Abstract
Systemic infection induces conserved physiological responses that include both resistance and ‘tolerance of infection’ mechanisms1. Temporary anorexia associated with an infection is often beneficial2,3 reallocating energy from food foraging towards resistance to infection4 or depriving pathogens of nutrients 5. It imposes, however, a stress on intestinal commensals, as they also experience reduced substrate availability and impacting host fitness due to the loss of caloric intake and colonization resistance (protection from additional infections)6. We hypothesized that the host might utilize internal resources to support the gut microbiota during the acute phase of the disease. Here we show that systemic exposure to Toll-like receptor (TLR) ligands causes rapid α1,2-fucosylation of the small intestine epithelial cells (IEC), which requires sensing of TLR agonists and production of IL-23 by dendritic cells, activation of innate lymphoid cells and expression of α1,2-Fucosyltransferase-2 (Fut2) by IL-22-stimulated IECs. Fucosylated proteins are shed into the lumen and fucose is liberated and metabolized by the gut microbiota, as shown by reporter bacteria and community-wide analysis of microbial gene expression. Fucose affects the expression of microbial metabolic pathways and reduces the expression of bacterial virulence genes. It also improves host tolerance of the mild pathogen Citrobacter rodentium. Thus, rapid IEC fucosylation appears to be a protective mechanism that utilizes the host's resources to maintain host-microbial interactions during pathogen-induced stress.
To maintain the healthy gut microbiota during a systemic response induced by microbial products the host may use its internal resources. L-fucosylation could present an example of such a resource: L-fucose attached to glycoproteins and glycolipids is accessible for microbial, but not for host energy harvest7,8. Constitutive α(1,2)fucosylation affects microbial community in a diet-dependent manner9, serves as a substrate for pathogens during antibiotic exposure10 and for microbes colonizing the ileum of newborns or adult germ-free (GF) animals11-13. Under normal conditions, however, the small intestine (SI) of specific pathogen-free (SPF) BALB/c mice is largely free of surface fucose. In contrast, a systemic injection of agonists of Toll-like receptors (TLRs) such as lipopolysaccharide (LPS, TLR4 ligand) (Fig. 1), CpG DNA (TLR9 ligand), or Pam3CSK4 (TLR2 agonist), led to ubiquitous α(1,2)fucosylation of the SI in mice of different genetic backgrounds, which started within a few hours after LPS exposure and lasted several days (Extended Data Fig.1a-c). It did not result in differentiation of IECs into functional M cells 14 that are permanently fucosylated and are involved in microbial sensing and translocation (Extended Data Fig. 1d). Induced fucosylation was independent of the gut microbiota (observed in GF mice), and was not induced by oral LPS (Extended Data Fig. 1e).
Figure 1. MyD88-dependent fucosylation of SI IECs by systemic stimulation of TLRs.
All panels: Ulex europaeus Agglutinin 1(UEA-1, binds α(1,2)-fucosylated substrates) staining in the proximal 1/3 of SI of mice untreated or 24 hours after i.p. LPS injection, or 6 hours after injection of IL-22 (MyD88−/− mouse). Scale bars=100 μm. Staining of tissue from mutant mice was always accompanied by staining of wild-type controls, and is representative of at least two independent experiments for each genotype.
Global deletion of the TLR signaling adaptor molecule MyD88 prevented IEC fucosylation and its conditional deletion from dendritic cells (DCs), but not IECs, abrogated the process (Fig. 1). The inducible fucosylation pathway was similar to induction of anti-microbial peptides by a systemic microbial signal15: it required MyD88-expressing DCs, production of IL-23, the transcriptional regulator RORγt and IL-22 (Fig. 1, Extended Data Fig. 2a), and was induced by a direct injection of IL-22 into MyD88−/− mice (Fig. 1). IEC fucosylation in mice lacking T cells (Fig. 1) suggested that ILCs were a sufficient source of IL-22. Salmonella enterica subsp.Typhimurium, known to spread systemically, induced SI IEC fucosylation (Extended Data Fig. 2b). The α(1,2)fucosyltransferase responsible for fucosylation of IECs in SI was identified as fucosyltransferase 2 (Fut2) (Fig. 2a), inducible by stress conditions16,17 and constitutively expressed in the stomach and large intestine18. Genetic ablation of the Fut2 gene blocked IEC fucosylation in response to LPS (Fig. 2b, c). The overall chain of events is shown in Extended Data Fig. 3.
Figure 2. Consequences of the loss of Fut-2-dependent fucosylation.
a, Expression of mouse α(1,2)fucosyltransferase genes (Fut2, Fut1 and Sec1) and control villin-encoding gene, vil1 in the gut (cec, cecum; col, colon) 24 hours after LPS injection (semi-quantitative RT-PCR). b, Intestinal fucosylation (green) of Fut2-sufficent and -deficient mice. Red, propidium iodide. Scale bars=100 μm. c, FACS histograms of SI IECs from PBS (left) or LPS-injected (right), Fut2+ (top) or Fut2− (bottom) mice. d, Food consumption in LPS-treated Fut2+ (n=5, black bars) and Fut2− mice (n=3, open bars) (mean±s.e.m.) Representative of 3 experiments. e, Dependence of weight recovery after LPS challenge on the presence of microbiota and expression of Fut2 (mean±s.e.m. of percent of starting body weight, data combined from 4 experiments). *P<0.05, one-way ANOVA. abx, antibiotics (ampicillin and vancomycin).
LPS injection caused marked sickness in BALB/c Fut2-deficient (−/−) and Fut2-sufficient (+/+ or +/−) littermates hours after injection: mice displayed measurable anorexia, adipsia, reduced activity, diarrhea, and weight loss. Food consumption, weight loss, and production of inflammatory cytokines and antimicrobial peptides (Fig.2d,e and Extended Data Fig 4a,b) were similar in both groups. Although Fut2−/− mice have been shown to be healthy under normal SPF conditions19, they were significantly behind the controls in body weight recovery (Fig. 2e) after LPS injection. Importantly, food deprivation without LPS challenge did not cause SI fucosylation and the weight recovery was identical in Fut2-sufficient and Fut2-deficient animals (Extended Data Fig.1a and 4c). Thus, fucosylation of SI was a response to activation by microbial stimuli and not to anorexia per se.
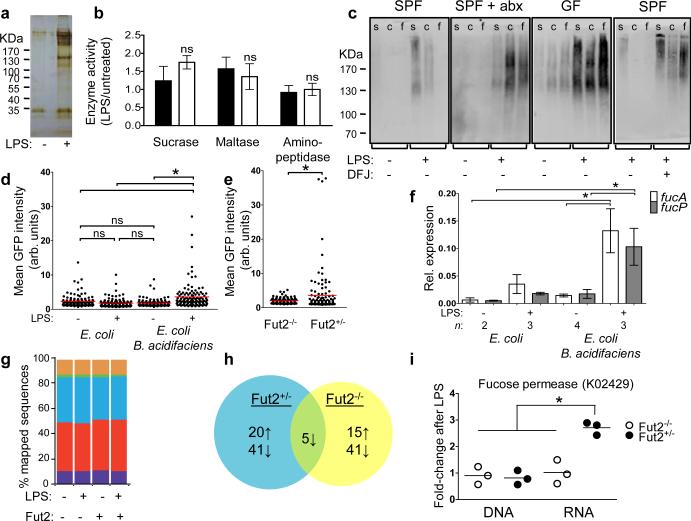
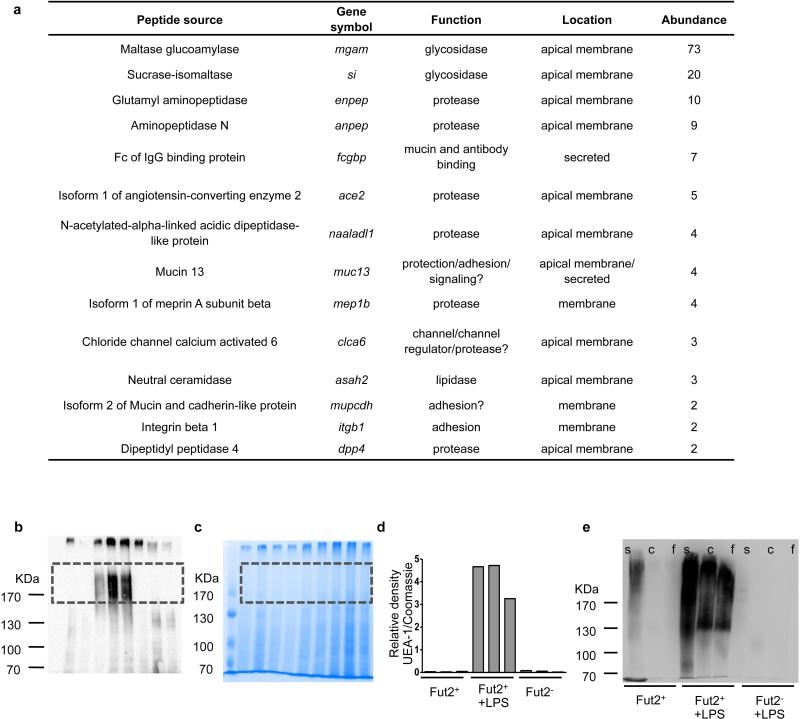
To understand the reason for slow weight recovery in Fut2−/− mice, we tested whether fucosylation affected the function of host IEC proteins identified by direct sequencing as secreted mucins and digestive enzymes (Fig. 3a). Fucosylation did not change activity of several enzymes (Fig. 3b). Thus, changes in enzymatic activity are unlikely to explain the slow weight recovery in Fut2−/− mice, although the role of less abundant fucosylated proteins cannot be excluded.
Figure 3. Commensals utilize fucose detached from proteins fucosylated by Fut2 upon systemic challenge with LPS.
a, Silver-stained SDS-PAGE of UEA-1-precipitated SI IEC protein from control or LPS-treated mouse. b, Ratios of digestive enzymes activities in SI IECs of LPS-treated to untreated Fut2-sufficient (black bars) or Fut2-deficient (open bars) mice two days after LPS injection. Mean±s.e.m. of 4 combined experiments, 4 mice/group. c, SDS-PAGE of intestinal contents blotted on nitrocellulose and stained with UEA-1-peroxidase complexes. s, SI; c, cecum; f, feces; abx, antibiotic treated mice; DFJ, deoxyfuconojirimycin. d, e, Fucose-sensitive GFP reporter expression in gnotobiotic mice colonized with the indicated strains (d, ) or SPF mice (e). Dots are values for individual bacteria, lines are means, n=120; *P<0.05 [one-way ANOVA with Bonferroni post-test (d), two-tailed Student's t test (e)]; representative of 3 independent experiments. f, fucA and fucP gene expression relative to housekeeping gene rpoA (Quantitative RT-PCR) in E. coli tested as in d. *P<0.05 by ANOVA with Bonferroni's post-test. g, Stable relative abundance of bacterial phyla across treatment groups and genotypes, as indicated by shotgun sequencing of community DNA. Phyla with a mean RPKM (reads per kilobase per million mapped reads) >40,000 are shown including Actinobacteria (purple), Bacteroidetes (red), Firmicutes (blue), Fusobacteria (green), Proteobacteria (orange), and Tenericutes (yellow). 16S rRNA gene sequencing confirms these observations. Extended Data Fig. 7 shows 16S rRNA gene sequencing results. h, Differentially expressed KEGG orthologous groups following LPS treatment (paired glm edgeR analysis; q<0.05, >2-fold change; see Supplemental Information Table 2 for complete list). i, Increased gut microbial expression of fucose permease (fucP; KO2429) in Fut2-sufficient mice (mean±s.e.m.; *P<0.01, Mann-Whitney test).
The beneficial effect of fucosylation was dependent on the microbiota: the weight recovery after LPS challenge was delayed in GF mice and in wild-type SPF mice treated with antibiotics (Fig. 2e). Antibiotics did not have direct effects on the host's responses to LPS (Extended Data Fig. 4a,b,d-g) and did not further impair the recovery of Fut2−/− mice (Fig. 2e). Thus, Fut2 and an intact gut microbiota were both necessary for efficient recovery of body weight following LPS challenge.
To serve as a substrate to the bacteria residing in the large intestine20, fucosylated proteins (Extended Data Fig. 5a-d) must be released into the SI lumen. After LPS challenge Fut2-dependent fucosylation of luminal proteins was detectable at much higher levels in germ-free mice, SPF mice treated with antibiotics or with a fucosidase inhibitor, than in SPF controls (Fig. 3c, Extended Data Fig. 5e). Thus, fucose is available, released and used by microbes in the large intestine.
Furthermore, we employed a reporter system, in which E. coli expressed green fluorescent protein (GFP) driven by the promoter of the E. coli fucose metabolism operon21,22 (Fig. 3d, e and Extended Data Fig. 6a). Because E. coli lacks α(1,2)fucosidase that cleaves fucose off substrates, in GF mice monocolonized with the reporter E. coli it did not upregulate GFP, even after LPS injection (Fig. 3d). Thus, free fucose was not readily available for reporter bacteria in the gut and required bacterial fucosidase activity, which was sensitive to antibiotics (Extended Data Fig. 6b). A commensal bacterium with α(1,2)fucosidase activity,Bacteroides acidifaciens was isolated from our mouse colony (Extended Data Fig. 6c-e). In LPS-injected GF mice co-colonized with the reporter E. coli and B. acidifaciens, the reporter strain expressed significantly more GFP (Fig. 3d), as well as genes for fucose import (fucP) and metabolism (fucA) (Fig. 3f). In LPS-treated SPF mice Fut2 was required for GFP reporter expression (Fig. 3e and Extended Data Fig. 6f). These findings made it clear that fucose can serve as a substrate for the microbiota under conditions of stress applied to the host and underscored the interdependence between members of the gut microbial community23.
To confirm these findings in mice with a complex gut microbiota, we profiled microbial community structure, gene abundance, and transcriptional activity before and after LPS treatment of Fut2+/− and Fut2−/− mice (Supplemental Information Table 1). Analyses of community structure based on 16S and shotgun DNA sequencing revealed that the gut microbiota was largely robust to host genotype and LPS exposure (Fig. 3g, Extended Data Fig. 7a,b). We did not detect (a) significant clustering of microbial communities based on genotype or LPS treatment [P>0.05 for both comparisons; permutational multivariate analysis of variance (PERMANOVA) of unweighted UniFrac distances]; (b) species-level operational taxonomic units (OTUs) significantly associated with host genotype prior to or following LPS treatment (all were q>0.05; ANOVA); or (c) significant differences in overall microbial diversity (Extended Data Fig. 7c). However, we were able to detect a significantly increased abundance of B. acidifaciens following LPS treatment in Fut2-sufficient mice (P<0.05, LDA score>4; LefSe analysis of 16S profiles), consistent with its ability to utilize fucosylated glycans. At the same time, LPS markedly altered community-wide gene expression in both Fut2+/− and Fut2−/− mice (Fig. 3h) with multiple orthologous groups differentially expressed upon LPS treatment: 61 in Fut2+/− mice and 56 in Fut2−/− animals. These changes were not due to altered community structure: only 1 differentially expressed orthologous group (K05351, xylulose reductase) also exhibited significant changes in gene abundance (Supplemental Information Table 2). As expected, we detected a significant upregulation of fucose permease (fucP; K02429) in Fut2-sufficient mice after exposure to LPS (Fig. 3i), and increased expression of metabolic modules for anaerobic respiration, protein and ATP synthesis, isoprenoid biosynthesis, and amino sugar import, in addition to pathways for aminoglycan degradation (Extended Data Fig. 7d). Thus, intact host fucosylation appears to affect gut microbial metabolism.
Importantly, LPS challenge led to the significantly increased expression of microbial virulence genes in Fut2-negative but not Fut2-sufficient mice, including RtxA (K10953) and hemolysin III (K11068) (Supplemental Information Table 2). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways potentially involved in microbial pathogenesis (defined as flagellar synthesis, chemotaxis, plant/pathogen interaction and Vibrio cholera infection) were over-represented in Fut2-deficient mice (Extended Data Fig. 7d). We hypothesized that fucosylation induced by systemic microbial challenge might limit the effects of additional exogenous or endogenous pathogens. We tested this by infecting Fut2-sufficient and Fut2-deficient mice with a non-lethal intestinal pathogen, Citrobacter rodentium. Four days after infection, mice were treated with LPS. Infected Fut2-negative mice lost significantly more weight than Fut2-sufficient animals compared to respective LPS-treated non-infected controls (Fig. 4a). Thus, infection with a non-lethal pathogen further reduced the overall fitness of Fut2-deficient mice in response to LPS. C. rodentium did not induce SI IEC fucosylation and did not colonize the SI (Extended Data Fig. 8), indicating that systemic challenge by a microbial product was required to reveal the role of inducible fucosylation.
Figure 4. Host fucosylation increases tolerance of a pathogen.
a, Difference in % weight loss between LPS-injected C. rodentium-infected and uninfected mice (mean±s.e.m.; *P=0.01 ***P=0.0001, two-tailed Student's t test; combined from 6 experiments). b, Fecal CFUs of C. rodentium from Fut2+ or Fut2− mice (mean±s.e.m., data combined from 6 experiments). c, Luminescence of thoroughly washed mid-colon of mice infected with pler-lux+ C. rodentium. n=4 for d12 and 8 for d7; mean±s.e.m. d, Average crypt depth in uninfected (n=3) or infected mice (n=4) at day 12 p.i., with or without LPS injection on day 4 p.i., mean±s.e.m.. In a-d black bars and circles – Fut2-positive; open bars and circles – Fut2-negative mice. e, Representative hematoxylin/eosin staining of distal colon of LPS-treated mice at day 12 p.i. Scale bars=100 μm. Bottom row - magnified boxed regions.
The fitness of infected animals can be maintained via either decreased pathogen burden (resistance), or by an increase in pathogen tolerance without a change in pathogen burden. We quantified the abundance of C. rodentium in the feces of infected mice and adherence to IEC of C. rodentium expressing luciferase24,25 (Fig. 4b,c). No differences in pathogen loads were found between Fut2-sufficient and Fut2-deficient animals treated with LPS. Thus, fucosylation of the SI upon systemic treatment with LPS likely enhances tolerance of the pathogen. Fut2-negative mice infected with C.rodentium and injected with LPS had more pronounced colonic hyperplasia (a trade mark of this infection) compared to Fut2-sufficient mice or mice which did not receive LPS (Fig. 4d,e).
Thus, inducible IEC fucosylation might be viewed as an emergency measure taken by the host to support the gut commensals. Fucose used by microbes as an energy source may contribute to protection of the host from endogenous opportunistic pathogens, or it could increase tolerance of infection by regulating bacterial genes responsible for quorum sensing26 or virulence27. Fucose can also serve as a substrate for microbial production of the short chain fatty acid propionate (Extended Data Fig. 9), which is primarily produced by members of the Bacteroidetes phylum28. Whether this process contributes to overall fitness of the animals under infection-induced stress remains to be elucidated. Of note, around 20% of humans lack a functional FUT2 gene, which is linked to Crohn's disease29 and to lethality from sepsis in premature infants30. Overall, fucosylation of SI in response to systemic microbial exposure can be considered a type of ‘tolerance of infection’ response. It is interesting, however, that a very similar pathway regulates secretion of antimicrobial proteins - a resistance mechanism15. Thus, the mechanisms of resistance and tolerance to pathogens could be evolutionarily linked to increase the fitness of the host.
Methods
Mice
BALB/cJ, C57BL/6J, NOD/LtJ, C3H/HeN, RAG1−/− (B6.129S7-Rag1tm1Mom/J), Fut2−/− (B6.129X1-Fut2tm1Sdo/J), Villin-cre (B6.SJL-Tg(Vil-cre)997Gum/J), and CD11c-cre (C57BL/6J-Tg(Itgax-cre,-EGFP)4097Ach/J) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Fut2−/− mice were backcrossed greater than 7 generations to BALB/c. Knock-out mice produced litters of mixed genotypes kept cohoused to homogenize their gut microbiota. B6 MyD88−/− mice were a generous gift of Shizuo Akira (Osaka University). B6 MyD88 floxed mice were described 31. Rorc−/− 32, and IL-23p19−/− 33 were provided by Yang-Xin Fu (The University of Chicago). IL-22−/− mice 34 were maintained at the Memorial Sloan-Kettering Cancer Center. Mice were housed in a specific pathogen-free facility and used in accordance with institutional guidelines for animal welfare. 6-12 week old male and female mice were used for randomization purposes. The numbers of mice per group were chosen as the minimum needed to obtain biologically significant results, based on previous experience. Evaluations were made in a blind fashion. Functional experiments were done with Fut2-negative mice on the BALB/c genetic background using Fut2-sufficient littermates as controls.
Metabolic studies
Mice were single-housed in TSE Systems LabMaster cages (Chesterfield, MO) to monitor physical activity (x/y and z axes by infrared beams crossed), drinking, and feeding.
Fucosylation activation in vivo
LPS from Salmonella enterica serotype typhimurium (#L6511, Sigma–Aldrich, St. Louis, MO) was injected i.p. at 1 μg/g body weight, or gavaged to GF mice at 1 mg in 400 μl sterile PBS. CpG (100 μg CpG-B ODN 1826, Coley Pharmaceutical Group Ltd., Ottawa, Canada) and Pam3CSK4 (100 μg, Invivogen, San Diego, CA), were injected i.p. Carrier-free recombinant mouse IL-22 (Biolegend, San Diego, CA) was diluted in 1% BSA/PBS and 1.5 μg given i.p.
Antibiotic treatment
1 g/L ampicillin and 200 mg/L vancomycin (Sigma-Aldrich) were 0.22 μm filtered and added to autoclaved drinking water starting two days before LPS treatment (day −2) and lasting for the duration of the experiment.
Lectin staining
For whole-mount staining, small intestine was removed, a 1 cm piece from the upper third was excised, opened, cleaned of mucus in cold PBS, and incubated with Ulex europaeus aggluitinin-1 (UEA-1) conjugated to FITC, TRITC or atto-594 (Vector Laboratories, Burlingame, CA or Sigma-Aldrich) for 15 min on ice. Tissue was placed lumen side up on a slide for microscopy. For IL-22−/− experiments, whole tissue was fixed in 4% paraformaldehyde before proceeding with whole-mount staining. Fixation did not affect staining pattern or intensity.
For staining of sections, tissues were fixed in 2% paraformaldehyde overnight at 4 °C, cryoprotected in 20% sucrose/PBS overnight at 4 °C, and embedded in OCT compound (Sakura Finetek, Torrance, CA), and 10 μm sections stained with UEA-1-FITC (1 μg/ml, Vector) for 30 min at RT, and incubated for 20 min at 37 °C in propidium iodide (0.5 μg/ml, Sigma-Aldrich) with RNase A (10 μg/ml, Sigma-Aldrich) to label nuclei.
For single cell analysis by FACS, small intestine (with Peyer's patches and mesenteric fat removed) was divided into equal thirds, opened longitudinally and washed with cold PBS. Tissue was cut into 1 cm pieces and shaken in 10 mM EDTA/1 mM DTT/PBS at 37 °C for 20 min, and filtered through nylon mesh. Single cell suspensions were pelleted and fixed in 5 ml of 1% paraformaldehyde overnight at room temperature. Cells were then stained with UEA-1-FITC (1 μg/ml, Vector), and gated on the FSC/SSC high epithelial cell population.
Light microscopy
Fluorescence microscopy of whole-mounts and sections used a Leica DM LB microscope (Leica Camera AG, Solms, Germany) and Spot RT Slider camera and software (Diagnostic Instruments, Sterling Heights, MI). Confocal microscopy of bacteria was performed with an Olympus DSU spinning disk microscope (Olympus, Center Valley, PA) and Slidebook software (3I, Santa Monica, CA). All images in an experiment were taken using the same exposure settings.
Scanning electron microscopy
Tissues were fixed in 2% paraformaldehyde/2% glutaraldehyde/0.1 M cacodylate buffer, transferred to cacodylate buffer overnight and processed with the OTOTO procedure 35. To dehydrate, samples were passed through increasing concentrations of acetone in water, followed by hexamethyldisilazane (Electron Microscopy Sciences, Hatfield, PA). Samples were mounted with colloidal silver paste (Electron Microscopy Sciences) and imaged with an FEI Nova NanoSEM 230 (FEI, Hillsboro, OR) at 5 kV.
Identification of fucosylated proteins
To visualize fucosylated proteins in the lumen contents, the intestinal contents were gently removed by squeezing and homogenized at 1 g sample per 5 ml of Tris-Triton-X100 buffer (150 mM NaCl, 50 mM TRIS pH 8, 1% Triton X100, and protease inhibitor tablet [Roche, Mannheim, Germany]) on ice and spun at 17,000 g for 20 min at 4°C. Supernatant proteins were separated by 6% non-reducing SDS-PAGE, transferred to PVDF membrane (Bio-Rad, Hercules, CA), blocked with 0.5% gelatin in 0.05% tween-20/PBS, and visualized with UEA-1-HRP (1 μg/ml, Sigma-Aldrich).
For sequencing of fucosylated epithelial cell proteins, small intestine epithelial cells were isolated as described above and lysed in Tris-Triton-X100 buffer. Lysates were pre-cleared by incubating with unconjugated agarose beads (Vector) twice for 45 min at 4 °C. Cleared lysates were then incubated with washed UEA-1-conjugated beads (Vector) for 45 min. Beads were washed 5 times to remove unbound protein. To elute UEA-1-bound protein, beads were incubated with 200 mM L-fucose (Sigma-Aldrich) for 30 min. Eluted proteins were separated on a 4-15% gradient SDS-PAGE gel (Bio-Rad), silver-stained (Thermo Fisher Scientific, Rockford, IL), and bands were excised for identification by mass spectrometry at the Taplin Biological Mass Spectrometry Facility, Harvard Medical School.
Isolation of fucosidase-positive bacteria
BHIS agar plates (Brain heart infusion agar [Becton, Dickinson, and Co., Sparks, MD] with added thioglycolic acid, menadione, and hematin/histidine [Sigma-Aldrich]) were pre-reduced in an anaerobic chamber with 2.5% hydrogen atmosphere at 37 °C, and spread with 40 μl of 5-bromo-4-chloro-3-indolyl-α-L-fucopyranoside (50 mM in DMF) (Carbosynth, Berkshire, UK). Fecal pellets were homogenized in reduced 0.01% thioglycolic acid/PBS, plated, and grown for 3 days at 37 °C anaerobically, then 4 °C aerobically to develop color. Blue colonies were identified by sequencing of their 16S rDNA genes using primers 8F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1391R (5’-GACGGGCGGTGWGTRCA-3’), and sequencing of their gyrB genes using primers gyrB F (5’-GAAGTCATCATGACCGTTCTGCAYGCNGGNGGNAARTTYGA-3’) and gyrB R (5’- AGCAGGGTACGGATGTGCGAGCCRTCNACRTCNGCRTCNGTCAT-3’) for amplification and gyrB FS (5’- GAAGTCATCATGACCGTTCTGCA-3’) and gyrB RS (5’- AGCAGGGTACGGATGTGCGAGCC-3’) for sequencing36. B. thetaiotaomicron VPI-5482 and B. uniformis (a gift of Dr. Cathryn Nagler) were used as controls.
Measurement and inhibition of fucosidase activity
Fecal pellets were weighed and homogenized in 10 μl PBS per 1 mg sample, and centrifuged at 17,000 g for 10 min. 50 μl of this supernatant was incubated for 1 h at 37 °C with or without adding 0.5 μl of 50 mM 4-methylumbelliferyl fucopyranoside (in DMSO, Sigma or Gold Biotechnology, St. Louis, MO). The reaction was diluted 100-fold in 0.2 M glycine-NaOH buffer, pH 10.5, and fluorescence measured at 365 nm excitation, 445 nm emission. Fluorescence of the no-substrate control was subtracted from the substrate-containing reaction. The amount of cleaved substrate and fucosidase activity was then calculated by comparison to a standard curve of 4-methylumbelliferone (Sigma) in glycine-NaOH buffer. Deoxyfuconojirimycin (DFJ; Enzo Life Sciences, Farmingdale, NY) was dissolved in PBS and gavaged to mice in 100μl (5μmol total).
Reporter E. coli
The pXDC94 plasmid 21, containing the ptac promoter controlling mCherry expression and a multiple cloning site upstream of the promoterless GFP gene, was a gift of Howard Shuman (The University of Chicago). The promoter region upstream of the E. coli fucPIK genes 22 was amplified using primers that added restriction sites (underlined) at the 5’ and 3’ ends of the amplicon, respectively (fucproF, 5’ TATGGTACCGGATTCATTTTCAAATAAAA-3’; fucproR, 5’-TATCCCGGGTAGCTACCTCTCTCTGATTC-3’). The PCR product and vector were digested with XmaI and KpnI (New England Biolabs, Ipswich, MA), gel purified, ligated, and introduced into E. coli K-12 strain BW25113 (Yale Coli Genetic Stock Center) by electroporation. Correct expression of mCherry and GFP was verified by growth in minimal medium 37 with 10 mM glucose and the indicated concentrations of fucose.
E. coli K12 bacteria carrying the pXDC94-fucPIK-pro reporter were grown overnight in LB with shaking at 37 °C, centrifuged at 5,000 g, resuspended to ~109 CFU/ml in PBS, and 400 μl gavaged to mice that had received LPS i.p. 6 h earlier (SPF mice), or at the time of gavage (gnotobiotic mice). For dual colonization with B. acidifaciens, 100 μl of stationary-phase culture was gavaged at the same time as E. coli. 24 h after gavage, mice were sacrificed and their cecum and colon contents homogenized in PBS. Bacteria were enriched by centrifuging at 200 g for 5 min, centrifuging the supernatant at 5,000 g for 5 min, and resuspending the pellet in 750 μl PBS. This was underlayed with 300 μl Histopaque-1119 (Sigma-Aldrich) and centrifuged for 1 min at 11,600 g. The interface containing mostly bacteria was washed with PBS, fixed in 2% paraformaldehyde for 20 min at RT, washed and resuspended in 50-100 μl PBS. 4 μl of bacterial suspension was placed on a slide and coverslipped. Random fields were selected using red fluorescence only, and an image was acquired in both the red and green channels. Fluorescence of individual bacteria were measured in ImageJ 1.41 software 38 by gating on areas of red fluorescence and measuring the mean green pixel intensity within the gated area.
Estimation of total bacterial loads
Fresh fecal pellets were placed in an anaerobic chamber and mashed in 500 μl of reduced PBS containing 0.01% thioglycolic acid. Serial dilutions were made in reduced PBS and plated on pre-reduced Brucella blood agar plates (Becton, Dickinson, and Co.; for anaerobic counts), removed from the anaerobic chamber, and plated on TSA/5% sheep's blood plates (Becton, Dickinson, and Co.; aerobic counts). Total colonies were counted after 2 days (aerobic) or 3 days (anaerobic) incubation at 37 °C.
For 16S copy number, fecal pellets were weighed, DNA isolated by a bead beating and phenol/chloroform extraction method 39, and qPCR performed and copy number determined as described 40.
S. typhimurium and C. rodentium infection
For S. typhimurium infection, mice were gavaged with 20 mg streptomycin (Sigma) in 100 μl sterile water, 24 h before infection. S. typhimurium strain SL1344 was grown overnight in LB with streptomycin (50 μg/ml) and gavaged to mice at 5×108 CFU in 100 μl volume.
Citrobacter rodentium strains DBS100 or DBS120 pler-lux were grown in LB overnight at 37 °C, then diluted 1:100 and grown for 2.5 hours, centrifuged and resuspended in 0.01 volumes PBS, and mice were gavaged with ~5×109 bacteria in 100 μl LB. To determine mouse colonization levels, a fresh fecal pellet or SI contents (gently squeezed to remove, except last 3 cm of ileum) was weighed, mashed in 500 μl of PBS, serially diluted, and plated on MacConkey (Becton, Dickinson, and Co.) or LB agar with 50 μg/ml kanamycin. For luciferase measurements, the fecal homogenate was adjusted to 10 mg in 1 ml PBS in an eppendorf tube and light measured in a Triathler scintillation counter (Hidex, Turku, Finland), before plating dilutions in PBS on agar with kanamycin. Colon-adherent bacteria were measured in a 1 cm piece from the middle of the colon. The piece was opened longitudinally, washed in 1 mM DTT/PBS by vortexing for 10 s, then washed in PBS, and placed in an eppendorf in 500 μl PBS for light measurement as before. The piece was then mashed and dilutions plated on agar with kanamycin.
Histology
Distal colon from C. rodentium infected mice or uninfected controls was fixed in neutral formalin, then kept in 70% EtOH until being embedded in paraffin, and 5 μm sections cut and stained with hematoxylin/eosin. Well-oriented crypts were photographed and their lengths measured in ImageJ, and the mean taken for each mouse.
Serum cytokine ELISA
Serum concentrations were measured by ELISA according to the manufacturer's instructions (IL-1β: eBioscience, San Diego, CA; IL-6 and TNF-α: Becton, Dickinson and Co.).
Short-chain fatty acid measurements
For gavage experiments, food was removed and SPF mice were gavaged at 0, 3, and 6 hours with 300 μl of 0.1 M L-fucose or 0.1 M D-galactose (Sigma-Aldrich) in autoclaved tap water, or water only. At 8 hours, mice were sacrificed and cecal contents removed and kept at −80 °C until processing.
Concentrations of the short-chain fatty acids in cecal contents were measured using the direct injection gas chromatography – mass spectrometry (GC-MS) method adapted from 41-43.
Cecal contents were extracted in two steps. Deionized water was added to the samples in the amount of 5 ml per 1 g of sample, followed by brief vortex-mixing and sonication for 15 min. In the second step acetonitrile (#1103, BDH, Radnor, PA) containing 20 mM of tetradeuteroacetic acid (#16621, Acros Organics, New Jersey, USA) was added to the aqueous extracts in the amount of 5 ml per 1 g of initial sample, followed by the second round of brief vortex-mixing and sonication for 15 min.
Extracted samples were centrifuged for 5 min at 17,000 g and 0.02 ml of clear supernatant was mixed with 0.98 ml of acetonitrile containing 20 mM of formic acid (#94318, Fluka, Buchs, Switzerland) and 0.05 mM of 2-ethylbutyric acid (#109959, Aldrich, Milwaukee, USA). The mixtures were briefly vortex-mixed and centrifuged for 5 min at 17,000 g and 1 μl of the obtained supernatants were analyzed by direct injection GC-MS on an Agilent 6890N GC system equipped with a Mass Selective (MS) Detector 5973 (Agilent Technologies).
The instrument was used in a splitless mode with an installed double taper inlet liner (#23308, Sky by Restek) and fused-silica column with polyethylene glycol stationary phase (INNOWax #19091N-133, J&W Scientific, Agilent Technologies) 30 m × 0.25 mm ID coated with 0.25 μm film.
Helium carrier gas was supplied at 1.00 mL/min flow rate. The injection port temperature was 260 °C. The initial oven temperature of 60 °C was maintained for 2 min, then increased to 150 °C at 10 °C/min and further to 250 °C at 25 °C/min and held at that temperature for 4 min bringing the total duration of the run to 19 min. The MS detector temperature was set to 245 °C.
GC-MS data was analyzed using the MSD ChemStation D.01.02.16 software (Agilent Technologies). Tetradeuteroacetic and 2-ethylbutyric acids served as internal standards.
Enzymatic activity of digestive enzymes
Epithelial cells were dissociated from the upper half of the small intestine as described above. Hematopoietic cells were removed with anti-CD45-biotin (clone 30-F11, Biolegend) and streptavidin magnetic beads with a MACS LS column (Miltenyi Biotec, Bergisch Gladbach, Germany). Epithelial cells were diluted in 96-well plates in triplicate in appropriate substrate solutions (for sucrase, 60 mM sucrose in PBS; for maltase, 30 mM maltose in PBS; for aminopeptidase, 5 mM leu-pNA (Enzo Life Sciences) in 50 mM TRIS, pH 7.4). Cells were incubated for 30-60 min at 37 °C. For the aminopeptidase assay, absorbance was measured at 405 nm. For the sucrase and maltase assays, plates were centrifuged and 5 μl of supernatant was used for a colorimetric assay to measure liberated glucose (Cayman Chemical, Ann Arbor, MI).
Metagenomic library preparation and sequencing
Methods for microbial community DNA and mRNA sequencing were as previously described 44,45. Fecal pellets collected from mice before or after LPS treatment (pooled from 2 and 3 days post-injection) were kept at −80°C until DNA and RNA isolation using the guanidinium thiocyanate/cesium chloride gradient method 46 as described, except that crude particles were removed by centrifugation prior to overlaying the gradient. RNA was subjected to DNase treatment (Ambion), purification using MEGAClear columns (Ambion), and rRNA depletion via subtractive hybridization (MICROBExpress, Ambion, in addition to custom depletion oligos). The presence of genomic DNA contamination was assessed by PCR with universal 16S rDNA primers. cDNA was synthesized using SuperScript II and random hexamers (Invitrogen, Carlsbad, CA), followed by second strand synthesis with RNaseH and E.coli DNA polymerase (New England Biolabs). Samples were prepared for sequencing with an Illumina HiSeq instrument after enzymatic fragmentation (NEBE6040L/M0348S). Libraries were quantified by quantitative PCR (qPCR) according to the Illumina protocol. qPCR assays were run using ABsoluteTM QPCR SYBR Green ROX Mix (Thermo Scientific) on an Mx3000P QPCR System instrument (Stratagene, La Jolla, CA). The size distribution of each library was quantified on an Agilent HS-DNA chip (Agilent, Santa Clara, CA).
16S rRNA gene sequencing and analysis
Community DNA was PCR-amplified using universal bacterial primers targeting variable region 4 of the 16S rRNA gene with the following thermocycler protocol: denature at 94°C for 3 min, 35 cycles of 94ºC for 45 sec, 50°C for 30 sec, and 72°C for 90 sec, with a final extension at 72°C for 10 min 45,47,48. Triplicate reactions for each sample were pooled and amplification was confirmed by 1.5% gel electrophoresis. 16S rRNA gene amplicons were cleaned with the Ampure XP kit (Agencourt, Danvers, MA) and quantified using the Quant-iT Picogreen ds DNA Assay Kit (Invitrogen). Barcoded amplicons from multiple samples were pooled and sequenced using the Illumina HiSeq platform 47. 16S rRNA gene sequences were analyzed using the QIIME (Quantitative Insights Into Microbial Ecology) 44 software package along with custom Perl scripts. Datasets were randomly subsampled prior to clustering analyses at a depth that retained all of the individual samples (180,000 sequences/sample). All sequences were used for the comparison of the relative abundance of bacterial taxonomic groups. Operational taxonomic units (OTUs) were picked at 97% similarity against the Greengenes database 49 (constructed by the nested_gg_workflow.py QiimeUtils script on 4 Feb 2011), which we trimmed to span only the 16S rRNA region flanked by our sequencing primers (positions 521-773). The LefSe package was used to identify taxonomic groups significantly associated with each treatment50. LefSe was run on the sub-sampled datasets, after filtering out species-level OTUs with <100 sequences or present in only 1 sample. Statistical analyses were also performed using the QIIME scripts “otu_category_significance” (ANOVA) and “compare_categories.py” (PERMANOVA).
Reference genome database
A custom database was constructed from draft and finished reference genomes obtained from human-associated microbial isolates [538 genomes from the Human Microbiome Project Data Analysis and Coordination Center (www.hmpdacc.org), in addition to the Eggerthella lenta DSM2243 reference genome. All predicted proteins from the reference genome database were annotated with KEGG orthologous groups (KOs) using the KEGG database (version 52; BLASTX e-value<10-5, Bit score>50, and >50% identity) 51. For query genes with multiple matches, the annotated reference gene with the lowest e-value was used. When multiple annotated genes with an identical e-value were encountered after a BLAST query, we included all KOs assigned to those genes. Genes from the database with significant homology (BLASTN e-value<10−20) to non-coding transcripts from the 539 microbial genomes were excluded from subsequent analysis.
Metagenomic sequence analysis
DNA- and RNA-seq analysis was performed with our recently described pipeline 45. Briefly, high-quality reads (see Extended Data Table 3 for sequencing statistics) were mapped using SSAHA2 52, to our custom 539-genome database and the Illumina adaptor sequences (SSAHA2 parameters: “-best 1 -score 20 -solexa”). The number of transcripts assigned to each gene was then tallied and normalized to reads per kilobase per million mapped reads (RPKM). To account for genes that were not detected due to limited sequencing depth, a pseudocount of 0.01 was added to all samples. Samples were clustered in Matlab (version 7.10.0) based on gene expression or abundance. Genes were grouped by KOs by calculating the cumulative RPKM for each sample for genes present in at least 6 samples. We used HUMAnN, a recently developed software package for metabolic reconstruction from metagenomic data 53, followed by LefSe analysis to identify metagenomic biomarkers 50. A modified version of the “SConstruct” file was used to input KO counts into the HUMAnN pipeline for each RNA-seq dataset. We then ran LefSe on the resulting KEGG module abundance file using the “-o 1000000” flag. We used the edgeR package 54 to identify orthologous groups with significantly altered abundance or expression. Prior to analysis, we calculated the cumulative number of sequencing reads assigned to each sample for each KO (without RPKM normalization). We then used a paired glm analysis to determine consistent changes within each animal following LPS treatment. Default parameters were used, with one exception: for the estimateGLMTrendedDisp step “min.n” was set to 50/300 for the genomes and KO, respectively. Significance was accepted at a false discovery rate (FDR) < 0.05 and >2-fold change.
PCR and RT-PCR
RNA was isolated from mouse tissues and gut contents by the guanidinium thiocyanate/cesium chloride gradient method 46. RNA was DNase treated (Sigma) and reverse transcribed with Superscript III (Invitrogen). Primer sequences were as follows: fut1F 5’-CAAGGAGCTCAGCTATGTGG-3’, fut1R 5’-GACTGCTCAGGACAGGAAGG-3’; fut2F 5’-ACAGCCAGAAGAGCCATGGC -3’, fut2R 5’-TAACACCGGGAGACTGATCC-3’; sec1F 5’-ATCCAAGCAGTGCTCCAGC-3’, sec1R 5’-CAATATTCGCCCATCTGGTTC-3’; villinF 5’-GCTTGCCACAACTTCCTAAG-3’, villinR 5’-CTTGCTTGAAGTAGCTCCGG-3’. Quantitative RT-PCR was performed on an Applied Biosystems (Foster City, CA) StepOnePlus instrument with Universal Sybr Green Universal Supermix (Bio-Rad), and the following primers27: E. coli fucA F 5’-GGCGCGCAAGGAATAGAA-3’, E. coli fucA R 5’-GATCCCCGCTATTCACTACATGA-3’; E. coli fucP F 5’-CCAAATACGGTTCGTCCTTCA-3’, E. coli fucP R 5’-ACCCATGACCGGAGTGACAA-3’; E. coli rpoA F 5’-GCGCTCATCTTCTTCCGAAT-3’, E. coli rpoA R 5’-CGCGGTCGTGGTTATGTG-3’.
Statistics
Statistical analyses were performed with GraphPad Prism 5 software (GraphPad Software, Inc, La Jolla, CA).
Extended Data
Extended Data Figure 1. Requirements and kinetics for SI fucosylation induced by systemic injection of TLR ligands.
a, Systemic injection of bacterial TLR ligands induces small intestine fucosylation, but simple starvation does not.
UEA-1 staining (as in Fig. 1) after i.p. injection of CpG DNA, or Pam3CSK4, or food deprivation for 24 h of BALB/c SPF mouse. b, LPS injection causes SI fucosylation in various inbred mouse strains. SPF mice of the indicated strains were injected with LPS i.p. and SI was stained with UEA-1 after 24 hours, as in Fig. 1. Representative of at least 2 independent experiments. c, Fucosylation peaks at 8 h after LPS injection and is still detectable at 96 h. d, M cells can be readily detected by SEM and UEA-1 staining of the domes of the Peyer's patches, but are rare in the villi and are not massively induced in the villi by LPS injection. UEA-1 staining and SEM were performed on adjacent pieces from proximal 1/3 of SI. Scale bars=100 μm for UEA-1 staining, 50 μm for SEM images. Representative of at least 2 independent experiments. e, SI fucosylation does not require the presence of endogenous microbiota (LPS injection in GF mouse) and is not induced by oral administration of LPS (1 mg).
Extended Data Figure 2. MyD88-dependent pathway for fucosylation of SI IECs in response to systemic stimulation of TLRs.
a, FACS analysis of IECs from three segments of small intestine from the indicated mice. Cells are gated on the FSC/SSC high epithelial cell population. At least 2 mice per mutant genotype were stained along with two control mice in the experiments shown.
b, SPF mice were pretreated with 20 mg streptomycin and orally infected with Salmonella typhimurium. SI was stained at 24 hours p.i. MyD88 expression was necessary in CD11c+ cells but not villin+ IECs for S. typhimurium-induced fucosylation.
Extended Data Figure 3. A proposed model for the mechanisms linking inducible fucosylation to the gut microbiota.
Systemic microbial agonists activate TLRs on CD11c+ DCs, causing secretion of the cytokine IL-23, which in turn stimulates RORγt-dependent innate lymphoid cells (ILCs) to secrete IL-22. IL-22 causes small intestine epithelial cells to upregulate the α(1,2)fucosyltransferase 2 enzyme (Fut2). Fucosylated proteins are either secreted into the lumen or expressed on the cell surface and later shed into the lumen. Fucosidase-expressing bacteria (blue) liberate fucose residues, which they can utilize and share with other bacteria lacking the fucose-cleaving enzyme. Bacterial metabolism of fucose potentially produces metabolites such as SCFAs. Fucose also directly or indirectly downregulates virulence gene expression by pathobionts (red) or bona fide pathogens27.
Extended Data Figure 4. Consequences of LPS injection in Fut2-sufficient and Fut2-deficient BALB/c mice.
a, Inflammatory cytokines IL-1β, IL-6 and TNF-α were measured by ELISA in sera of mice before or 2 h after injection with LPS (4 h for IL-1β). abx, mice on antibiotic water for 2 days prior to injection. Bars are mean±s.e.m.; n.d., not detected.
b, Expression of RegIIIγ (also regulated by the MyD88-IL23-IL22 pathway). Measurement by qPCR of reg3g gene expression in mid-SI tissue, relative to gapdh (ddCt method). Numbers indicate mean fold change±s.e.m. in LPS-treated vs. untreated mice. Differences between LPS-treated Fut2+ and abx or Fut2− levels are not significant (P>0.05, two-tailed Student's t test).
c, Weight loss and recovery is not different in Fut2+/− and Fut2−/− mice after simple starvation (mean±s.e.m., P>0.05 at all time points, two-tailed Student's t test).
d, Lack of direct toxic effect of antibiotics (abx) measured as the weight loss of BALB/c GF animals treated with LPS i.p (mean±s.e.m., P>0.05 by two-tailed Student's t test at all timepoints).
e-g, Similar total bacterial loads in Fut2+/− and Fut2−/− mice before and after LPS injection and antibiotic treatment. Total bacterial loads in feces were estimated by plating on aerobic (e) and anaerobic (f) non-selective media, and by quantitative PCR for 16S gene copies (g). There were no significant differences between Fut2-sufficient (filled circles) and Fut2-deficient (open circles) mice before or after LPS treatment (two-tailed Student's t test). Circles - individual mice. Horizontal lines - means. Red circles- antibiotic-treated mice.
Extended Data Figure 5. Fucosylated protein in IECs and gut contents.
a, Proteins α(1,2)fucosylated in IECs after LPS injection identified by UEA-1 precipitation and mass spectrometry. Abundance is the number of peptide fragments attributed to each gene. b, IECs from Fut2+ untreated, Fut2+ LPS-treated, or Fut2− untreated mice were isolated, and lysates separated by SDS-PAGE. α(1,2)fucosylated proteins were detected by blotting with UEA-1 lectin conjugated to HRP. c, Identical gel stained with Coomassie blue for total protein content. d, Relative density of the boxed area of each lane from b divided by the relative density in c. e, UEA-1 staining of luminal proteins as in Fig. 3c. Blot is overexposed to show absence of luminal fucosylated proteins in the LPS-treated, Fut2− mouse.
Extended Data Figure 6. Generation of fucose-sensing reporter bacterial strains.
a, Reporter E. coli were grown to stationary phase in minimal medium containing 10mM glucose and the indicated concentrations of L-fucose (* indicates promoterless vector), and GFP fluorescence was measured.
b, Fucosidase activity is dramatically reduced after 2 days of antibiotics (abx) treatment but recovers after cessation of treatment. Measurement of total α-L-fucosidase activity in feces. Fecal supernatant was assayed for cleavage of 4-methylumbelliferyl-fucopyranoside substrate by fluorescence. n = 5 SPF abx-treated, 3 GF mice.
c, Fecal homogenates were plated anaerobically on BHIS agar containing 5-bromo-4-chloro-3-indolyl α-L-fucopyranoside, which forms a blue precipitate upon cleavage of the fucosyl residue. Both blue and white colonies are present.
d, Pure cultures of Bacteroides species were streaked on the same medium as in c. B. uniformis (left) is not predicted to carry an α-L-fucosidase gene, and remains white; B. acidifaciens (middle) and B. thetaiotaomicron (right) both express fucosidase activity and develop blue colonies.
e, Loss of B. acidifaciens from the feces of mice treated with antibiotics (Abx) in water (PCR for the gyrB gene). C-, water control. C+, B. acidifaciens genomic DNA.
f, Summary of reporter E. coli experiments in SPF mice (representative experiment is shown in Fig. 3e). Points are mean GFP fluorescence from all reporter bacteria measured in each of 3 independent experiments (n = 65 bacteria per mouse; *P<0.05, Student's t test).
Extended Data Figure 7. Microbial community structure is impacted by co-housing yet robust to host fucosylation and LPS exposure, whereas microbial gene expression depends on Fut2.
a, Stable relative abundance of bacterial phyla across treatment groups and genotypes, as indicated by 16S rRNA gene sequencing. Values represent the mean abundance of phyla found at >1% relative abundance in at least one sample.
b, Unweighted UniFrac analysis of the gut microbiota of Fut2-deficient (no outline) and Fut2-sufficient (black outline) mice. Points are colored based on kinship and labeled by timepoint (before or after LPS exposure). Results are based on 180,000 randomly selected 16S rRNA gene sequences/sample.
c, Microbial diversity as measured by the Shannon Diversity Index (n = 178,100 sequences/sample). Values are mean±s.e.m. (n = 3 Fut2+, 4 Fut2− mice/timepoint).
d, KEGG modules and pathways expressed in microbiota at higher levels after LPS exposure in Fut2-positive (left) and Fut2-negative mice (right) (n = 3/group; Humann/LefSe analysis; LDA>2).
Extended Data Figure 8. Lack of indicible fucosylation and SI colonization in C rodentium infected mice.
a, C. rodentium causes no SI fucosylation in SPF mice at day 3, day 7, or day 12 p.i.
b, SI colonization by C. rodentium is low regardless of Fut2 expression and LPS treatment. SI contents were removed by gentle squeezing, homogenized in PBS, and plated. Means±s.e.m; n = 4. Dotted line shows the limit of detection.
Extended Data Figure 9.
Effect of exogenous fucose on cecal short-chain fatty acid (SCFA) levels. Cecal SCFAs were measured after gavaging starved mice with the indicated sugars (100 mM concentration). Fucose gavage leads to increased propionate production in SPF but not GF mice. Means±s.e.m.; **P<0.01, Student's two-tailed t test.
Supplementary Material
Acknowledgments
We thank C. Reardon and C. Daly for sequencing support, Dr. Honggang Ye for help with metabolic cage analysis, and Dr. Nathan F. Dalleska for assistance and use of GC-MS instrumentation in the Environmental Analysis Center at the California Institute of Technology and Dr. Gabriel Nuñez for luciferase-expressing C. rodentium. This work was supported by grants from the NIH (P50 GM068763 to PJT, AI96706 and AI42135 to EGP, T32 AI065382 to JMP, the Harvard Bauer Fellows Program, NSF grant EFRI-1137089 to RFI, Digestive Disease Research Core Center grant DK42086 and a Kenneth Rainin Foundation grant to AVC.
Footnotes
Author Contributions: JMP, MAK, MCA and EGP performed analysis of inducible fucosylataion in mice including mutant strains; JMP and CFM produced DNA and RNA sequencing data and PJT analyzed these data; DS produced MyD88fl/fl mice; TVG produced GF BALB/c mice and performed cytokine ELISA analysis; SRB and RFI performed analysis of SCFA; RFI, EGP and PJT contributed to writing of the manuscript; AVC conceived the project, analyzed the results and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Author Information: The DNA and RNA shotgun sequencing data have been deposited in the Gene Expression Omnibus (GEO) database and are accessible through GEO Series accession number GSE60874; 16S rRNA gene sequencing reads are deposited in MG-RAST with the accession number 10494. The authors declare no competing financial interests.
Supplementary information is linked to the online version of the paper at www.nature.com/nature
References
- 1.Ayres JS, Schneider DS. Tolerance of infections. Annu Rev Immunol. 2012;30:271–294. doi: 10.1146/annurev-immunol-020711-075030. doi:10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- 2.Ayres JS, Schneider DS. The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS biology. 2009;7:e1000150. doi: 10.1371/journal.pbio.1000150. doi:10.1371/journal.pbio.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray MJ, Murray AB. Anorexia of infection as a mechanism of host defense. Am J Clin Nutr. 1979;32:593–596. doi: 10.1093/ajcn/32.3.593. [DOI] [PubMed] [Google Scholar]
- 4.Kyriazakis II, Tolkamp BJ, Hutchings MR. Towards a functional explanation for the occurrence of anorexia during parasitic infections. Animal behaviour. 1998;56:265–274. doi: 10.1006/anbe.1998.0761. doi:10.1006/anbe.1998.0761. [DOI] [PubMed] [Google Scholar]
- 5.Exton MS. Infection-induced anorexia: active host defence strategy. Appetite. 1997;29:369–383. doi: 10.1006/appe.1997.0116. doi:10.1006/appe.1997.0116. [DOI] [PubMed] [Google Scholar]
- 6.Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Current opinion in microbiology. 2011;14:82–91. doi: 10.1016/j.mib.2010.10.003. doi:10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Bocci V, Winzler RJ. Metabolism of L-fucose-1-14C and of fucose glycoproteins in the rat. The American journal of physiology. 1969;216:1337–1342. doi: 10.1152/ajplegacy.1969.216.6.1337. [DOI] [PubMed] [Google Scholar]
- 8.Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. doi:10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 9.Kashyap PC, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013;110:17059–17064. doi: 10.1073/pnas.1306070110. doi:10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. doi:10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umesaki Y, Tohyama K, Mutai M. Appearance of fucolipid after conventionalization of germ-free mice. Journal of biochemistry. 1981;90:559–561. doi: 10.1093/oxfordjournals.jbchem.a133506. [DOI] [PubMed] [Google Scholar]
- 12.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 13.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. doi:10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 14.Clark MA, Jepson MA, Simmons NL, Booth TA, Hirst BH. Differential expression of lectin-binding sites defines mouse intestinal M-cells. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1993;41:1679–1687. doi: 10.1177/41.11.7691933. [DOI] [PubMed] [Google Scholar]
- 15.Kinnebrew MA, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. doi:10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomsson KA, et al. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fucalpha1-2 glycosyltransferase. The Biochemical journal. 2002;367:609–616. doi: 10.1042/BJ20020371. doi:10.1042/BJ20020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmen JM, Olson FJ, Karlsson H, Hansson GC. Two glycosylation alterations of mouse intestinal mucins due to infection caused by the parasite Nippostrongylus brasiliensis. Glycoconjugate journal. 2002;19:67–75. doi: 10.1023/a:1022589015687. [DOI] [PubMed] [Google Scholar]
- 18.Domino SE, Zhang L, Lowe JB. Molecular cloning, genomic mapping, and expression of two secretor blood group alpha (1,2)fucosyltransferase genes differentially regulated in mouse uterine epithelium and gastrointestinal tract. The Journal of biological chemistry. 2001;276:23748–23756. doi: 10.1074/jbc.M100735200. doi:10.1074/jbc.M100735200. [DOI] [PubMed] [Google Scholar]
- 19.Domino SE, Zhang L, Gillespie PJ, Saunders TL, Lowe JB. Deficiency of reproductive tract alpha(1,2)fucosylated glycans and normal fertility in mice with targeted deletions of the FUT1 or FUT2 alpha(1,2)fucosyltransferase locus. Molecular and cellular biology. 2001;21:8336–8345. doi: 10.1128/MCB.21.24.8336-8345.2001. doi:10.1128/MCB.21.24.8336-8345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. doi:10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hovel-Miner G, Faucher SP, Charpentier X, Shuman HA. ArgR-regulated genes are derepressed in the Legionella-containing vacuole. Journal of bacteriology. 2010;192:4504–4516. doi: 10.1128/JB.00465-10. doi:10.1128/JB.00465-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Yen MR, Saier MH., Jr. Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli. Journal of bacteriology. 2010;192:2013–2019. doi: 10.1128/JB.01085-09. doi:10.1128/JB.01085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. doi:10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. doi:10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barba J, et al. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. Journal of bacteriology. 2005;187:7918–7930. doi: 10.1128/JB.187.23.7918-7930.2005. doi:10.1128/JB.187.23.7918-7930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. Journal of bacteriology. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. doi:10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacheco AR, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. doi:10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Vadder F, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. doi:10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 29.McGovern DP, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. doi:10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrow AL, et al. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. The Journal of pediatrics. 2011;158:745–751. doi: 10.1016/j.jpeds.2010.10.043. doi:10.1016/j.jpeds.2010.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinridders A, et al. MyD88 signaling in the CNS is required for development of fatty acid-induced leptin resistance and diet-induced obesity. Cell metabolism. 2009;10:249–259. doi: 10.1016/j.cmet.2009.08.013. doi:10.1016/j.cmet.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eberl G, et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. doi:10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 33.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. doi:10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 34.Zenewicz LA, et al. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647–659. doi: 10.1016/j.immuni.2007.07.023. doi:10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malick LE, Wilson RB. Modified thiocarbohydrazide procedure for scanning electron microscopy: routine use for normal, pathological, or experimental tissues. Stain technology. 1975;50:265–269. doi: 10.3109/10520297509117069. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto M, Ohkuma M. Identification and classification of the genus Bacteroides by multilocus sequence analysis. Microbiology. 2011;157:3388–3397. doi: 10.1099/mic.0.052332-0. doi:10.1099/mic.0.052332-0. [DOI] [PubMed] [Google Scholar]
- 37.Boronat A, Aguilar J. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. Journal of bacteriology. 1979;140:320–326. doi: 10.1128/jb.140.2.320-326.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. doi:10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. doi:10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleming SE, Traitler H, Koellreuter B. Analysis of volatile fatty acids in biological specimens by capillary column gas chromatography. Lipids. 1987;22:195–200. doi: 10.1007/BF02537302. [DOI] [PubMed] [Google Scholar]
- 42.Tangerman A, Nagengast FM. A gas chromatographic analysis of fecal short-chain fatty acids, using the direct injection method. Analytical biochemistry. 1996;236:1–8. doi: 10.1006/abio.1996.0123. doi:10.1006/abio.1996.0123. [DOI] [PubMed] [Google Scholar]
- 43.Zhao G, Nyman M, Jonsson JA. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomedical chromatography : BMC. 2006;20:674–682. doi: 10.1002/bmc.580. doi:10.1002/bmc.580. [DOI] [PubMed] [Google Scholar]
- 44.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. doi:10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. doi:10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 47.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. doi:10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108 Suppl. 2011;1:4516–4522. doi: 10.1073/pnas.1000080107. doi:10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. doi:10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. doi:10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. doi:10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome research. 2001;11:1725–1729. doi: 10.1101/gr.194201. doi:10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS computational biology. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. doi:10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. doi:10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.