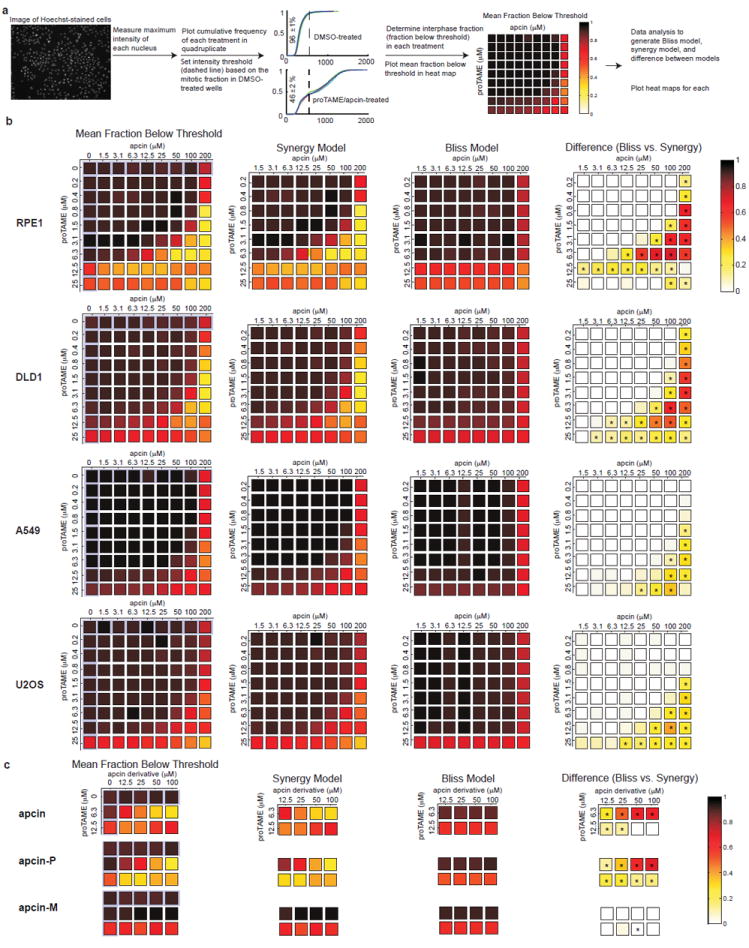
a, Summary of the fixed-cell imaging assay and data processing methods to determine synergy between apcin and proTAME. See methods for detailed description of the assay. b, First column: Primary data plotted as a heat map displaying mean fraction below threshold for each drug treatment concentration in each of four cell lines. This threshold is established based on the mitotic index of DMSO-treated cells. Note that a high value in this column indicates a low mitotic index. Effects of single drugs alone are highlighted in pale purple. Second column (labeled “Synergy Model”): Calculated effect of the combination of drugs based on a model that permits synergistic interaction between proTAME and apcin. Because this panel shows calculated values of combination effects, the effects of individual drugs are not shown. Note that the synergy model closely parallels the actual data shown in the first column. Third column: Calculated effect of the combination of drugs based on a model that permits only multiplicative interaction between proTAME and apcin (Bliss Model). Because this panel shows calculated values of combination effects, the effects of individual drugs are not shown. Note that the Bliss model does not closely parallel the actual data shown in the first column. Fourth column: Heat map of the difference between the Synergy and Bliss model predictions shows the degree of synergy at each drug dose combination (same as Fig. 4a for RPE1 cells). Asterisk indicates P<0.05 for analysis of four technical replicates. See Supplementary Information for details of the statistical analysis. c, Activity of apcin derivatives in the fixed cell assay described in a, using RPE1 cells.
Extended Data Figure 8. Apcin and proTAME synergize to block mitotic exit in human cells, as measured in a live cell assay.
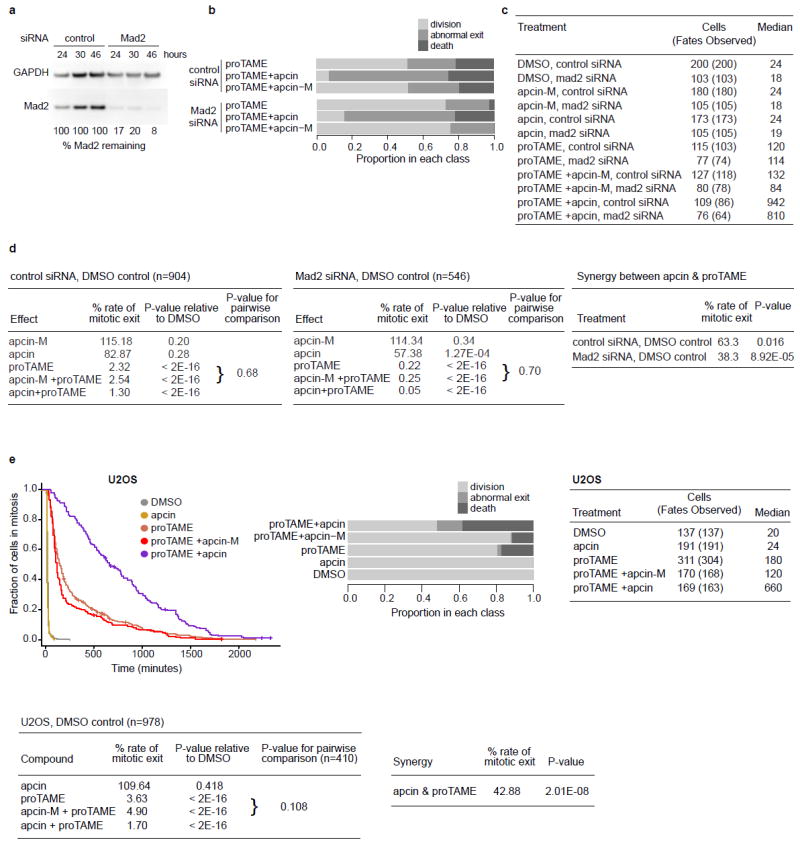
a, Western blot of Mad2 knockdown in RPE1 cells by siRNA from one of the experiments shown in Fig. 4b. b, Analysis of cell fate in RPE1 cells for the experiment shown in Fig 4b. c, Combined data from two independent experiments in RPE1 cells shown in Fig. 4b. “Cells” is the total number of cells analyzed, while “Fates Observed” is the subset of cells whose fate was observed, excluding cells that migrated out of view during the movie or were still arrested at the end of imaging. The median is the time on the x-axis of the Kaplan-Meier curve corresponding to 0.5 on the y-axis “Fraction of cells in mitosis”. d, Statistical modeling of data from experiment in Fig. 4b (RPE1 cells). The two tables on the left show rate of mitotic exit relative to DMSO control for each of two siRNA treatment subgroups (control siRNA, top; Mad2 siRNA, bottom), the P-value for the comparison to DMSO (Cox proportional hazards model, see supplemetary methods), and the P-value for the comparison of proTAME +/- apcin-M. To determine P-values for the pairwise comparisons of proTAME vs. apcin-M + proTAME, we fit a Cox proportional hazards model similar to that described in the methods but with just an apcin-M effect and analyzed just the subset of cells that were treated with either proTAME only or apcin-M + proTAME. The table labeled “Synergy between apcin and proTAME” shows the rate of mitotic exit with both compounds (apcin + proTAME), relative to what would be predicted by a multiplicative combination of the effects of each compound alone. The magnitude of the synergy is roughly doubled when checkpoint activity is reduced by Mad2 siRNA. e, Synchronized U2OS H2B-GFP cells were treated with apcin or apcin-M (25 μM) and/or proTAME (12 μM). Cells were then imaged every 6 or 10 minutes for 45 hours. Mitotic duration and cell fate were determined by manual inspection of the videos and plotted as Kaplan-Meier curves. The hatch marks on the Kaplan-Meier curved indicate mitotic duration endpoints of censored cells. Graphs include the combined results of five independent experiments. U2OS model differs from the model used to test RPE1 data in that the U2OS analysis is not stratified by either date or person, and the U2OS data does not include an effect of apcin-M alone (in the absence of proTAME). Pairwise comparison between proTAME and proTAME+apcin-M tested using a Cox proportional hazards model stratified by date, using data from only the experimental blocks in which both proTAME and proTAME+apcin were tested.
Extended Data Figure 9. Model of the effects of apcin and TAME on formation of the APC/C-Cdc20-substrate ternary complex.

a, Schematic drawing of core APC/C subunits. Not all subunits are indicated, and not all known interactions between subunits are illustrated for sake of simplicity. Light blue oval and polygon indicate binding sites for Cdc20 on core APC/C subunits. b, In the absence of substrate, Cdc20 (green) can bind to the APC/C via the C-box (labeled “C”), which interacts with APC8, and the IR-tail (labeled “IR”), which interacts with APC3 (Cdc27). c, Binding of substrates (red) that contain a D-box (labeled “D”) can promote formation of a co-receptor interaction between the WD-40 domain of Cdc20 and Apc10. d, The RING-containing subunit APC11 can recruit the E2 enzyme to conjugate ubiquitin to the substrate. e, TAME binds APC3 to interfere with the IR-tail binding site. f. Apcin binds to the leucine pocket of the WD-40 domain of Cdc20. g, In the presence of TAME (labeled “T”), the IR-binding site is disrupted, but Cdc20 can still be recruited to the APC/C through the C-box interaction and co-receptor interaction. h, Apcin (labeled “A”) can disrupt the D-box interaction between the substrate and Cdc20, but Cdc20 can still interact through the C-box and IRtail interactions. i, Combined use of apcin and TAME disrupts both interactions, cooperatively disrupting the interaction between APC/C, Cdc20, and substrate.
Extended Data Table 1. Data collection and refinement statistics for Cdc20-apcin structure.
| Data collection | |
|---|---|
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 41.04, 87.18, 48.04 |
| β (°) | 112.72 |
| Resolution range (Å) | 40.00 – 2.10 (2.13 – 2.10) |
| Rmerge (%)a | 8.20 (38.9) |
| I/σ(I) | 13.7 (1.8) |
| Data completeness (%) | 92.8 (58.5) |
| Redundancy | 3.5 (1.6) |
| Energy (eV) | 12,684.1 |
| Unique reflections | 16,859 (522) |
| Wilson B-value (Å2) | 27.8 |
|
| |
|
Refinement statistics
| |
| Resolution range (Å) | 28.58 – 2.10 (2.23 – 2.10) |
| No. of reflections Rwork/Rfree | 16,859/857 (1,915/100) |
| Data completeness (%) | 92.4 (67.0) |
| Atoms (non-H protein/solvent/inhibitor) | 2,430/81/27 |
| Rwork (%) | 16.5 (21.4) |
| Rfree (%) | 21.3 (29.1) |
| R.m.s.d. bond length (Å) | 0.002 |
| R.m.s.d. bond angle (°) | 0.73 |
| Mean B-value (Å2) (protein/solvent/inhibitor) | 29.6/30.0/51.1 |
| Ramachandran plot (%) (favored/additional/disallowed)b | 96.5/2.9/0.6 |
| Maximum likelihood coordinate error | 0.20 |
| Missing residues, by chain | A: 161 – 164 |
Data for the outermost shell are given in parentheses.
As defined by the validation suite MolProbity.
Supplementary Material
Acknowledgments
We thank Wade Harper (Harvard Medical School) for providing constructs for WD40- containing proteins, Tim Gahman (Ludwig Institute, San Diego CA) for assistance with apcin synthesis, and Diana Tomchick (UT Southwestern) for assistance with structure refinement. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02- 06CH11357. This work was supported by grants from the National Institutes of Health (GM085004 to X.L and GM066492 to R.W.K.) and by a grant from the Lynch Foundation to R.W.K.
Footnotes
Author Contributions
K.L.S. and X.Z. performed experiments to identify Cdc20 as the target of apcin. K.L.S. characterized binding of apcin to Cdc20, and evaluated effects of Cdc20 mutations on apcin binding and cyclin proteolysis. W.T. purified Cdc20 and performed crystallization and structure determination of the Cdc20-apcin complex. H.Y. and X.L. contributed to structure determination and data analysis. N.D. and X.Z. characterized effects of apcin and TAME on substrate degradation, ubiquitylation, and Cdc20 binding to APC/C in Xenopus extract. N.D. evaluated binding of apcin to Cdc20 using the thermal shift assay. M.Z. characterized effects of apcin and proTAME on mitotic index in fixed cell assays. K.L.S. and X.Z. characterized effects of apcin and proTAME by live cell imaging. F.S. developed the high-throughput mitotic index assay. K.L.S., X.Z., J.M., K.L.P. and F.S. analyzed time-lapse videos. T.B.S. developed statistical models and performed statistical analysis. R.W.K. conceived of the project, assisted with experimental design and data analysis, and wrote the manuscript, with assistance from all authors.
Structure coordinates have been deposited in the Protein Data Bank under the accession number 4N14.
The authors declare competing financial interests.
References
- 1.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 2.Primorac I, Musacchio A. Panta rhei: The APC/C at steady state. J Cell Biol. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang HC, Shi J, Orth JD, Mitchison TJ. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 2009;16:347–358. doi: 10.1016/j.ccr.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manchado E, et al. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55α,δ phosphatase. Cancer Cell. 2010;18:641–654. doi: 10.1016/j.ccr.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 5.Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Passmore L, et al. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passmore LA, Barford D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 2005;6:873–878. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 10.Eytan E, Moshe Y, Braunstein I, Hershko A. Roles of the Anaphase-Promoting Complex/Cyclosome and of its activator Cdc20 in functional substrate binding. Proc Natl Acad Sci U S A. 2006;103:2081–2086. doi: 10.1073/pnas.0510695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34:68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buschhorn B, et al. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J, et al. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol Cell. 2013;50:649–660. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng X, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng X, King RW. An APC/C inhibitor stabilizes cyclin B1 by prematurely terminating ubiquitylation. Nat Chem Biol. 2012;8:383–392. doi: 10.1038/nchembio.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verma R, et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science. 2004;306:117–120. doi: 10.1126/science.1100946. [DOI] [PubMed] [Google Scholar]
- 18.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 19.Tian W, et al. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc Natl Acad Sci U S A. 2012;109:18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwab M, Neutzner M, Möcker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the Anaphase-Promoting Complex mediate binding to the activator protein Cdh1. Curr Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 22.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18:543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Thornton BR, et al. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Zon W, et al. The APC/C recruits cyclin B1-Cdk1-Cks in prometaphase before D box recognition to control mitotic exit. J Cell Biol. 2010;190:587–602. doi: 10.1083/jcb.200912084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolthuis R, et al. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 26.Di Fiore B, Pines J. How cyclin A destruction escapes the spindle assembly checkpoint. J Cell Biol. 2010;190:501–509. doi: 10.1083/jcb.201001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes MJ, et al. Early mitotic degradation of Nek2A depends on Cdc20- independent interaction with the APC/C. Nat Cell Biol. 2006;8:607–614. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- 28.Kimata Y, Baxter JE, Fry AM, Yamano H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol Cell. 2008;32:576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Sedgwick GG, et al. Mechanisms controlling the temporal degradation of Nek2A and Kif18A by the APC/C-Cdc20 complex. EMBO J. 2013;32:303–314. doi: 10.1038/emboj.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lara-Gonzalez P, Taylor S. Cohesion fatigue explains why pharmacological inhibition of the APC/C induces a spindle checkpoint-dependent mitotic arrest. PLoS One. 2012 doi: 10.1371/journal.pone.0049041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 31.Murray A. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 32.Salic A, King RW. Identifying small molecule inhibitors of the ubiquitinproteasome pathway in Xenopus egg extracts. Methods Enzymol. 2005;399:567–585. doi: 10.1016/S0076-6879(05)99038-1. [DOI] [PubMed] [Google Scholar]
- 33.Martinez Molina D, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 34.Dimova NV, et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ, et al. Phaser crystollagraphic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D: Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sigoillot F, et al. A time-series method for automated measurement of changes in mitotic and interphase duration from time-lapse movies. PloS One. 2011 doi: 10.1371/journal.pone.0025511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


