Abstract
Objectives
Horizontal transfer of integrons is one of the important factors that can contribute to the occurrence of multidrug-resistant (MDR) bacteria. This study aimed to determine the prevalence of integrons among MDR Escherichia coli strains isolated from stool specimens and investigate the associations between the existence of integrons and MDR properties in the southwest of Iran.
Methods
There were 164 E. coli strains isolated from January 2012 to June 2012. Fecal specimens identified as E. coli by the conventional methods. Subsequently the antibiotic resistance was assessed using Clinical and Laboratory Standard Institute criteria. The presence of class 1–3 integrons and embedded gene cassettes was verified using specific primers by multiplex polymerase chain reaction assay.
Results
Among a total of 164 studied samples, 69 (42.07%) isolates were multidrug resistant. Class 1 and class 2 integrons were present in 78.26% and 76.81% MDR isolates, respectively. For the first time in Iran, class 3 integron was observed in 26.09% MDR isolates. Significant correlations were identified between: class 1 integron and resistance to amikacin, gentamicin, chloramphenicol, ampicillin, tetracycline, nalidixic acid, and co-trimoxazole; class 2 integron and resistance to aminoglycosides, co-trimoxazole, cefalexin, ampicillin, and chloramphenicol; and class 3 integron and resistance to gentamicin, kanamycin, and streptomycin.
Conclusion
Our results indicate that integrons are common among MDR isolates and they can be used as a marker for the identification of MDR isolates. Therefore, due to the possibility of a widespread outbreak of MDR isolates, molecular surveillance and sequencing of the integrons in other parts of the country is recommended.
Keywords: class 3 integron, diarrheagenic Escherichia coli, Iran, multidrug resistance
1. Introduction
The appearance of resistance to various antimicrobial agents in pathogenic bacteria has become an important public health threat. Isolates are categorized as: pandrug resistance, extensive drug-resistance, multidrug resistance (MDR), and nonmultiresistant, if resistance is found to all, to all except 1 or 2, to ≥3, and to <3 antibiotic classes, respectively [1,2].
The phenomenon of MDR is a major health care problem among pathogenic bacteria such as Escherichia coli, where it is associated with increased mortality and morbidity, worldwide [3,4]. Although it is clear that antibiotic utilization is pivotal in the selection of bacterial resistance, the facile spread of resistance genes has been a fundamental force in the rapid evolution of resistance to a wide range of unrelated antibiotics among diverse bacteria.
The spread of antimicrobial resistance in bacteria is a complex process involving a diversity of different mechanisms. Susceptible bacteria may obtain resistance through mutations or the transfer of resistance genes located on mobile DNA elements such as integrons [5,6]. MDR in intestinal bacteria such as Escherichia coli is known to be associated with integrons [7].
Integrons were defined by Hall and Collis as DNA elements that function as gene-capture and expression systems [8,9,12]. This element contains three necessary components located within the 5′ conserved segment include: an integrase gene (IntI), which encodes a site-specific recombinase enzyme; an attI site [8], which is recognized by the integrase and acts as an acceptor for gene cassettes; and a promoter region (PC) [7,10,11]. Gene cassettes become a part of the integron when integrated [12–14].
Although integrons are not mobile, they can be transferred between bacteria by transposons or plasmids in which they are present. Accordingly, integrons are a major mechanism for the spread of multidrug resistance [15]. Three types of integrons, each with different int genes have been identified (IntI1, IntI2, and IntI3) that are known to be associated with antibiotic resistance [7].
Class 1 integrons have been reported in many Gram-negative bacteria, including Acinetobacter, Vibrio, Aeromonas, Proteus, Burkholderia, Alcaligenes, Campylobacter, Enterobacter, Citrobacter, Klebsiella, Mycobacterium, Pseudomonas, Serratia, Salmonella, Shigella, and Escherichia [14]. Class 2 integrons are embedded in the Tn7 family of transposons and have been found in Salmonella, Acinetobacter, Escherichia, Shigella, Aeromonas, and Morganella [13]. Class 3 integrons appear to be much less common and so are less involved in the spread of multidrug resistance. Class 3 integrons have been described in Acinetobacter spp., Citrobacter freundii, Pseudomonas aeruginosa, Escherichia coli, Serratia marcescens, Alcaligenes xylosoxidans, Pseudomonas putida, Salmonella spp., Klebsiella pneumonia, and Delftia spp. [13,14,16].
Several studies have investigated prevalence of integrons in MDR Escherichia coli isolates around the world. These studies found a substantial association between the presence of integrons and antibiotic resistance. However, there is not enough information available on spread of class 1–3 integrons and their association with MDR in diarrheagenic Escherichia coli in our area of research. This study aimed to assess the prevalence of three classes of integrons in a MDR diarrheagenic Escherichia coli strains isolated from children <5 years in the southwest of Iran and investigates associations between MDR and the existence of integrons.
2. Materials and methods
2.1. Type of study and microorganism identification
This descriptive cross-sectional study was performed on a total of 164 fecal samples were recovered from children aged between 1 month and 5 years, referred to Yasouj Hospital (Emam Sajad Hospital, Dena and Yazdanpanah private laboratories, Yasouj-Iran), in a period of 5 months (January 2012 to June 2012). Fecal samples were seeded on eosin methylene blue agar plates and incubated at 37°C for 24 hours. One colony from each sample with a typical Escherichia coli morphology was recovered and verified by standard biochemical tests (Triple Sugar Iron, Sulfide indole motility, Methyl Red/Voges-Proskauer, Lysine Iron Agar, citrate, urea). After identification, the isolates were subcultured in tryptic soy broth (Oxoid, UK) and were then stored at −70°C for further investigations.
2.2. Antibiotic susceptibility testing
Antimicrobial susceptibility of all isolates was determined using the standard Kirby–Bauer disk diffusion method according to Clinical and Laboratory Standard Institute guidelines [17].Antimicrobial agents tested were, ampicillin, nalidixic acid, cefalexin, sulfamethoxazole-trimethoprim, gentamicin, tetracycline, streptomycin, kanamycin, chloramphenicol, and amikacin (Padtanteb, Tehran-Iran). The E. coli ATCC 25922 strain was used as a reference isolate. Intermediate sensitivity was considered as resistance.
2.3. Polymerase chain reaction
Extraction of genomic DNA from isolates was performed by the boiling method [3]. Briefly, a single colony of each organism was inoculated from a blood agar plate into 5 mL of Luria–Bertani broth (Sigma–Aldrich, Munich, Germany) and incubated for 20 hours at 37°C. Cells from the overnight culture were harvested by centrifugation at 12,000 × g for 5 minutes. Then the supernatant was decanted and the pellet was resuspended in 300–400 μL of sterile distilled water. Later the cells were lysed via heating at 95°C for 10 minutes and any cell debris was removed by centrifugation for 5 minutes at 12,000 × g. The supernatant was stored at −20°C and used as the source of the template for amplification.
The presence of class 1–3 integrons in all MDR E. coli isolates was tested by multiplex polymerase chain reaction (PCR) using primers specific for integrases genes of the integron, intI1, intI2, and intI3 (Table 1). The size of variable regions of class 1 and 2 integrons was determined by PCR assay. Primer sequences, sizes of PCR products, and PCR conditions are shown in Table 1. A tube containing PCR reaction without any DNA template was used as a negative control. All primers were obtained by Cinnagene Co. (Tehran, Iran).
Table 1.
Primers and PCR conditions used in this study.
| Gene | Primer sequence | Size of product (bp) | PCR conditions | Reference |
|---|---|---|---|---|
| IntI1-F | GGT CAA GGA TCT GGA TTT CG | 436 bp | 5 min at 94°C; 32 cycles of 1 min at 94°C, 1 min at 60°C, 2 min at 72°C; 10 min at 72°C | Machado, 2005 |
| IntI-R | ACA TGC GTG TAA ATC ATC GTC | |||
| IntI2-F | CAC GGA TAT GCG ACA AAA AGG | 788 bp | 5 min at 94°C; 32 cycles of 1 min at 94°C, 1 min at 60°C, 2 min at 72°C; 10 min at 72°C | Machado, 2005 |
| IntI2-R | TGTA GCA AAC GAG TGA CGA AAT G | |||
| IntI3-F | AGT GGG TGG CGA ATG AGT G | 600 bp | 5 min at 94°C; 32 cycles of 1 min at 94°C, 1 min at 60°C, 2 min at 72°C; 10 min at 72°C | Machado, 2005 |
| IntI3-R | TGT TCT TGT ATC GGC AGG TG | |||
| 5'CS | GGC ATC CAA GCA GCA AG | Variable | 5 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 58°C, 2 min at 72°C; 10 min at 72°C | Machado, 2005 |
| 3'CS | AAG CAG ACT TGA CCT GA | |||
| attI2-F | GAC GGC ATG CAC GAT TTG TA | Variable | 5 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 58°C, 2 min at 72°C; 10 min at 72°C | Machado, 2005 |
| orfX-R | GAT GCC ATC GCA AGT ACG AG |
The multiplex PCR assay was performed as follows. Each 50 μL of reaction mixture contained 0.5 μL dNTPs, 0.75 μL of each primer, 1.5 mM MgCl2, 2.5 μL 10× reaction buffer (10 mM Tris-HCl, 50 mM KCl), 0.25 μL of TaqDNA polymerase, and 5 μL of template DNA. The expected amplicons were analyzed by electrophoresis on 1.5% w/v agarose gel in TBE buffer.
2.4. Statistical analysis
Statistical analyses were performed by SPSS software version 15 (SPSS Inc., Chicago, IL, USA). The Chi-square test was used to calculate the association between antibiotic resistance and integration existence. The significance level was defined as p < 0.05.
3. Results
E. coli isolates were collected from 164 clinical specimens of patients referred to Yasouj Hospital. The isolates were obtained from children aged <5 years. Sixty-nine isolated strains (42.07%) were designated as MDR, with 23 (33.33%) isolates from female and 46 (66.67%) from male patients. The percentage of the resistant isolates to the tested antimicrobials is presented in Figure 1. Of the 69 MDR E. coli isolates, 54 (78.26%) isolates were identified as being positive for class 1 integron. PCR amplification of the integron cassette region occurred in 48 (69.56%) class 1 integron-containing isolates (Table 2). Class 2 integron was detected in 53 (76.81%) isolates. The integron cassette region could not be amplified by PCR in 24 (45.28%) of the class 2 integron-containing isolates (Table 3). The frequency of simultaneous occurrence of integrons is depicted in Figure 2. For the first time in Iran, class 3 integron was observed in 18 (26.09%) isolates. A significant correlation was revealed between class 1 integron and the gene cassettes with resistance to amikacin (p = 0.027), gentamicin (p = 0.040, p = 0.001), chloramphenicol (p = 0.026), ampicillin (p = 0.018, p = 0.000), tetracycline (p < 0.001), nalidixic acid (p = 0.026), and co-trimoxazole (p = 0.032, p = 0.015); and also between class 2 integron and gene cassette with resistance to kanamycin (p = 0.006), streptomycin (p = 0.006), amikacin (p = 0.005), gentamicin (p = 0.000), cefalexin (p = 0.029), co-trimoxazole (p = 0.021), ampicillin (p = 0.029), and chloramphenicol (p = 0.009). In addition, a significant association was found between class 3 integron and resistance to gentamicin (p = 0.031), kanamycin (p = 0.039), and streptomycin (p = 0.007).
Figure 1.
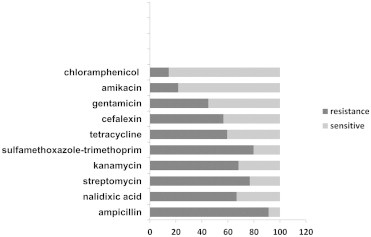
The percentages of antimicrobial resistance detected among multidrug-resistant E. coli isolates.
Table 2.
Sizes of variable regions of integron class I cassettes in intI1 positive isolates.
| Pattern of integron I cassettes bands (pb) | No. of isolates (%) |
|---|---|
| 750 | 2 (3.70) |
| 800 | 16 (29.63) |
| 1000 | 2 (3.70) |
| 1200 | 2 (3.70) |
| 1500 | 25 (46.30) |
| 2000 | 1 (1.85) |
| Without PCR product | 6 (11.12) |
| Total no. of intI1 positive isolates | 54 (100) |
Table 3.
Sizes of variable regions of integron class 2 cassettes in intI2 positive isolates.
| Pattern of integron 2 cassettes bands (pb) | No. of isolates (%) |
|---|---|
| 800 | 11 (20.76) |
| 1300 | 2 (3.77) |
| 1500 | 2 (3.77) |
| 2000 | 14 (26.42) |
| Without PCR product | 24 (45.28) |
| Total no. of intI2 positive isolates | 53 (100) |
Figure 2.
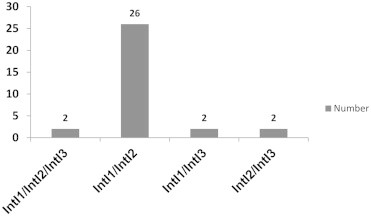
Frequency of simultaneous occurrence of integrons in multidrug-resistant isolates.
4. Discussion
Acceleration of the frequency and spectrum of antimicrobial resistant infections in recent years is a major public health concern. Investigations have suggested that regardless of antibiotic consumption pattern, resistance genes could be transferred between bacterial populations. The acquisition of resistance genes by horizontal transfer is currently thought to play a major role in the development of MDR strains [18].This study aimed to investigate the role of class 1–3 integrons in antibiotic resistance MDR diarrheagenic E. coli isolates.
In this study, MDR E. coli isolates with resistance to three or more different antibiotics were common. Sixty-nine isolates (42.07%) had the MDR phenotype, which is similar to the rate of MDR reported in E. coli isolates by Rezaee et al [3]. MDR E. coli isolates in our study were highly resistant to ampicillin (91.30%). High-level resistance to ampicillin (100%) among E. coli strains isolated from children has also been documented in Marvdasht, Iran [19]. Also, in the present study, resistance to sulfamethoxazole-trimethoprim, streptomycin, kanamycin, nalidixic acid, tetracycline, cefalexin, gentamicin, amikacin and chloramphenicol in MDR E. coli isolates were 79.71%, 76.81%, 68.12%, 66.67%, 59.42%, 56.52%, 44.93%, 21.74%, and 14.49, respectively.
However, Jones et al [20] reported a resistance to ampicillin 100%, cefalexin and streptomycin 89%, kanamycin and chloramphenicol 84%, gentamicin and tetracycline 79%, nalidixic acid 68%, and amikacin 68%. In another article by Murshed et al [21], resistance rates to cefalexin, co-trimoxazole, gentamicin, chloramphenicol, tetracycline, nalidixic acid, and ampicillin were noted as 79%, 75%, 54%, 50%, 77%, 83%, and 96%, respectively.
The various percentages of resistance in different parts of the world are due to differences in the prevalence of antibiotic consumption in each country [22]. In this study frequency of integrons of class 1, 2 and 3 were estimated as 78.26%, 76.81%, and 26.09%, respectively. The integron prevalence was relatively higher than in other investigations; for example, Jones et al [20] reported that 47% of MDR isolates carried class 1 and 2 integrons, whereas no integron class 3 was detected. Furthermore, research by Farshad et al [15] found frequencies of 25.6% for class 1 integron and 41.10% for class 2 integron, whereas Rezaee et al [3] reported 26.03% and 5.08% of class 1 and 2 integrons, respectively. In other research by Ranjbaran et al [23], a class 1 and 2 integron prevalence of 86% and 8% was reported, respectively. In contrast to investigations in Australia [24], Korea [25], France [26], Spain [27], China [28], Iran [3,15,23,29], Taiwan [9], Malaysia [30], and Pakistan [31], that did not detect any class 3, in the current study we found class 3 integron at a frequency of 26.09% in E. coli isolates, for the first time in Iran. In addition, here we studied the existence and the sizes of variable regions of class 1 and 2 integrons, using their specific primers by PCR technique, as we detected 48 (69.56%) and 29 (42.03%) isolates containing the gene cassette among 54 (78.26%) and 53 (76.81%) class 1 and 2 integron-bearing isolates, respectively. The gene cassettes ranged between 750 base pairs and 2000 base pairs, and amplicons of 1500 base pairs and 2000 base pairs were the most common gene cassette regions harbored in class 1 and 2 integrons, respectively. Moreover Machado et al 2007 [32] and Bakhshi et al 2012 [33] published similar results confirming our investigation.
As previously noted, the presence of integrons is closely related to resistance to quinolones, aminoglycoside compounds, trimethoprim, chloramphenicol, and β-lactam antibiotics [28,34–37]. We also detected a substantial correlation between class 1 integron, embedded gene cassette and resistance to amikacin, gentamicin, chloramphenicol, ampicillin, tetracycline, nalidixic acid, and co-trimoxazole, and also between class 2 integron, correspondent gene cassette and resistance to aminoglycosides, co-trimoxazole, cefalexin, ampicillin, and chloramphenicol. We also found a significant relationship between integron class 3 and the resistance to gentamicin, kanamycin, and streptomycin. We also confirmed that among a total of 69 MDR strains in this research, none of them contained unaccompanied class 3 integron, but it was found with high frequency co-occurring with class 1 and 2 integrons (Figure 2).
Considering the results obtained from our study, it was determined that some strains, despite being a MDR and having the integrase gene do not encompass the variable region. As Dawes et al [38] and Japoni-Nejad et al [39] reported, the differences in PCR results of integron identification, in the samples, could be due to variation in the 3′ region of the integron and the primer-binding site or the extensive size of the gene cassette. Also, as can be seen from Tables 2 and 3 and Figure 2, there are some strains containing multiple integrons and gene cassettes; therefore, it is concluded that there are multiple integrons present at the different chromosomal regions of the isolates.
In conclusion, recent investigations suggest that integrons are common among MDR isolates and they can be used as a marker for the identification of MDR isolates. This could lead to a serious threat of an outbreak of antimicrobial resistance development, which complicates the treatment of infections in the future, therefore precautionary measurements must be adopted to prevent the spread of these integrons. Eventually, considering the number of isolates bearing class 1 and class 2 integrons, lacking any gene cassette, it is suggested that the integron structures should be further investigated at the preserved regions in future researches.
Conflicts of interest
All contributing authors declare no conflicts of interest.
Acknowledgments
The authors acknowledge all staff members of the Biotechnology Research Center, Islamic Azad University, Shahrekord branch, Iran for their help and kind support during this study.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Magiorakos A., Srinivasan A., Carey R. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012 Mar;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 2.Japoni-Nejad A., Sofian M., Belkum A. Nosocomial outbreak of extensively and pan drug resistant Acinetobacter baumannii in tertiary hospital in central part of Iran. Jundishapur J Microbiol. 2013 Oct;6(8):e9892. [Google Scholar]
- 3.Rezaee M.A., Sheikhalizadeh V., Hasani A. First report of class 1 and class 2 integrons in multidrug-resistant (MDR) Escherichia coli strains isolated from clinical specimens in northern west of Iran. Jpn J Infect Dis. 2012;65(3):256–259. [Google Scholar]
- 4.Phongpaichit S., Wuttananupan K., Samasanti W. Class 1 integrons and multidrug resistance among Escherichia coli isolates from human stools. Southeast Asian J Trop Med Public Health. 2008 Mar;39(2):279–287. [PubMed] [Google Scholar]
- 5.Barlow R.S., Pemberton J.M., Desmarchelier P.M. Isolation and characterization of integron-containing bacteria without antibiotic selection. Antimicrob Agents Chemother. 2004 Mar;48(3):838–842. doi: 10.1128/AAC.48.3.838-842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Normark B.H., Normark S. Evolution and spread of antibiotic resistance. J Intern Med. 2002 Aug;252(2):91–106. doi: 10.1046/j.1365-2796.2002.01026.x. [DOI] [PubMed] [Google Scholar]
- 7.Bass L., Liebert C.A., Lee M.D. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob Agents Chemother. 1999 Dec;43(12):2925–2929. doi: 10.1128/aac.43.12.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire A.J., Brown D.F., Gray J.J. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting Gram-negative bacteria and its use in molecular epidemiology. Antimicrob Agents Chemother. 2001 Apr;45(4):1002–1009. doi: 10.1128/AAC.45.4.1022-1029.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinué L., Saénz Y., Somalo S. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J Antimicrob Chemother. 2008 Nov;62(5):934–937. doi: 10.1093/jac/dkn331. [DOI] [PubMed] [Google Scholar]
- 10.Leverstein-van Hall M.A., Blok H.E., Donders A.R.T. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J Infect Dis. 2003;187:251–259. doi: 10.1086/345880. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz F., Hafner D., Geisel R. Increased prevalence of class i integrons in Escherichia coli, Klebsiella species, and Enterobacter species isolates over a 7-year period in a German university hospital. J Clin Microbiol. 2001 Oct;39(10):3724–3726. doi: 10.1128/JCM.39.10.3724-3726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collis C.M., Hall R.M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992 Mar;174(5):1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fluit A.C., Schmitz F.J. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004 Apr;10(4):272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 14.Yu G., Li Y., Liu X. Role of integrons in antimicrobial resistance: A review. Afr J Microbiol Res. 2013 Apr;7(15):1301–1310. [Google Scholar]
- 15.Farshad S., Japoni A., Hosseini M. Low distribution of integrons among multidrug resistant E. coli strains isolated from children with community-acquired urinary tract infections in Shiraz, Iran. Pol J Microbiol. 2008;57(3):193–198. [PubMed] [Google Scholar]
- 16.Avila A. 2013. Prevalence and characterization of integrons in multidrug-resistant non-clinical enteric bacterial isolates. Presented to the faculty of the Department of Biological Sciences California State University, Sacramento. [Google Scholar]
- 17.Wikler M.A. Clinical and Laboratory Standards Institute; Wayne: 2011. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement. [Google Scholar]
- 18.Eslami G., Seyedjavadi S., Goudarzi H. Distribution of integrons among multidrug resistant E. coli and Klebsiella strains. Pejouhesh. 2010;34(1):61–65. [Google Scholar]
- 19.Kargar M., Homayoon M. Survey of enterohemorrhagic Escherichia coli (EHEC) and its antibiotic resistance among children less than 5 years in Marvdasht. Med Sci J Islamic Azad Univ Tehran Med Branch. Winter. 2009;19(4):268–273. [In Persian] [Google Scholar]
- 20.Jones L., McIver C., Rawlinson W. Polymerase chain reaction screening for integrons can be used to complement resistance surveillance programs. Commun Dis Intell Q Rep. 2003;27(Suppl.):S103–S110. [PubMed] [Google Scholar]
- 21.Murshed M., Shahnaz S., Abdul Malek M. Detection of resistance gene marker intl1 and antimicrobial resistance pattern of E. coli isolated from surgical site wound infection in Holy Family Red Crescent Medical College Hospital. Bangladesh J Med Microbiol. 2010;4(2):19–23. [Google Scholar]
- 22.WHO . World Health Organization; Geneva: 2012. The evolving threat of antimicrobial resistance: options for action.http://whqlibdoc.who.int/publications/2012/9789241503181_eng.pdf Available from: [accessed] [Google Scholar]
- 23.Ranjbaran M., Zolfaghari M., Japoni-Nejad A. Molecular investigation of integrons in Escherichia coli and Klebsiella pneumoniae isolated from urinary tract infections. J Mazand Univ Med Sci. 2013;23(105):20–27. [Google Scholar]
- 24.White P., McIver C., Rawlinson W. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001 Sep;45(9):2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H., Lee J., Kang H. Changes in gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J Clin Microbiol. 2003 Dec;41(12):5429–5433. doi: 10.1128/JCM.41.12.5429-5433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skurnik D., Menac'h A., Zurakowski D. Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob Agents Chemother. 2005 Jul;49(7):3062–3065. doi: 10.1128/AAC.49.7.3062-3065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machado E., Canton R., Baquero F. Integron content of extended-spectrum-β-lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob Agents Chemother. 2005 May;49(5):1823–1829. doi: 10.1128/AAC.49.5.1823-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su J., Shi L., Yang L. Analysis of integrons in clinical isolates of Escherichia coli in China during the last six years. FEMS Microbiol Lett. 2006 Jan;254(1):57–80. doi: 10.1111/j.1574-6968.2005.00025.x. [DOI] [PubMed] [Google Scholar]
- 29.Japoni A., Gudarzi M., Farshad S. Assay for integrons and pattern of antibiotic resistance in clinical Escherichia coli strains by PCR-RFLP in southern Iran. Jpn J Infect Dis. 2008 Jan;61(1):85–88. [PubMed] [Google Scholar]
- 30.Ibrahim N., Wajidi M., Yusof M. The integron prevalence of extended-spectrum betalactamase producing enterobacterial isolates in a Malaysian teaching hospital. Trop Biomed. 2011 Dec;28(3):668–671. [PubMed] [Google Scholar]
- 31.Muhammad I., Uzma M., Yasmin B. Prevalence of antimicrobial resistance and integrons in Escherichia coli from Punjab, Pakistan. Braz J Microbiol. 2011 Apr–Jun;42(2):462–466. doi: 10.1590/S1517-83822011000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machado E., Ferreira J., Novais A. Preservation of integron types among Enterobacteriaceae producing extended-spectrum β-lactamases in a Spanish Hospital over a 15-year period (1988 to 2003) Antimicrob Agents Chemother. 2007 Jun;51(6):2201–2204. doi: 10.1128/AAC.01389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakhshi B., Fallahzad S., Pourshafie M. The occurrence of atypical enteropathogenic Escherichia coli strains among children with diarrhea in Iran. J Infect Chemother. 2013 Aug;19(4):615–620. doi: 10.1007/s10156-012-0526-0. [DOI] [PubMed] [Google Scholar]
- 34.Chang C., Chang L., Chang Y. characterisation of drug resistance gene cassettes associated with class 1 integrons clinical isolates of Escherichia coli from Taiwan. ROC. J Med Microbiol. 2000 Dec;49(12):1097–1102. doi: 10.1099/0022-1317-49-12-1097. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Freijo P., Fluit A., Schmitz F. Class 1 integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998 Dec;42(6):689–696. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- 36.Rijavec M., Erjavec M., Avgustin J. High prevalence of multidrug resistance and random distribution of mobile genetic elements among uropathogenic Escherichia coli (UPEC) of the four major phylogenetic groups. Curr Microbiol. 2006 Aug;53(2):158–162. doi: 10.1007/s00284-005-0501-4. [DOI] [PubMed] [Google Scholar]
- 37.Singh R., Schroeder C., Meng J. Identification of antimicrobial resistance and class 1 integrons in Shiga toxin-producing Escherichia coli recovered from humans and food animals. J Antimicrob Chemother. 2005 Jul;56(1):216–219. doi: 10.1093/jac/dki161. [DOI] [PubMed] [Google Scholar]
- 38.Dawes F., Kuzevski A., Bettelheim K. Distribution of class 1 integrons with is26-mediated deletions in their 3′-conserved segments in Escherichia coli of human and animal origin. PLoS ONE. 2010 Sep;5(9):e12754. doi: 10.1371/journal.pone.0012754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Japoni-Nejad A., Farshad Sh, Van Belkum A. Novel cassette array in a class 1 integron in clinical isolates of Acinetobacter baumannii from central Iran. Int J Med Microbiol. 2013 Dec;303(8):645–650. doi: 10.1016/j.ijmm.2013.09.005. [DOI] [PubMed] [Google Scholar]


