Abstract
Background and objective
Intermountain Healthcare has a long history of using coded terminology and detailed clinical models (DCMs) to govern storage of clinical data to facilitate decision support and semantic interoperability. The latest iteration of DCMs at Intermountain is called the clinical element model (CEM). We describe the lessons learned from our CEM efforts with regard to subjective decisions a modeler frequently needs to make in creating a CEM. We present insights and guidelines, but also describe situations in which use cases conflict with the guidelines. We propose strategies that can help reconcile the conflicts. The hope is that these lessons will be helpful to others who are developing and maintaining DCMs in order to promote sharing and interoperability.
Methods
We have used the Clinical Element Modeling Language (CEML) to author approximately 5000 CEMs.
Results
Based on our experience, we have formulated guidelines to lead our modelers through the subjective decisions they need to make when authoring models. Reported here are guidelines regarding precoordination/postcoordination, dividing content between the model and the terminology, modeling logical attributes, and creating iso-semantic models. We place our lessons in context, exploring the potential benefits of an implementation layer, an iso-semantic modeling framework, and ontologic technologies.
Conclusions
We assert that detailed clinical models can advance interoperability and sharing, and that our guidelines, an implementation layer, and an iso-semantic framework will support our progress toward that goal.
Keywords: Medical Records Systems, Computerized/Standards; Semantics; Health Information Systems/Standards; Information Storage and Retrieval; Vocabulary, Controlled
Background and significance
Beginning in the 1970s, Intermountain Healthcare's HELP system stored structured patient data decorated with controlled terminology codes called ‘PTXT’ (Pointer-to-Text) codes. In the 1990s, building on their PTXT experience, Intermountain began developing ASN.1-based clinical event models to represent the various data elements that would be stored in their clinical data repository.1–4 These models would prove to be an early example of what is today known as detailed clinical models (DCMs).3–5 We at Intermountain now have hundreds of millions of data instances stored against these structures.
A DCM provides a specification—at a very granular level—of ‘valid data’ for a particular clinical concept (eg, a valid instance of a pneumonia finding or a valid instance of a heart rate measurement). An example of a DCM is shown in figure 1. Intermountain's latest rendition of DCMs (succeeding the ASN.1 clinical events) are called clinical element models (CEMs).6 7 The CEM strategy defines generic structures which are used for all models. The structures specify the data type of an element, its permissible values or range, whether the element is single valued or composed of multiple values, permissible qualifying information, etc. Typically in systems, if this information is maintained at all, it is distributed across master files, database schema definitions, programming logic, and documentation and is often inconsistent, across applications and subsystems.
Figure 1.
An example detailed clinical model (for a heart rate measurement).
We use these structures to model every element we seek to store in our electronic health record (EHR)—orders, observations, procedures, patients, encounters, referrals, etc. We expect CEMs to govern storage of all these elements, enhancing interoperability between applications and most notably, facilitating decision support. The ultimate objective, though, is that DCMs be shareable across platforms and institutions, promoting interoperability on a larger scale. The Clinical Information Modeling Initiative (CIMI)8 seeks to gain consensus across the modeling community on common formats and models.
Objective
Ensuring that all modelers (within an institution or across institutions) model consistently would reap dividends in accumulating a common set of shareable models. We at Intermountain have found, though, that agreeing on a syntax and fundamental modeling ‘building blocks’ (ie, our generic CEM structures) is insufficient to result in consistent models. Even given a single syntax and set of structures, multiple options sometimes exist for modeling a clinical element, none of which are ‘bad’ or ‘incorrect’. We strive to develop modeling ‘guidelines’ to foster consistency between models and to facilitate implementation of the models in a system. In this discussion, we attempt to explicate the lessons we have learned in the process, with the hope that they can be applied by others who are creating and implementing DCMs and ultimately facilitate CIMI's goals.
Materials and methods
The language we have used to express CEMs is the Clinical Element Modeling Language (CEML).6 7 We are currently developing a new version of CEML with two expression forms—XML-based and ‘tag-free’. We are also in the process of developing a CEML-specific authoring tool. We have a browser (clinicalelement.com) from which CEMs may be viewed and downloaded. We have a team of 10 modelers, all of whom have clinical backgrounds. We have approximately 5000 CEMs in CEML. What models we create and what we include in the models are directed by clinicians’ patient care requirements. We are presently in the process of developing a CEM-aware service layer atop our legacy systems.
Results
The discussion that follows presents principles that we have uncovered as we have created our CEMs.
Two major modeling issues
Creating DCMs is an inexact science. A modeler is often confronted with multiple alternative ways to create a model, each with its advantages and disadvantages. We have noticed that most modeling decisions distill down to one or both of two issues: (1) whether to precoordinate or postcoordinate, and (2) whether to put information in the information model or in the terminology model. These two issues will be discussed below.
Precoordination versus postcoordination
A model can represent its topic in either precoordinated or postcoordinated fashion.9 10 In precoordination, the meanings of multiple logical concepts are combined (coordinated) into a single attribute/value pair (where the value represents a composition of simpler concepts) before (‘pre’) storage or message transmission. In postcoordination, the meanings of the concepts are kept as multiple separate attribute/value pairs (where each attribute/value represents a simple concept) and are combined (coordinated) as needed (in a query or a display, for instance) after (‘post’) storage or transmission of an instance. We have found no definitive and universally applicable set of rules for determining when to precoordinate and when to postcoordinate. In general, though, postcoordination results in data in a form more consumable by decision support. For example, a model of an infection with explicit attributes for ‘infecting organism’ and ‘infected organ system’, such that ‘acute meningococcal pericarditis’ were stored as the combination of ‘acute inflammation’ with organ system=‘cardiovascular’, site=‘pericardial structure’, and infecting organism=‘Neisseria meningitides’ makes orthogonal queries for ‘all current patients with infections where the infecting organism=Neisseria meningitides’ or ‘all current patients with infections where organ system=cardiovascular’ easier than if ‘acute meningococcal pericarditis’ were modeled using a single precoordinated code. (It should be noted that robust terminology structure and services could also support such queries. In terminology, the concept ‘acute meningococcal pericarditis’ could have properties or relationships that indicate it is an infection of the ‘cardiovascular’ system and that its infecting organism is ‘Neisseria meningitides’. Data query services have typically not been integrated with terminology services sufficient to navigate such terminology at runtime, but as such functionality becomes more available it could change the modeling decisions we make.)
Precoordination, on the other hand, may result in data that are more natural in their expression. For example, most clinicians would rather see ‘laparoscopic cholecystectomy’ on a history of procedures than ‘procedure: removal, target site: gall bladder, method: laparoscopy’. Also, in data entry, it is much easier to select the precoordinated term ‘laparoscopic cholecystectomy’ than to separately select the procedure, target site, and method.
We have found no definitive and universally applicable set of rules for determining when to precoordinate and when to postcoordinate. We have, though, identified several general principles useful in making a determination:
A CEM (and, we assert, every DCM) is at its essence a ‘key/value pair’. For example, the heart rate measurement CEM, is a ‘key’ (Heart Rate Measurement) and a ‘value’ (60 beats per minute, 70 beats per minute, etc.). The value may be supplemented by other attributes (body location, body position, etc.), where each attribute is itself another key/value pair.
Precoordination/postcoordination decisions may pertain to either the name part or the value part.
Clinical usefulness should be the primary criterion in making the decision between precoordination and postcoordination.
For the key part, the most clinically useful solution seems to most often be postcoordination.
For the value part, the most clinically useful solution seems to most often be precoordination.
Two examples may illustrate.
Glucose tolerance lab results
The ‘key’ part of a glucose tolerance lab result model may be precoordinated (ie, individual models named ‘glucose 1 h post 75 g glucose’, ‘glucose 2 h post 100 g glucose’, etc.) or postcoordinated (ie, a single ‘glucose’ model containing attributes for ‘challenge dose’ and ‘time post challenge’). These options are shown in figure 2. The postcoordinated approach appears to be more clinically useful—a patient's ‘glucose’ is the element most often queried and presented on reports or review screens, rather than a patient's ‘glucose 1 h post 75 g glucose’, ‘glucose 2 h post 100 g glucose’, etc.
Figure 2.

Meaning in precoordinated keys versus meaning in multiple attributes. (A) A precoordinated approach in which multiple separate models, each with a precoordinated 'key', are created. (B) A postcoordinated approach in which a single model is created and differences are captured in multiple attributes.
Body location
The ‘value’ part of the ‘heart rate body location’ model (which captures the location on the body at which a heart rate measurement is taken) may be precoordinated (ie, its values drawn from the value set ‘left wrist’, ‘right wrist’, ‘inside left elbow’, ‘inside right elbow’, etc.) or postcoordinated (ie, its values drawn from the simpler set of values such as ‘wrist’, ‘inside elbow’, etc. and the laterality separately captured in another attribute). These options are shown in figure 3. Reasonable values in this value set will not encompass every location in the body, but just a relatively few locations with laterality (‘left wrist’, ‘right wrist’, ‘inside left elbow’, ‘inside right elbow’, etc.). Rather than separate location and laterality as two different attributes, the guidance would say to simply precoordinate the values.
Figure 3.

Meaning in precoordinated values versus meaning in value+attribute. (A) A precoordinated approach in which the value of ‘BodyLocation’ is drawn from a set of precoordinated values. (B) A postcoordinated approach in which the value of ‘BodyLocation’ is captured by the combination of a value and a laterality.
What to put in the model versus what to put in terminology or some other knowledge representation
A model declares the structure (ie, the valid attributes and data type) and semantics of an element of data that will be stored in an electronic medical record. By including certain attributes, it implies what data will be stored in an instance of the model and, by omitting other attributes, implies what data will not be stored in an instance of the model. In modeling a drug administration, for example, a modeler must decide whether to include an attribute for the classification (‘sedative’, ‘antibiotic’, etc.) of the drug being administered. Including it implies that it is important information to store in an instance of a drug administration. This is option ‘A’ in figure 4. Option ‘B’ in the figure omits the attribute and instead assumes the hierarchy information can be looked up in terminology.
Figure 4.
An example of a model versus terminology choice.
The general rule we follow is:
Put ‘static knowledge’ in terminology or some other knowledge representation (not CEMs).
Put ‘instance data’ in the model.
‘Static’ knowledge is knowledge that is independent of the patient—that is, will not vary between patients or between instances of patient data. Usually such knowledge is unchanging or slowly changing. It is best maintained centrally in a terminology server or knowledge server where applications that need the information can call a service to look it up. Storing the knowledge in each instance of data is redundant and inefficient.
Examples of static knowledge are:
IS-A relationships or categorizations, for example, ‘amoxicillin’ is a ‘penicillin’, ‘en-US’ (American English) is a form of ‘en’ (English), etc.
IS-PART-OF or compositional relationships, for example, ‘acetaminophen 300 mg tablet’ has strength ‘300 mg’.
Other descriptive relationships, such as:
‘Lovastatin’ has an elimination half life of 1–3 hours
‘diabetes’ has possible symptom of ‘excessive thirst’
‘blood pressure’ is commonly measured using an ‘arterial line’.
Knowledge that may change frequently, but is static relative to the patient. For example, the drugs in a hospital's formulary at a given time may change frequently, but are independent of the patient.
In contrast, ‘instance data’ is data that is dependent on (will vary with) the patient or the patient's care. This kind of information needs to be captured by model attributes. For example:
The device that was used to take this heart rate measurement (various devices could have been used).
The actual route of administration of this medication administration (various routes could have been used).
The approach site of this procedure (various sites could have been used).
Model only logical attributes
We have recognized the importance of separating the logical models from their implementation and have deliberately omitted some attributes from our CEMs, recognizing them as ‘metadata’ or ‘model of use’10–12 attributes as opposed to logical, ‘model of meaning’10–12 attributes. Our task is to model the elements needed to provide care for a patient—what a clinician needs to record. This is different from both (1) modeling the underlying physiologic structure or process, and (2) modeling the classes and attributes of an EHR or other system. For instance, in modeling a ‘heart rate measurement’, we are neither modeling (1) the heart or the process of the heart beating nor (2) a software class or storage structure that will be used to transmit or store a heart rate measurement. We attempt to model those attributes that a clinician desires to capture in measuring a heart rate (the number of beats per minute, the patient position, the method or device used, the time of the measurement, etc.) for proper interpretation and hence proper care of the patient. (How to set the ‘boundaries’ of the model is out of scope for this discussion, but a brief discussion of the topic is found in the online supplementary material.) We recognize that implementation of the models in a system or application certainly imposes other requirements on the models. However, since those latter requirements vary significantly depending on the use case addressed by the system (clinical trials vs clinical care, messaging vs EHR, etc.) and on the particular implementation, we seek to isolate the logical clinical requirements from those IT requirements (see Discussion section). We do not include, for example, ‘patient identifier’ and identifiers for the source system of the data. These and other such attributes are certainly necessary for implementing the models in an EHR system. However, they are not attributes we would consider to be logical parts of a ‘heart rate measurement’. Rather than place them in our logical CEMs, we use other means to ‘add in’ such attributes in our software implementation. This separation has the benefit of making the models less subject to implementation and use case differences and hence potentially more shareable.
This same strategy of separation of logical concerns from implementation has been used in other modeling approaches, such as the Care Assessment Scales and other structures developed in the Health Level 7 (HL7) v3 Care Provision Domain.13
Iso-semantic models
Ideally, to best promote interoperability, we would define one and only one model for each clinical element addressed in healthcare. That, however, proves a difficult proposition. There are a number of situations in which arguments can be made to model the same clinical element a number of different ways. When multiple models store the same information in different ways, they are said to be ‘iso-semantic’. Ideally, the transform between instances of iso-semantic models would be perfectly lossless.
One situation in which multiple models are possible involves ‘assertion’ models versus ‘context’ or ‘usage’ models. An ‘assertion’ model is a very fine grained assertion of a condition that exists in the patient, for example, a ‘pneumonia’ assertion model used to assert that the patient has pneumonia. The ‘pneumonia’ model would have qualifiers and qualifier values specific to the disorder. Similarly, we create a ‘nausea’ model, a ‘dyspnea’ model, a ‘myocardial infarction’ model, etc.
The presence of ‘pneumonia’, however, may be noted in numerous contexts. For example:
as a discharge diagnosis
as a cause of death
as a complication of surgery
as a problem on a problem list.
One approach to capturing ‘pneumonia’ in the context of a discharge diagnosis would be to create an instance of the ‘pneumonia’ assertion model and link it to a patient encounter instance with a ‘semantic link’ of type ‘discharge diagnosis of’.
Alternatively, though, we may create a more generic model for ‘discharge diagnosis’, with its value bound to a large value set of conditions (‘pneumonia’, ‘fever’, ‘pancreatitis’, etc.). An instance of ‘pneumonia’ as a discharge diagnosis would be captured as an instance of the discharge diagnosis model with its value set to ‘pneumonia’. The same ‘discharge diagnosis’ model would be used for discharge diagnoses of ‘pneumonia’, ‘fever’, ‘pancreatitis’, etc. We call such a model a ‘context’ or ‘usage’ model. Similarly we could create a ‘cause of death’ context model, a ‘complication of surgery’ context model, and a ‘problem’ context model.
The motivation for creating these context models is common usage. The context models map so easily to common use cases that it is very tempting to create them in addition to the individual assertion models. We say that a specific assertion model for a condition (eg, the ‘pneumonia’ model) is ‘iso-semantic’ with a context model whose value is set to a particular value (eg, the ‘discharge diagnosis’ model with its value set to ‘pneumonia’).
Adding another dimension to the iso-semantic model issue is the variety of ongoing overlapping information modeling efforts. The archetypes of OpenEHR and ISO 13606,14–17 the models of the ISO 13940 Continuity of Care (ContSys) standard,18 the HL7 v3 template efforts,19 and the Dutch Detailed Clinical Models20 are examples. Iso-semantic models may exist within the same modeling approach (eg, two iso-semantic CEMs), but they may also exist across modeling approaches (eg, a CEM that is iso-semantic with an ISO model). For example, what we have called usage/context models have been modeled by the ISO 13940 standard (problem lists, health issues, and health issue threads)18 and the HL7 v3 Care Provision Domain (health concerns).13 These models could be viewed as iso-semantic with the individual CEM assertion models.
Discussion
Through experience, we have developed guidelines that assist modelers in making the decisions they encounter. This fosters consistency and best practices in our modeling efforts. However, there have been cases in which implementation or application concerns caused us to make decisions that ran contrary to our guidelines. For example:
We added a ‘drug category’ to a drug administration model even though, according to the principles outlined in the ‘What to put in the model versus what to put in terminology or some other knowledge representation’ section, a drug's category should rightfully be maintained in terminology and not stored in every instance of a model. We added the ‘drug category’ attribute as a performance denormalization for decision support, so the system would not need to look up the drug category with a call to the terminology server.
Even though our ‘precoordination/postcoordination’ guidelines would have suggested a postcoordinated ‘glucose tolerance test result’ model with attributes for ‘challenge dose’ and ‘time post challenge dose’ (figure 2, option B), we instead constructed precoordinated challenge lab models (eg, a model for ‘glucose 2 h post 100 g glucose load’) because the precoordinated lab models were more compatible with the LOINC terminology (which precoordinates lab test names). (In this use case, to be able to use LOINC, LOINC would need to support postcoordinated code expressions as SNOMED21 22 does. This seemed unlikely—or at least a very long term solution—so we did not approach LOINC. In other situations, if LOINC were simply missing a code that we needed, we would simply request that the LOINC maintainers add the code.)
In a medication administration model, guidelines would favor a postcoordinated approach, with separate attributes for the drug, form, and strength. However, an exchange partner needed a precoordinated code and we doubted that the partner could navigate the necessary terminology relationships to get from the model's multiple attribute values to the needed precoordinated code. Consequently, we included both the precoordinated code and the postcoordinated attributes in the model.
Most if not all such exceptions could be resolved by implementing a more robust ‘implementation layer’ and an ‘iso-semantic model framework’ as described below.
Implementation layer
Having an implementation layer essentially means that the logical models are not compiled directly into the artifacts used in an implementing system. Instead, ‘implementation models’ serve as a buffer between the logical models and the system implementation. The implementation models might contain attributes not found in the logical models, constrain logical models, combine/split the logical models into classes more convenient for the system, or add processing instructions.
We have made some forays into creating such a layer. We have built mechanisms to ‘add in’ certain attributes that have deliberately been omitted from the logical models (eg, patient id and sending system id) at the point the models are used in the EHR. Enhancing these mechanisms to be more robust and flexible will permit us to be more faithful to the guidelines.
With regard to the first example of a compromise to our guidelines (the ‘drug category’ example), for instance, we could omit that attribute from the logical model as per guidelines. If the implementing system needs a ‘drug category’ attribute in the model for performance denormalization, it can be added to the implementation model via a compile step. This is depicted in figure 5. Indeed, one of our assumptions stated earlier—that the model implies what is stored in a patient record—need not be strictly true if good logical/implementation separation exists.
Figure 5.
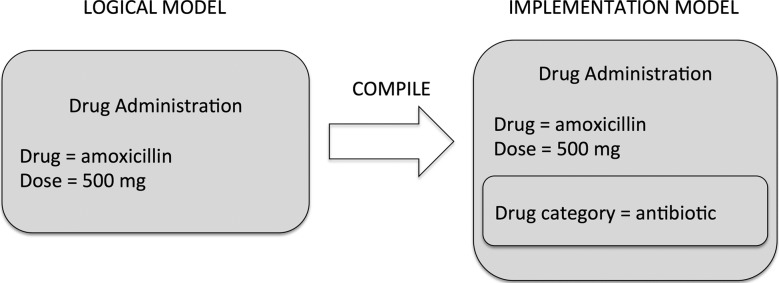
An example of an implementation model.
The layer must robustly address versioning, allowing the logical model to undergo revisions while preserving the implementation-specific modifications. As we advance our efforts to build a robust implementation layer, we will reduce the number of exceptions we make to our modeling guidelines. This kind of separation has been recognized by other modeling groups, such as HL7 v3's Patient Care Workgroup, under whose auspices the HL7 Care Provision structures13 have been developed.
Iso-semantic model framework
Rector10 suggests there be only one way of representing a given item of information or, alternatively, that there be a means of expressing transforms between models. When the former is not possible, an iso-semantic model framework would support the latter. It would allow for multiple models to be constructed for the same clinical element, as different use cases (eg, the needs of interfaces or user-facing applications) demand. When multiple models are called for, the goal should still be to designate just one of them as the ‘storage’ model (or the ‘exchange’ model if the use case is sharing between institutions). We refer to this designated model as a ‘canonical model’. Instances of the other models would be converted to instances of the canonical model before storage.
This strategy has significant architectural implications. It assumes mechanisms for:
marking members of a group of models as being ‘iso-semantic’
marking one of the members of the group as the ‘canonical’ model
detecting models that are potentially iso-semantic
expressing the ‘transforms’ from other members of the group to the ‘canonical’ model
a ‘transform engine’ that is able to read the transforms and transform an instance of one model to the appropriate iso-semantic canonical model.
Ontologic terminologies such as SNOMED CT,21 22 supplemented with more of the information contained in models, may be of use in detecting iso-semantic models and expressing transforms.
We have made some progress toward developing a framework. For instance, we transform a precoordinated model instance to a postcoordinated instance on storage via a ‘mapping’ stored in our knowledge repository. However, we have yet to develop a complete robust system. Combining an iso-semantic conversion framework with the implementation layer would allow the following with regard to the second and third examples of guideline exceptions previously described:
The canonical logical model for a glucose tolerance test result would be postcoordinated; if interface models that mapped better to LOINC were needed, they could be created as iso-semantic models. Those models could be used to accept interfaced data and the conversion to the canonical model could take place before storage. This is depicted in figure 6.
The canonical model for drug would bear just the postcoordinated drug, form, and strength attributes (and not the precoordinated clinical drug attribute). If a model containing the precoordinated attribute in addition were needed, it would be an iso-semantic implementation model.
Figure 6.
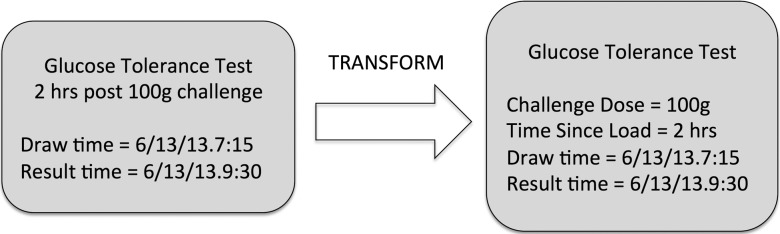
An example of an iso-semantic transform.
Ontologic technologies
Work on our detailed clinical models essentially began in the 1970s with PTXT structures followed by the ASN.1 clinical event models developed in the 1980s and 1990s. The CEM strategy originated in the early 2000s. Until now, we have refrained from expressing our models and terminology in semantic ontologic technologies (OWL/RDF) because of their complexity (and the consequent learning curve our clinically-oriented modelers would need to work through), concerns over suitability of ontologies to ‘modeling’ (eg, numeric values, cardinality, etc.), and the lack of good tooling. Adoption of the technologies by standards organizations, better tooling, and in general a ‘mainstreaming’ of the technologies has caused us to reconsider. We have begun to leverage OWL in organizing key terminology referenced by our models and colleagues have done considerable work attempting to represent CEMs in OWL.23–25 We recognize that expression of the model and terminology together, in an ontology, could provide benefit in the following areas mentioned in this text:
Decision of what to put in the model versus what to put in the terminology. Expressing both in an ontology avoids the classic information model/terminology model schism.
Detection of models that may be iso-semantic.
Mapping of iso-semantic models in an iso-semantic model framework.
Conclusion
We have described some of the guidelines we have gleaned for creating models consistently, described some of the challenges to using those guidelines, and proposed solutions for addressing exceptions, with the hope that our lessons learned will advance the DCM effort. Some argue that DCMs cannot possibly solve the problem of modeling all of biomedicine, with its diversity and complexity. The emerging field of genomics poses a particularly daunting challenge and it appears impossible to produce an exhaustive set of detailed clinical models to represent all data that healthcare will need. However, that need not preclude us from generating a set of models for commonly used elements that clinicians presently need to describe and document. We acknowledge that even within this set, different use case and implementation requirements will require variations in the models, but focus on robust implementation layering and iso-semantic frameworks can facilitate model sharing and thus promote the interoperability that we all seek.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the many individuals who have over the years participated in Intermountain's modeling efforts and shaped the thinking reflected in this manuscript.
Footnotes
Contributors: TAO wrote the manuscript. Through the years, all authors created models and engaged in many conversations that have evolved into the guidelines described. All authors reviewed the manuscript, some offering revisions and suggestions.
Funding: This manuscript was made possible in part by funding from the Strategic Health IT Advanced Research Projects (SHARP) Program (90TR002) administered by the Office of the National Coordinator for Health Information Technology. The contents of the manuscript are solely the responsibility of the authors.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Huff SM, Rocha RA, Solbrig HR, et al. Linking a medical vocabulary to a clinical data model using Abstract Syntax Notation 1. Methods Inf Med 1998;37:440–52 [PubMed] [Google Scholar]
- 2.Huff SM, Rocha RA, Bray BE, et al. An event model of medical information representation. J Am Med Inform Assoc 1995;2:116–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huff SM, Rocha RA, Coyle JF, et al. Integrating detailed clinical models into application development tools. Stud Health Technol Inform 2004;107(Pt 2):1058–62 [PubMed] [Google Scholar]
- 4.Coyle JF, Rossi Mori A, Huff SM. Standards for detailed clinical models as the basis for medical data exchange and decision support. Int J Med Inform 2003;69:157–74 [DOI] [PubMed] [Google Scholar]
- 5.Goossen W, Goossen-Baremans A, van der Zel M. Detailed clinical models: a review. Healthc Inform Res 2010;16:201–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coyle JF. The clinical element model—detailed clinical models [PhD dissertation]. Salt Lake City, Utah: University of Utah, 2013 [Google Scholar]
- 7.Coyle JF, Heras Y, Oniki T, et al. Clinical Element Model. November 14, 2008. http://clinicalelement.com (accessed 3 Jun 2014).
- 8.Mayo Clinic. Clinical Information Modeling Initiative wiki. Main Page. http://informatics.mayo.edu/CIMI. Published March 6, 2014. Accessed March 21, 2014
- 9.Parker CG, Rocha RA, Campbell JR, et al. Detailed clinical models for sharable, executable guidelines. Stud Health Technol Inform 2004; 107(Pt 1):145–8 [PubMed] [Google Scholar]
- 10.Rector AL. The interface between information, terminology, and inference models. Stud Health Technol Inform 2001;84(Pt 1):246–50 [PubMed] [Google Scholar]
- 11.Rector AL, Johnson PD, Tu S, et al. Interface of inference models with concept and medical record models. In: Quaglini S, Barahona P, Andreassen S, eds. Artificial Intelligence in Medicine Europe (AIME). Cascais, Portugal: Springer, 2001:314–23 [Google Scholar]
- 12.Rector A, Taweel A, Rogers J. Models and inference methods for clinical systems: A principled approach. In: Proc Medinfo-2004; San Francisco: North Holland; 79–83 [PubMed] [Google Scholar]
- 13.Health Level 7. Normative Edition HL7 V3 2013 Care Provision Domain. Ann Arbor: HL7 International, 2013 [Google Scholar]
- 14.Beale T. Archetypes and the EHR. Stud Health Technol Inform 2003;96:238–44 [PubMed] [Google Scholar]
- 15.Beale T. Archetypes: constraint-based domain models for future-proof information systems. In: Baclawski K, Kilov H. eds. Eleventh OOPSLA Workshop on Behavioral Semantics: Serving the Customer. Evanston, IL: Northeastern University, 2002:16–32 [Google Scholar]
- 16.ISO 13606–1:2008. Health informatics—Electronic health record communication—Part 1: Reference model
- 17.ISO 13606–2:2008. Health informatics—Electronic health record communication—Part 2: Archetype interchange specification
- 18.ISO/DIS 13940:2012. Health informatics—System of concepts to support continuity of care (draft)
- 19.Health Level 7. HL7 Templates Standard: Specification and Use of Reusable Information Constraint Templates, Release 1. DSTU: 2014 [Google Scholar]
- 20.Goossen WT. Using detailed clinical models to bridge the gap between clinicians and HIT. In: De Clercq, De Moor G, Bellon J, Foulon M, van der Lei J, eds. Collaborative Patient Centred eHealth. Amsterdam: IOS Press, 2008:3–10 [PubMed] [Google Scholar]
- 21.Schulz S, Suntisrivaraporn B, Baader F, et al. SNOMED reaching its adolescence: ontologists’ and logicians’ health check. Int J Med Inform 2009;78(Suppl 1):S86–94 [DOI] [PubMed] [Google Scholar]
- 22.International Health Terminology Standards Development Organization: Technical Implementation Guide. Jan 2014; http://ihtsdo.org/fileadmin/user_upload/doc/en_us/tig.html. (accessed 30 May 2014). [Google Scholar]
- 23.Tao C, Parker CG, Oniki TA, et al. An OWL meta-ontology for representing the Clinical Element Model. AMIA Annu Symp Proc 2011;2011:1372–81 [PMC free article] [PubMed] [Google Scholar]
- 24.Tao C, Jiang G, Oniki TA, et al. A semantic-web oriented representation of the clinical element model for secondary use of electronic health records data. J Am Med Inform Assoc 2013;20:554–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martínez-Costa C, Bosca D, Legaz-García MC, et al. Isosemantic rendering of clinical information using formal ontologies and RDF. Stud Health Technol Inform 2013;192:1085. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




