Abstract
Background. ZNRD1 was identified as a host protein required for the completion of the human immunodeficiency virus (HIV) lifecycle in a genome-wide screen using small interfering RNA gene silencing. Subsequently, a genome-wide association study (GWAS) of host determinants for HIV-1 disease identified an association of single nucleotide polymorphisms (SNPs) in the ZNRD1 region with CD4+ T-cell depletion.
Methods. We investigated the effects of SNPs in the ZNRD1 region on human immunodeficiency virus type 1 (HIV-1) infection and progression to clinical outcomes in 5 US-based HIV-1 longitudinal cohorts consisting of men who have sex with men, males with hemophilia, and injection drug users (IDUs) (n = 1865). SNP function was evaluated by electrophoretic mobility shift assay and promoter luciferase assay.
Results. A haplotype in the ZNRD1 gene showed significant association with a 35% decreased risk of HIV-1 acquisition (OR = 0.65, 95% CI, .47–.89), independent of HLA-C rs9264942, in European Americans. The SNP rs3132130 tagging this haplotype, located in the ZNRD1 5′ upstream region, caused a loss of nuclear factor binding and decrease in ZNRD1 promoter activity. ZNRD1 variants also affected HIV-1 disease progression in European- and African-American cohorts.
Conclusions. This study provides novel evidence that ZNRD1 polymorphism may confer host resistance to HIV-1 acquisition.
Keywords: HIV-1, infection, host susceptibility, AIDS, SNP, single nucleotide polymorphism, ZNRD1, genetic association
Individual variation in susceptibility to human immunodeficiency virus type 1 (HIV-1) acquisition, control of viral replication, and rate of disease progression is well documented [1–4]. The underlying biological mechanisms to individual susceptibility are multifactorial, with contributions from host genetic factors in addition to viral factors and environmental exposures [1–4]. Candidate gene association studies and genome-wide association studies (GWAS) have identified variation in host genes encoding HIV-1 coreceptor CCR5, and HLA- B (HLA-B*57), HCP5 (rs2395029), and HLA-C (rs9264942) that influence viral load and progression to AIDS [1, 2, 5–7]. However, only homozygosity for the CCR5 32 base-pair deletion (CCR5-Δ32), found on European ancestry chromosomes, has been securely identified as a resistant genetic factor to HIV acquisition [8, 9]. Other genetic factors associated with altered risk of HIV acquisition have not been consistently validated in independent studies [2, 10–13].
ZNRD1, a zinc ribbon domain-containing 1 protein, is a DNA-dependent RNA polymerase that catalyzes the transcription of DNA into RNA. ZNRD1 was first identified as one of 250 HIV-1 dependency factors required for HIV-1 replication in a genome-wide small interfering RNA (siRNA) knock-down experiment [14]. A separate siRNA knock-down experiment targeted only at ZNRD1 demonstrated a >50% reduction of R5 or X4-tropic HIV-1 replication in HeLa derived cells or lymphoid cells, likely through inhibition of viral transcription [15]. Subsequently, a GWAS of HIV-1 identified several SNPs near ZNRD1 on chromosome 6 that are associated with CD4+ T-cell counts [6]; however, it is still unresolved whether the ZNRD1 association is independent or tracks other variants in the human leukocyte antigen (HLA) region through linkage disequilibrium; it is also not known if ZNRD1 influences host susceptibility/resistance to HIV-1 acquisition.
We examined the effects of variation in the ZNRD1 gene on HIV-1 acquisition and disease progression in 1865 participants enrolled in 5 natural history, treatment-naive HIV-1 cohorts. We demonstrate by both regression and a more conservative stratified analysis robust statistical evidence for an association of ZNRD1 variation with HIV-1 acquisition. We further demonstrate that a haplotype-defining SNP shows differential transcriptional factor binding and altered ZNRD1 gene promoter activity.
MATERIALS AND METHODS
Study Participants
Details about the cohorts have been described elsewhere [16]. Study participants were enrolled in 5 US-based, treatment-naive natural history HIV/AIDS cohorts during 1978–1989: AIDS Link to the Intravenous Experience (ALIVE), an intravenous injection drug user cohort in Baltimore [17], consisting of mainly Africa Americans; Multicenter AIDS Cohort Study (MACS), a longitudinal prospective cohort of men who have sex with men (MSM) [18]; The San Francisco City Clinic Study (SFCC), a cohort of MSM [19]; Hemophilia Growth and Development Study (HGDS), a multicenter prospective study that enrolled children and adolescents with hemophilia who received contaminated blood products [20]; and The Multicenter Hemophilia Cohort Study (MHCS), a prospective study of hemophiliacs [21]. The latter 4 cohorts mainly comprise European Americans. Study protocols were approved by the Institutional Review Boards of participating institutions and informed consent was obtained from all study participants.
The study group comprises HIV-1 seroconverters (infected after study enrollment), seroprevalents (infected at study enrollment), at-risk seronegatives, and highly exposed uninfected. HIV-1 uninfected individuals were classified into 2 categories based on an individual's documented exposure levels to HIV-1. Highly exposed uninfected subjects were those with documented repeated exposure through sharing of injection equipment [17], anal receptive sex with multiple partners [22–24], or numerous transfusions with factor VIII replacement products prior to 1984, when viral inactivation procedures were initiated [25]. At-risk seronegatives subjects are those enrolled in the cohorts who remained HIV-seronegative despite ongoing or prior risk activities. The number of subjects studied in each category was as follows: seroconverters = 609 European Americans, 269 Africa Americans; at-risk seronegatives = 296 European Americans, 276 Africa Americans; highly exposed uninfected = 140 European Americans, 81 Africa Americans; and seroprevalents = 123 European Americans, 292 Africa Americans.
SNP Genotyping
Twelve SNPs in ZNRD1 region (chromosome 6 position 30 078 567 to 30 148 987, spanning 70 kb), and HLA-C rs9264942, known to be strongly associated with HIV viral load set point [6, 7] were genotyped using ABI TaqMan assays (Table 1, Figure 1). SNPs were selected if they were haplotype-tagging or altered protein coding, transcription, splicing, or microRNA binding (as predicted by SNPinfo web server [http://snpinfo.niehs.nih.gov]), or have published associations [6, 15, 26, 27, 29, 31].
Table 1.
SNPs in the ZNRD1/RNF39 and HLA-C Region Interrogated in the Study
| SNP no. | dbSNP ID | Position on Chr. 6p21.3 | SNP Location in Genes | Predicted SNP Functiona | Reported Association With CD4+ T-cell Counts |
|---|---|---|---|---|---|
| SNP1 | rs7758512 | 30 078 567 | 58 kb 5′ ZNRD1 | TFBS | Fellay et al [6] |
| SNP2 | rs9261174 | 30 104 834 | 30 kb 5′ ZNRD1 | TFBS | Fellay et al; Catano et al [26] |
| SNP3 | rs3869068 | 30 112 030 | 23 kb 5′ ZNRD1 | ZNRD1 expression [27] | Fellay et al [6] |
| SNP4 | rs3757327 | 30 135 391 | ZNRD1, −1889 promoter | TFBS | |
| SNP5 | rs3132130 | 30 135 430 | ZNRD1, −1850 promoter | TFBS | |
| SNP6 | rs10745 | 30 138 035 | ZNRD1, exon 3, Q101Q | Splicing | |
| SNP7 | rs9261269 | 30 138 092 | ZNRD1, intron 4 | TFBS, transcript QTL for HLA-A, HLA-F [28] | |
| SNP8 | rs1048412 | 30 140 473 | ZNRD1, 3′UTR | TFBS, splicing, miRNA | |
| SNP9 | rs8321 | 30 140 500 | ZNRD1, 3′UTR | TFBS, miRNA | Fellay et al [6]; Limou et al [29] |
| SNP10 | rs2301753 | 30 147 218 | RNF39, exon 4, A304E | Splicing, nsSNP | Fellay et al [6]; Limou et al [29] |
| SNP11 | rs2074480 | 30 148 788 | RNF39, intron 3 | Fellay et al [6] | |
| SNP12 | rs2074479 | 30 148 987 | RNF39, exon 3, S203P | Splicing, nsSNP | Fellay et al [6]; Van Manen et al [27] |
| SNP13 | rs9264942 | 31 382 358 | 35 kb 5′ HLA-C |
Abbreviations: nsSNP, nonsynonymous SNP; SNP, single nucleotide polymorphism; TFBS, transcription factor binding site.
a Predicated by SNPinfo web server (http://snpinfo.niehs.nih.gov).
Figure 1.
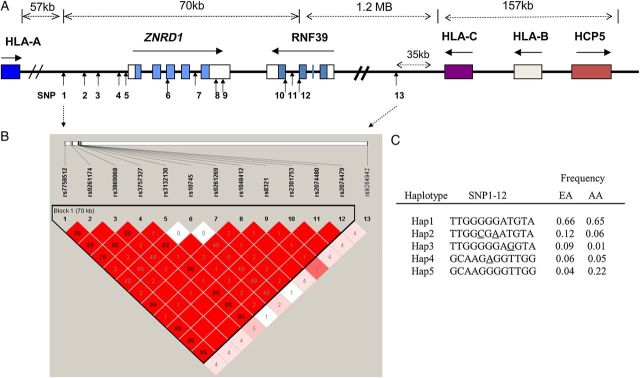
SNPs analyzed in the ZNRD1, RNF39, and HLA class I locus (6p21) region. A, Genomic structure and SNP locations. The colored blocks indicate exons, empty blocks, untranslated regions (UTR), horizontal arrows, the direction of transcription and vertical arrows, the positions of SNPs. B, Linkage disequilibrium matrix in European Americans (EA). Red block indicates D′ = 1.0, and the number in the blocks indicates the value of r2. The linkage disequilibrium block depicted by black triangle was based on the 95% Confidence interval criteria [30]. The intensity of the red color in each box is proportional to the strength of the linkage disequilibrium estimates (D′) for the SNP pair. C, Haplotype configuration and frequency in EA and in African American (AA). The haplotype-tagging SNPs are underlined. Abbreviation: SNP, single nucleotide polymorphism.
Genetic Association Analysis
The genetic effects of SNPs and haplotypes on HIV-1 infection susceptibility were assessed by comparing frequencies between the HIV-1 infected group composed of seroconverters and seroprevalent persons and the HIV-1 uninfected group composed of highly exposed uninfected and seronegatives. To avoid frailty bias in the seroprevalent group, which had an overrepresentation of long-term nonprogressors, potentially leading to the enrichment progression resistant factors, we restricted the HIV-positive group to HIV-1 seroconverters, which consist of unselected patients with natural disease progression spectrum in a sub-group comparison. As the effect of resistance factors may be dependent on the exposure level, we compared seroconverters with highly exposed uninfected and seronegative groups of high or low HIV-1 exposure levels separately. Odds ratios (OR) and P values for were obtained by using a conditional logistic regression test.
Kaplan–Meier survival statistics and the Cox proportional hazards model were used to assess the effects of SNPs and haplotypes on the rate of progression to AIDS among seroconverters. Two endpoints reflecting AIDS progression were considered for seroconverters: (1) HIV-1 infection plus a decline of CD4+ T-cell counts to <200 cells/mm3 (CD4 <200); (2) the 1987 Centers for Disease Control and Prevention (CDC) definition of AIDS (AIDS-87): HIV-1 infection plus AIDS-defining illness [32].
Electrophoretic Mobility Shift Assays (EMSA)
The EMSA was performed in HeLa and Jurkat cell nuclear extracts using oligonucleotides carrying ZNRD1 rs3132130 G/C with an Infrared EMSA Kit (LI-COR).
Gene Promoter Reporter Assay
DNA fragments of 1965 bp in the ZNRD1 5′ upstream region (spanning −1689 to +276 based on transcription start site (TSS)) carrying rs3132130 G or C at position −1565 were cloned into a pLightSwitch promoter reporter vector (SwitchGear Genomics) harboring Renilla luminescent reporter gene (RenSP). HeLa and 293 cells were transfected in triplicates. Promoter activity of Renilla luciferase activity was measured 24 hours later using the LightSwitch Dual Assay System and was normalized by the Cypridina luciferases activity (Cluc) from a cotransfected pTK-CLuc Vector (SwitchGear Genomics).
A more detailed description of the Materials and Methods is available in the Supplementary Data.
RESULTS
Linkage Disequilibrium of ZNRD1 Region SNPs
SNPs in the ZNRD1/RNF39 region showed weak to strong correlation with each other (r2 = 0.02–1.0; Figure 1); however, all had low correlation (r2 = 0.01–0.05) with the HLA-C rs9264942, previously shown to be strongly associated with control of HIV viral load (VL) [6], 1.2 MB downstream of the ZNRD1/RNF39 region. These 9 SNPs formed 5 common haplotypes with frequencies >3.0% in European Americans. Haplotypes of ZNRD1/RNF39 region were inferred by the tag SNPs 2 (rs9261174), 4 (rs3757327), 5 (rs3132130), 6 (rs10745), and 9 (rs8321). We performed association analyses on haplotypes, with and without adjustment for VL-affecting HLA-C rs9264942 allele.
ZNRD1 Haplotype and HIV-1 Acquisition
To determine the effect of ZNRD1 haplotypes on the risk to HIV-1 acquisition, we compared the haplotype frequencies between HIV-1 uninfected persons belonging to HIV-1 risk groups representing a range of exposures (n = 296) and HIV-1 infected persons (n = 732). A subgroup of 140 individuals comprised a group of highly exposed uninfected subjects with documented repeated parenteral or mucosal exposures to HIV-1. The HIV-1 infected group included 609 seroconverter cases with midpoint imputed dates of infection and 123 seroprevalent cases with unknown infection dates.
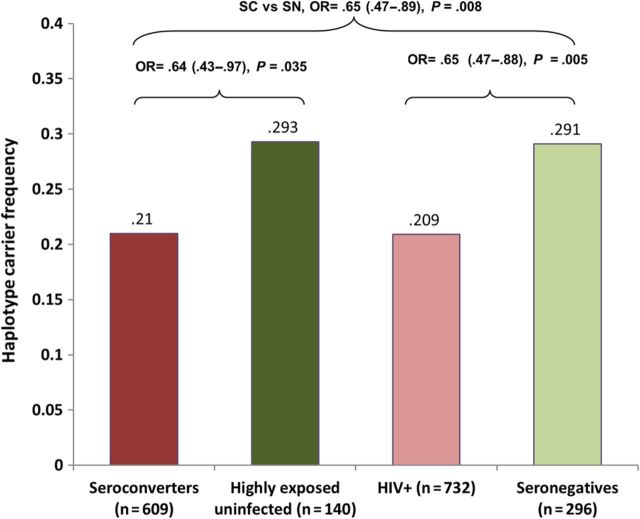
Among the 5 haplotypes, Hap2 showed evidence of association with resistance to HIV-1 acquisition (Figure 2). We first compared the HIV-infected group to the HIV-uninfected group and observed a statistically significant increase of Hap2 frequency in the uninfected group (odds ratio [OR] = 0.65, 95% confidence interval [CI], .47–.88, P = .005). To avoid distortions in allele frequencies due to potential frailty bias in the seroprevalent group, we performed a more conservative analysis comparing only seroconverters to the highly exposed uninfected group and found a similar effect size (OR = 0.64, P = .035). The Hap2 frequency was consistently higher in the HIV-1 uninfected group in all 3 subgroup comparisons: seroconverters vs highly exposed uninfected, seroconverters vs seronegatives, and all HIV-1 infected vs all uninfected, confirming that the association was not due to bias in 1 particular group. To address the issue of false discovery due to multiple comparisons, we performed a permutation test; the 5 haplotypes were included in a Fisher test with permutation resampling implemented in PROC MULTTEST of SAS software [33]. The permutation adjusted P value was .014 comparing the HIV-1 uninfected group and the HIV-1 infected group.
Figure 2.
The impact of ZNRD1 haplotype Hap2 tagged by rs3132130 on risk of HIV-1 acquisition among European Americans. Haplotype frequencies of individuals carrying 1 or 2 copies (dominant model) of Hap2 were compared between HIV-1 seroconverters and highly exposed uninfected, seroconverters and at-risk exposed seronegatives, and HIV + (HIV positives from both seroconverters and seroprevalents) and seronegatives. P values were from a χ2 test in a dominant model.
As HLA-C rs9264942 was not associated with infection (HIV-1 infected vs uninfected, OR = 1.18, 95% CI, .81–1.72, P = .40), the Hap2 association with HIV-1 infection status is not a result tracking by linkage disequilibrium with rs9264942. The effect size of the Hap2 remained similar even after stratifying on rs9264942 carriage. In addition, no epistatic interaction was observed among ZNRD1 haplotypes and rs9264942 in a Fisher test with permutation resampling. No other allele in the nearby HLA locus has been reported to affect HIV-1 acquisition [5], suggesting that the ZNRD1 associations are likely independent. Based on the haplotype configuration, the Hap2 association signal might stem from SNP5 (rs3132130 in the promoter region) or SNP7 (rs9261269 in intron 4) that are unique to Hap2 (Figure 1). The Hap 2 frequency is 2-fold higher in European Americans (12%) than in African Americans (6%); in African Americans, Hap2 was also associated with lower risk of infection but was not significant (OR = 0.82, 95% CI, .44–1.54).
ZNRD1 Haplotype and HIV-1 Disease Progression
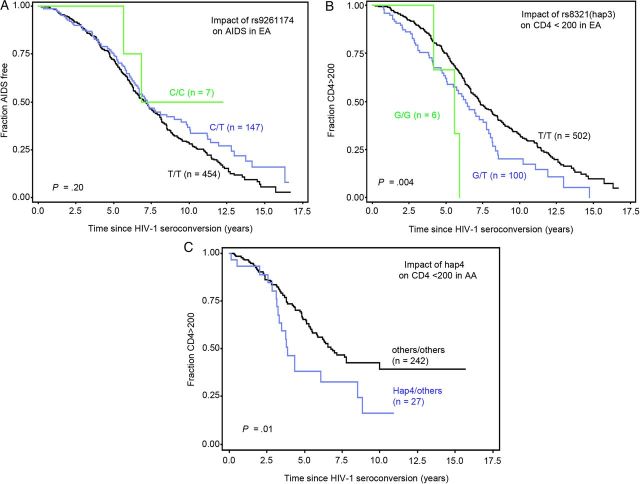
To assess the impact of ZNRD1 haplotypes on disease progression, the time from seroconversion to 2 AIDS progression endpoints: CD4+ T-cell dropping to <200/mm3 (CD4 < 200) and AIDS diagnosis with the CDC 1987 case definition (AIDS-87), were evaluated using Kaplan–Meier survival statistics and the Cox proportional hazards model. SNP rs9261174 was previously reported to affect progression to CD4 < 350 [6] or initiation of highly active antiretroviral therapy (HAART) with CD4 < 500 [7] (P = 3.9E − 07 or 1.8E − 08). In the current study, rs9261174 showed a nonsignificant protective trend on AIDS-87 (Cox model, RH = 0.81, 95% CI, .63–1.05, P = .11; survival analysis, P (log–rank test) = .20, Figure 3A), a more stringent and definitive endpoint reflecting AIDS progression; the direction of the effect is similar to those reported [6, 7]. However, the effect size diminished after adjusting for HLA-C rs9264942 (P = .36).
Figure 3.
Survival curve analysis of the impact of ZNRD1 SNPs on AIDS progression. A, Impact of ZNRD1 rs9261174 on clinical AIDS in EA; B, Impact of ZNRD1 rs8321(Hap3) on CD4 < 200 in EA; C, Impact of ZNRD1 hap4 on CD4 < 200 in AA. The x axis represents the time to progression endpoints since HIV-1 seroconversion (years), and the y axis is the percentage of individuals surviving for the outcome endpoint. P values were obtained from a log–rank test. Abbreviations: AA, African American; EA, European American; HIV-1, human immunodeficiency virus type 1; SNP, single nucleotide polymorphism.
Among 5 haplotypes tested in a dominant Cox model, Hap3 was associated with accelerated progression to the CD4 < 200 endpoint in EA (RH = 1.50, P = .005, Table 2). With conditioning on HLA-C rs9264942, plus inclusion of 6 other genetic factors securely associated with progression to AIDS (Table 2, footnote), the Hap3 effect on CD4 < 200 remained statistically robust. In contrast, Hap3 had no effect using the clinical AIDS-87 endpoint. Conversely, rs9264942 had a strong effect on AIDS-87 and death but had only marginal effect on CD4 < 200 [34]. These suggest independent effects from the 2 genes. SNP9 (rs8321) tagging Hap3 showed a clear separation of survival curves along the clinical course, with T/G heterozygotes having accelerated CD4+ T-cell depletion compared to the common rs8321 T/T genotype (P (log–rank test] = .004 and P (Wilcoxon test) = .003, Figure 3B). In a Cox model, the accelerating effect of the T/G genotype was significant after adjustment for the other genetic factors (RH = 1.39, 95% CI, 1.04–1.87, P = .027) and with additional adjustment for rs9264942 (RH = 1.36, 95% CI, 1.01–1.83, P = .04). In African Americans, a different haplotype, Hap4, was associated with accelerated CD4+ T-cell depletion (RH = 1.90, P = .01; Table 2, Figure 3C).
Table 2.
Impact of Haplotypes of ZNRD1 and RNF39 on AIDS Progressiona
| Haplotype | Population | CD4 < 200a | P Value | AIDS-87a | P Value |
|---|---|---|---|---|---|
| RH (95% CI) | 95% CI | ||||
| Hap1 | European American | .93 (.65–1.33) | .69 | .96 (.66–1.39) | .83 |
| African American | 1.22 (.63–2.34) | .56 | .91 (.41–2.05) | .82 | |
| Hap2 | European American | 1.13 (.87–1.46) | .38 | .95 (.71–1.28) | .95 |
| African American | .71 (.40–1.27) | .25 | .86 (.40–1.83) | .69 | |
| Hap3 | European American | 1.50 (1.13–1.98) | .005 | 1.08 (.79–1.47) | .65 |
| European American adj.b | 1.53 (1.15–2.02) | .003 | 1.11 (.81–1.52) | .52 | |
| African American | NA | NA | |||
| Hap4 | European American | .76 (.55–1.06) | .1 | .81 (.56–1.17) | .26 |
| African American | 1.94 (1.16–3.25) | .012 | 1.44 (.69–2.99) | .33 | |
| African American adj.c | 2.05 (1.21–3.48) | .008 | |||
| Hap5 | European American | 1.24 (.80–1.91) | .33 | 1.09 (.68–1.74) | .72 |
| African American | 1.19 (.80–1.77) | .38 | .94 (.54–1.61) | .81 |
Abbreviations: CI, confidence interval; NA, not analyzed due to rarity (1%) in African American.
a Results were for a dominant genetic model from a Cox proportional hazards model analysis; Hap3 was tagged by rs8321.
b Adjusted for the covariates HLA-C rs9264942, HLA-B*57, B*27, class I homozygosity, CCR5-Δ32, CCR5-P1, and CCR2–64I.
c Adjusted for the covariates HLA-B*57 and class I homozygosity (see Supplementary data).
SNP rs3132130 in the 5′ Upstream Region of ZNRD1 Alters Transcription Factor Binding
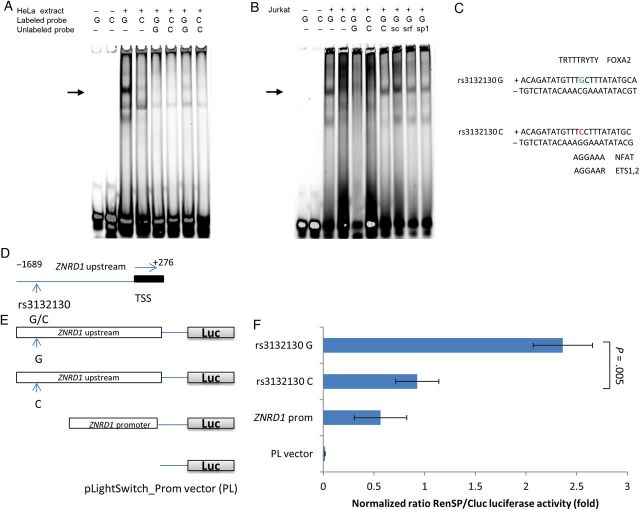
Because the Hap2-tagging SNP, rs3132130 G/C, is located in the ZNRD1 5′ upstream region (−1565 to transcription start site (TSS)) or the putative promoter region, we assessed whether this SNP affects DNA binding with transcription factors. We carried out electrophoretic mobility shift assays (EMSA) using fluorescently labeled oligonucleotide probes containing the alternative alleles (Figure 4A). Using HeLa cell nuclear extract stimulated with serum, the oligonucleotide probe containing the G allele formed a major DNA-protein complex. However, this DNA-protein complex was nearly absent when using an oligonucleotide probe containing the variant C allele present in Hap2 (Figure 4A, lanes 3 and 4). Addition of excessive unlabeled corresponding competitor G or C probes abolished the DNA-protein complex (lanes 5 and 6), in a dose-dependent manner (Figure 4A, lanes 7 and 8 vs 5 and 6), indicating the binding specificity. Similar results were also observed in Jurkat T-cell nuclear extract, and the G allele binding specificity of the top band was further confirmed by lack of inhibition from various nonspecific competitors (Figure 4B). These results indicate that certain transcription factors may bind to the rs3132130 G variant allele but not to the rs3132130 C minor allele.
Figure 4.
Functions of rs3132130 SNP in the ZNRD1 5′ upstream region. A, In vitro allele-specific DNA-protein interaction of rs3132130 detected by EMSA. Nuclear factors extracted from HeLa nuclear extract (4 hr serum response) were incubated with allele-specific labeled probes. Reaction conditions and probes containing the rs3132130 G or C (minor) allele for each lane are indicated over each lane. The position of the allele-specific DNA protein complex is indicated by an arrow. Lanes 1 and 2 contained no nuclear extract. To ascertain specific binding, 200× (lanes 5 and 6) or 50× (lanes 7 and 8) unlabeled probes were used as competitor. B, EMSA of rs3132130 G/C in Jurkat nuclear extract. To determine the specificity of the intense bands present in the rs3132130 G lane of the EMSA (lane 3), 200-fold excess of unlabeled rs3132130 C oligo (lane 7), a scambled (sc) oligo with similar base composition as the rs3132130 oligo (lane 8), SRF (one of the predicted binding factors, lane 9) and unrelated SP1 consensus oligos (lane 10, sequences in Supplementary Table 1) were used as competitors against labeled rs3132130 G oligo. Two-fold amounts of C- allele binding reactions were loaded to reveal possible bands (lane 4 and 6), which also led to high background smears. These data are representative of multiple independent EMSA experiments. C, Sequence context of SNP rs3132130 and matches to the consensus-binding motifs of selected transcription factors. SNP sites are colored. D, Schematic representation of the regions of the ZNRD1 promoter and its variants cloned into gene constructs. The black box indicates the untranslated region. E, Schematic representation of RenSP luciferase constructs containing the ZNRD1 promoter and its variants. Empty vector, pLightSwitch_Prom vector. (PL). F, Relative luciferase activity of the ZNRD1 promoter carrying the rs3132130. Promoter activity was shown as the normalized ratio of RenSP/Cluc luciferases. The relative luciferase activity is the value obtained with ZNRD1 fragments carrying RenSP vectors divided by the value from the cotransfected constitutive internal control Cluc vector. The data shown are mean ± standard deviations (SD) of triplicate samples; similar results were obtained in 3 separate experiments. Abbreviations: SNP, single nucleotide polymorphism; TSS, transcription start site.
We performed an in-silico search to assess the binding potential of rs3132130 with transcription factors. The SNPInspector (Figure 4C) predicted that the transversion of G to C may result in loss of the transcription factor binding sites for the family of Fork head domain factors (FOXA1, FOXA2, FOXD3), which are transcriptional activators, and serum response factor (SRF). On the other hand, it may result in the generation of transcription factor binding sites for the family of NFAT (nuclear factor of activated T cells), ETSF (human and murine ETS1 factors), IRFF (interferon regulatory factors), ISGF3G (interferon-stimulated transcription factor 3, gamma).
SNP rs3132130 in the ZNRD1 5′ Upstream Region Alters ZNRD1 Promoter Activity
To test whether rs3132130 influences ZNRD1 promoter activity, we performed luciferase gene promoter assays. Reporter constructs containing the 1.9-kb ZNRD1 promoter sequence with the 2 SNP rs3132130 G or C allele (Figure 4D and 4E) showed that the normalized luciferase activity of the rs3132130 C allele carrying sequence was decreased by 61.8% and 61.3%, compared with that of the G allele carrying sequence, in HeLa cells (Figure 4F) and HEK293 cells (data not shown), respectively (P = .005 and .006). This indicates that rs3132130 is within the ZNRD1 promoter region and the C minor allele of the rs3132130 has a transcription rate 2-fold lower than that of the G major allele.
DISCUSSION
We investigated genetic variants in ZNRD1 in well-characterized, treatment-naive HIV-1 natural history cohorts for association with infection status and rate of progression to clinical outcomes. The findings from this case-cohort study provide evidence that the ZNRD1 gene may influence HIV acquisition and rate of CD4+ T-cell decline. The differential impact of ZNRD1 haplotypes on CD4+ T-cell depletion and susceptibility to HIV-1 acquisition provides additional support that ZNRD1 may be important in modulating HIV-1 pathogenesis [6, 14].
This is the first report to our knowledge that ZNRD1 variation affects host resistance to HIV-1 acquisition. ZNRD1 Hap2 containing rs3132130 in the 5′ upstream region was enriched in the exposed but uninfected group compared to those who were HIV-1 infected. Our results suggest that the ZNRD1 associations are independent of the nearby HLA alleles; HLA-C rs9264942 had no impact on infection and none of the other alleles in the HLA B/C locus had an apparent impact on HIV-1 acquisition [5]. Based on an odds ratio of 0.65 and a population frequency of 12% for the Hap2 allele among European Americans, the corresponding population attributable fraction in providing protection to European Americans is 6.0%, which is substantial from a public health point of view. For comparison, the population attributable fraction contributed by CCR5-Δ32 homozygosity (frequency = 1% in European Americans), conferring near complete resistance to HIV-1 acquisition, is 1.0% [8, 9].
We showed that SNP rs3132130, located in the ZNRD1 5′ upstream region affected nuclear factor binding, as revealed by EMSA. Using the luciferase promoter reporter assays, we further demonstrated that the upstream region of ZNRD1 bearing the SNP rs3132130 processes promoter activity, which was significantly lower in the minor allele C-bearing than the common G-bearing fragments. The longer C-bearing fragment had a promoter activity comparable to a shorter standard ZNRD1 promoter. These results suggest that the C allele-bearing promoter had lower promoter activity, possibly due to loss of certain transcription activator factor binding as revealed in EMSA. It is reasonable to predict that the C allele-bearing promoter could produce a lower level of ZNRD1 gene expression than the G allele-bearing promoter. It is thus biologically plausible that lower expression of ZNRD1, which enhances HIV-1 replication, would be unfavorable for HIV-1 infection. This functional mechanism is in support of the population genetic epidemiological finding that rs3132130C carriers had decreased risk of HIV-1 acquisition. However, it remains to be determined whether the in vitro regulatory effect identified leads to in vivo gene expression changes in the relevant target cells such as CD4+ T cells, mucosal macrophage and epithelial cells.
Investigating unique genetic and immunological features among highly exposed uninfected individuals may uncover host factors underlying the resistance to HIV-1. Highly exposed uninfected are persons who are at high-risk for HIV-1 acquisition but remain seronegative [1–4, 35, 36]. HIV-1 is obligated to use cellular proteins (HIV-1 dependency factors) to complete its replication cycle at every stage, including viral entry, transcription, viral integration, virion maturation, and budding. On the other hand, the host activates innate and acquired immune defense systems (restriction factors, such as APOBEC3, TRIM5, HLA) to combat retroviral infections. Genetic changes in the key pathways in viral-host interaction that disrupt HIV-1 dependency factors or strengthen the defending factors would reasonably contribute to host HIV-1 resistance. This is best exemplified by a Δ32 knock-out homozygous genotype in the HIV-1 coreceptor CCR5 leading to near-complete resistance to HIV-1 infection, which was identified in the same exposed uninfected groups as examined in this study [8, 9]. With the exception of CCR5 Δ32, no new HIV-1 genetic variants that affect HIV-1 acquisition have been revealed by GWAS [10, 12, 37]. In one GWAS, HIV-1 negative African adults recruited from sexually transmitted infection clinics, considered as at-risk for HIV-1 exposure, were compared with HIV-1 positive controls [10]. Another GWAS compared HIV-1 exposed uninfected hemophiliacs with HIV-1 positive homosexuals, assuming resistant genes would largely be the same in patients with different transmission routes [37]. A multicenter GWAS focused on HIV-1 acquisition compared HIV-1 infected individuals to population controls identified only signals of association in the HLA-B region; however, this result was likely due to enrichment of progression protective HLA-B alleles among the HIV-1 controller group [12]. When the study was limited to only seroconverters, these associations were not observed [12]. Because these GWAS did not identify common variants reaching genome-wide significance, it was suggested that low frequency variants or variants with low effect sizes might explain the variation in HIV acquisition susceptibility.
Our study was a case-cohort design where every HIV-1- individual belonged to 1 or more HIV-1 risk groups and our HIV-1-positive cases comprised only seroconverters to limit bias. The enrichment of HIV-1 resistant variants rs3132130/rs9261269 was found in the exposed, uninfected populations (highly exposed uninfected and at-risk seronegatives). The limitation of our study includes the relatively small sample sizes in highly exposed uninfected group, resulting in decreased power, potential bias introduced by combining various cohorts, and the modest strength of association. The role of ZNDRI should be regarded as a candidate for further verification in larger population groups and by further functional studies to evaluate causality.
In addition, the impact of SNPs in or near the ZNRD1 region on disease progression has been reported in several studies [6, 7, 26, 27, 29, 31] (Table 1), although its association with HIV-1 acquisition has not been tested. In the first HIV-1 GWAS, Fellay et al found a group of correlated SNPs including rs9261174 (SNP2) located near ZNRD1 among top hits for CD4 T-cell depletion [6, 7]. Van Manen et al observed that rs2074479 was associated with the CD4+ T-cell count but not with viral load or disease progression to AIDS [27]. Limou et al performed GWAS in the French GRIV cohort of extreme phenotypes and found that rs8321 was associated with CD4+ T-cell drop [29]. Catano et al found that rs9261174 affected the rate of CD4+ T-cell loss in the WHMC cohort [26]. By genotyping a subset of the alleles that contribute to HLA-A10, this report suggested the ZNRD1 association was attributable to linkage disequilibrium with the HLA-A10 serogroup [26]; this finding was not supported by subsequent studies in other well-characterized HIV cohorts [7, 31]. Ballana et al showed that rs1048412 was associated with long-term nonprogression [15]. Our results, together with others, indicated that ZNRD1 variation mainly affects CD4+ T-cell depletion rather than the development of frank AIDS, but the mechanism underpinning this association requires further study.
This study provides the first evidence that ZNRD1 variation may affect host susceptibility to HIV-1 acquisition. This result is supported by prior functional findings that ZNRD1 is necessary for HIV-1 replication in cell culture [14, 15] and the functional evidence of the associated SNPs. These results, together with the observation that ZNRD1 silencing does not affect cell viability, suggest that the ZNRD1 gene or its protein may be a druggable target for antiretroviral therapy to prevent acquisition or to limit replication by HIV.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The funding sources had no role in study design, interpretation, and in the writing of the article.
Financial support. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The ALIVE study was funded by National Institute on Drug Abuse (R01-DA04334 and R01-DA12568). The Hemophilia Growth and Development Study is funded by the National Institutes of Health, National Institute of Child Health and Human Development, R01 HD41224. The MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute; and the National Heart, Lung, and Blood Institute: UO1-AI-35042, 5-M01-RR-00052 (GCRC), UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613, and UO1-AI-35041. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–74. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 2.An P, Winkler CA. Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet. 2010;26:13. doi: 10.1016/j.tig.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shearer G, Clerici M. Historical perspective on HIV-exposed seronegative individuals: has nature done the experiment for us? J Infect Dis. 2010;202(suppl 3):S329–32. doi: 10.1086/655974. [DOI] [PubMed] [Google Scholar]
- 4.Piacentini L, Biasin M, Fenizia C, Clerici M. Genetic correlates of protection against HIV infection: the ally within. J Intern Med. 2009;265:110–24. doi: 10.1111/j.1365-2796.2008.02041.x. [DOI] [PubMed] [Google Scholar]
- 5.Carrington M, Martin MP, van Bergen J. KIR-HLA intercourse in HIV disease. Trends Microbiol. 2008;16:620–7. doi: 10.1016/j.tim.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellay J, Ge D, Shianna KV, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene: Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–62. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 9.Ioannidis JP, Rosenberg PS, Goedert JJ, et al. Effects of CCR5-Delta32, CCR2–64I, and SDF-1 3′A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann Intern Med. 2001;135:782–95. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- 10.Petrovski S, Fellay J, Shianna KV, et al. Common human genetic variants and HIV-1 susceptibility: a genome-wide survey in a homogeneous African population. AIDS. 2011;25:513–8. doi: 10.1097/QAD.0b013e328343817b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingappa JR, Petrovski S, Kahle E, et al. Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PloS One. 2011;6:e28632. doi: 10.1371/journal.pone.0028632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaren PJ, Coulonges C, Ripke S, et al. Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls. PLoS Pathog. 2013;9:e1003515. doi: 10.1371/journal.ppat.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sironi M, Biasin M, Cagliani R, et al. A common polymorphism in TLR3 confers natural resistance to HIV-1 infection. J Immunol. 2012;188:818–23. doi: 10.4049/jimmunol.1102179. [DOI] [PubMed] [Google Scholar]
- 14.Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 15.Ballana E, Senserrich J, Pauls E, et al. ZNRD1 (zinc ribbon domain-containing 1) is a host cellular factor that influences HIV-1 replication and disease progression. Clin Infect Dis. 2010;50:1022–32. doi: 10.1086/651114. [DOI] [PubMed] [Google Scholar]
- 16.An P, Duggal P, Wang LH, et al. Polymorphisms of CUL5 are associated with CD4+ T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3:e19. doi: 10.1371/journal.pgen.0030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlahov D, Graham N, Hoover D, et al. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. JAMA. 1998;279:35–40. doi: 10.1001/jama.279.1.35. [DOI] [PubMed] [Google Scholar]
- 18.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 19.Buchbinder SP, Katz MH, Hessol NA, O'Malley PM, Holmberg SD. Long-term HIV-1 infection without immunologic progression. AIDS. 1994;8:1123–8. doi: 10.1097/00002030-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 20.Hilgartner MW, Donfield SM, Willoughby A, et al. Hemophilia growth and development study: design, methods, and entry data. Am J Pediatr Hematol Oncol. 1993;15:208–18. doi: 10.1097/00043426-199305000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Goedert JJ, Kessler CM, Aledort LM, et al. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N Engl J Med. 1989;321:1141–8. doi: 10.1056/NEJM198910263211701. [DOI] [PubMed] [Google Scholar]
- 22.Detels R, Liu Z, Hennessey K, et al. Resistance to HIV-1 infection: Multicenter AIDS Cohort Study. J Acquir Immune Defic Syndr. 1994;7:1263–9. [PubMed] [Google Scholar]
- 23.Detels R, Jacobson L, Margolick J, et al. The multicenter AIDS Cohort Study, 1983 to … . Public Health. 2012;126:196–8. doi: 10.1016/j.puhe.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imagawa DT, Lee MH, Wolinsky SM, et al. Human immunodeficiency virus type 1 infection in homosexual men who remain seronegative for prolonged periods. N Eng J Med. 1989;320:1458–62. doi: 10.1056/NEJM198906013202205. [DOI] [PubMed] [Google Scholar]
- 25.Kroner BL, Rosenberg PS, Aledort LM, Alvord WG, Goedert JJ. HIV-1 infection incidence among persons with hemophilia in the United States and Western Europe, 1978–1990: Multicenter Hemophilia Cohort Study. J Acquir Immune Defic Syndr. 1994;7:279–86. [PubMed] [Google Scholar]
- 26.Catano G, Kulkarni H, He W, et al. HIV-1 disease-influencing effects associated with ZNRD1, HCP5 and HLA-C alleles are attributable mainly to either HLA-A10 or HLA-B*57 alleles. PLoS One. 2008;3:e3636. doi: 10.1371/journal.pone.0003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Manen D, Kootstra NA, Boeser-Nunnink B, Handulle MA, van't Wout AB, Schuitemaker H. Association of HLA-C and HCP5 gene regions with the clinical course of HIV-1 infection. AIDS. 2009;23:19–28. doi: 10.1097/QAD.0b013e32831db247. [DOI] [PubMed] [Google Scholar]
- 28.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–7. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limou S, Le Clerc S, Coulonges C, et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199:419–26. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Trachtenberg E, Bhattacharya T, Ladner M, Phair J, Erlich H, Wolinsky S. The HLA-B/-C haplotype block contains major determinants for host control of HIV. Genes Immun. 2009;10:673–7. doi: 10.1038/gene.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and Prevention. Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. Council of State and Territorial Epidemiologists; AIDS Program, Center for Infectious Diseases. MMWR. 1987;36(suppl 1):1S–15S. [PubMed] [Google Scholar]
- 33.Westfall PH, Zaykin DV, Young SS. Multiple tests for genetic effects in association studies. Methods Mol Biol. 2002;184:143–68. doi: 10.1385/1-59259-242-2:143. [DOI] [PubMed] [Google Scholar]
- 34.Thomas R, Apps R, Qi Y, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–4. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lederman MM, Alter G, Daskalakis DC, et al. Determinants of protection among HIV-exposed seronegative persons: an overview. J Infect Dis. 2010;202(suppl 3):S333–8. doi: 10.1086/655967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pancino G, Saez-Cirion A, Scott-Algara D, Paul P. Natural resistance to HIV infection: lessons learned from HIV-exposed uninfected individuals. J Infect Dis. 2010;202(suppl 3):S345–50. doi: 10.1086/655973. [DOI] [PubMed] [Google Scholar]
- 37.Lane J, McLaren PJ, Dorrell L, et al. A genome-wide association study of resistance to HIV infection in highly exposed uninfected individuals with hemophilia A. Hum Mol Genet. 2013;22:1903–10. doi: 10.1093/hmg/ddt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.