Abstract
The majority of trait-associated loci discovered through genome-wide association studies are located outside of known protein coding regions. Consequently, it is difficult to ascertain the mechanism underlying these variants and to pinpoint the causal alleles. Expression quantitative trait loci (eQTLs) provide an organizing principle to address both of these issues. eQTLs are genetic loci that correlate with RNA transcript levels. Large-scale data sets such as the Cancer Genome Atlas (TCGA) provide an ideal opportunity to systematically evaluate eQTLs as they have generated multiple data types on hundreds of samples. We evaluated the determinants of gene expression (germline variants and somatic copy number and methylation) and performed cis-eQTL analyses for mRNA expression and miRNA expression in five tumor types (breast, colon, kidney, lung and prostate). We next tested 149 known cancer risk loci for eQTL effects, and observed that 42 (28.2%) were significantly associated with at least one transcript. Lastly, we described a fine-mapping strategy for these 42 eQTL target–gene associations based on an integrated strategy that combines the eQTL level of significance and the regulatory potential as measured by DNaseI hypersensitivity. For each of the risk loci, our analyses suggested 1 to 81 candidate causal variants that may be prioritized for downstream functional analysis. In summary, our study provided a comprehensive landscape of the genetic determinants of gene expression in different tumor types and ranked the genes and loci for further functional assessment of known cancer risk loci.
INTRODUCTION
Genome-wide association studies (GWAS) have successfully identified thousands of variants associated with hundreds of human traits. The vast majority of trait-associated loci are located outside of protein coding regions. This observation presents the next set of challenges in human genetics—to understand the mechanism of the locus and to identify the causal variant of risk loci. Expression quantitative trait loci (eQTLs) represent an organizing principle to address both of these issues.
eQTLs refer to genomic locations harboring genetic variants that associate with transcript levels. Many studies have unequivocally demonstrated that a substantial proportion of transcripts are under genetic control (1–3).
A practical value of eQTLs is their ability to implicate candidate genes that are influenced by non-protein coding trait-associated alleles. eQTL-based strategies provide an intermediate molecular layer between germline variants and traits. A transcript that is correlated with a variant in a relevant tissue or cell type becomes a strong candidate for downstream functional evaluation. Thus, eQTLs provide a logical strategy to evaluate the biology of non-protein-coding risk loci and indeed is one of the primary motivations behind large-scale genotype by gene expression databases (4,5).
When connecting trait-associated alleles with transcripts, identifying the particular cell type where the risk allele is exerting its effect remains a challenge. Evidence suggests that many eQTLs are tissue specific; however, data continue to emerge on this topic (5–7). Performing eQTL analysis for cancer risk loci raises another issue—should experiments be performed in normal and/or tumor tissue? Studying normal tissue is intuitively appealing because risk loci increase the risk of developing cancer and large-scale databases such as Genotype-Tissue Expression (GTEx) are just becoming available. On the other hand, the Cancer Genome Atlas (TCGA) has extensive data on multiple cancer types, including mRNA expression on hundreds of tumor samples. Performing eQTL analysis in the tumor tissue, however, is technically challenging because of the acquired somatic alterations that can also influence transcript levels. We recently described a method designed to overcome this challenge and to perform eQTL studies using gene expression from tumor samples (8).
eQTL–target gene correlations can also serve as an organizing principle for fine mapping. Similar to fine mapping in a case–control setting, the goal is to identify variants that have a statistical signal at least as strong as the initially reported variant. Since any eQTL–target gene association represents a relationship between a regulatory element and its target gene, epigenetic data can be incorporated into the study design to prioritize candidate variants that directly affect regulatory elements. Among the known epigenetic marks, DNaseI hypersensitivity (DHS) is a time-tested technique used for annotating regulatory elements as it marks areas of accessible chromatin (9). Fine mapping and DHS can be combined to prioritize a set of candidate loci for further testing as has been recently described (10).
In this study, we systematically performed eQTL analyses in five tumor types. First, we performed a genome-wide cis-eQTL analysis for both mRNAs and miRNAs. We then tested 149 independent risk alleles (across all five tumor types) for cis-associations with both mRNA and miRNA transcripts. Lastly, a fine mapping strategy was introduced in order to prioritize candidate causal alleles for future functional evaluation.
RESULTS
cis-acting eQTLs of mRNA in breast, colon, kidney, lung and prostate tumors
To identify the genetic determinants of gene expression in cancers, we performed cis-eQTL analyses for five common tumor types in TCGA using RNA-seq data (Table 1). Realizing that somatic alterations in the tumor genome can affect gene expression, we adjusted the expression levels for somatic copy number changes and CpG methylation variation as previously described (see Materials and Methods) (8). For each SNP locus, we evaluated the association between the corresponding germline genotypes and the transcript abundances of mRNAs within 500 kb upstream and 500 kb downstream of the locus (see Materials and Methods). A cis-association was determined by significant correlation between the germline genotypes and transcript levels at a false discovery rate (FDR) of 0.1 (see Materials and Methods). Our analyses identified 1306–5285 unique cis-associations (595–981 genes) across the five tumor types (Table 2, Supplementary Material, Table S1). The unadjusted P-values range from 4.28 × 10−7 to 1.74 × 10−296. In order to evaluate whether the analyses are confounded by systematic biases, we compared the distribution of the test P-values to that of the expected P-values using Q–Q plots (Supplementary Material, Fig. S1). The resulting λ statistics for all the analyses ranged from 0.923 to 1.02, demonstrating no systematic bias (see Materials and Methods).
Table 1.
Number of samples and risk loci included in the analyses
| Cancer type | Sample size, mRNA-analysis | Sample size, miRNA-analysis | Number of risk loci tested |
|---|---|---|---|
| Breast invasive carcinoma (ER+) | 391 | 227 | 53 |
| Colon adenocarcinoma | 121 | 193 | 19 |
| Kidney renal clear cell carcinoma | 163 | 102 | 3 |
| Lung adenocarcinoma | 183 | 26 | 5 |
| Prostate adenocarcinoma | 145 | 66 | 69 |
Table 2.
Summary of eQTL-mRNA associations in cis in five cancer types
| Cancer type | Number of associations in cis | Number of eQTLs | Number of associated genes | P-values | FDR |
|---|---|---|---|---|---|
| Breast invasive carcinoma (ER+) | 5285 | 4652 | 981 | 1.74 × 10−296–4.28 × 10−7 | 1.74 × 10−289–0.0999 |
| Colon adenocarcinoma | 1324 | 1161 | 602 | 8.88 × 10−147–1.59 × 10−7 | 8.63 × 10−140–0.0994 |
| Kidney renal clear cell carcinoma | 2250 | 1184 | 860 | 2.25 × 10−148–2.00 × 10−7 | 2.08 × 10−141–0.0997 |
| Lung adenocarcinoma | 1306 | 1197 | 595 | 5.79 × 10−192–1.59 × 10−7 | 5.82 × 10−185–0.0997 |
| Prostate adenocarcinoma | 1642 | 3466 | 1089 | 7.96 × 10−178–5.09 × 10−7 | 3.93 × 10−171–0.0999 |
Our data reaffirmed that the cis-associations tend to occur between SNPs and nearby transcripts. 24.5% of the cis-associations occur between a SNP locus and the target gene located closest to the risk SNP; 44.0% of the cis-associations occur between SNP locus and one of the five closest genes (Supplementary Material, Fig. S2).
cis-acting eQTLs of miRNA in human cancers
We next evaluated the genetic determinants of miRNA expression. We performed cis-eQTL analyses for the 1523 known miRNAs, of which the expression levels in the five aforementioned tumor types were available from TCGA RNA-sequencing data. Applying an FDR of 0.1, we identified 53–81 unique cis-associations (19–53 miRNAs) from the five tumor types, respectively (unadjusted P-value ranging from 7.09 × 10−7 to 1.08 × 10−133, Table 3, Supplementary Material, Table S2).
Table 3.
Summary of eQTL-miRNA associations in cis in five cancer types
| Cancer type | Number of associations in cis | Number of eQTLs | Number of associated miRNAs | P-values | FDR |
|---|---|---|---|---|---|
| Breast invasive carcinoma (ER+) | 74 | 74 | 19 | 2.28 × 10−40–2.90 × 10−7 | 6.13 × 10−35–0.0907 |
| Colon adenocarcinoma | 71 | 71 | 28 | 1.08 × 10−133–3.23 × 10−7 | 3.16 × 10−128–0.0873 |
| Kidney renal clear cell carcinoma | 81 | 76 | 28 | 1.25 × 10−22–7.09 × 10−7 | 3.31 × 10−17–0.0979 |
| Lung adenocarcinoma | 77 | 48 | 53 | 1.38 × 10−25–7.04 × 10−7 | 4.16 × 10−20–0.0944 |
| Prostate adenocarcinoma | 53 | 43 | 38 | 4.93 × 10−41–5.73 × 10−7 | 1.49 × 10−35–0.0984 |
Cancer risk loci as genetic determinants of gene expression
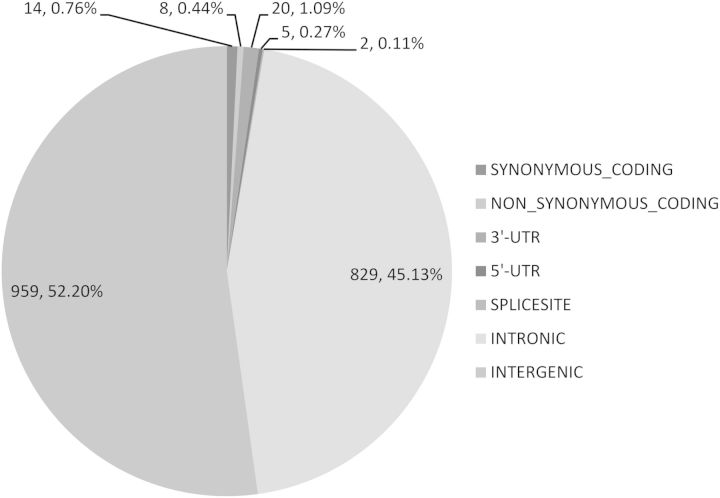
We next evaluated a set of 149 independent cancer risk loci from recent GWAS with P-values <1 × 10−7 (Table 1, Materials and Methods) (11–13). For each of the risk loci, we retrieved all the correlated variants (r2 ≥ 0.7) from the 1000 genomes project and assessed the distribution of this set of variants across the genome (Fig. 1, Materials and Methods). Consistent with previous studies, only a small fraction (1.1%) of the risk-associated variants (or their proxies) directly altered protein coding sequences, whereas the majority of variants were intergenic (52.2%) and intronic (45.1%) (13).
Figure 1.
The genomic distribution of the risk loci of five TCGA cancer types and their correlated (r2 ≥ 0.7) variants. Similar to prior observations, the majority of variants (98.8%) are outside of known protein coding region.
Forty-two of the 149 risk loci (28.2%) had 1–12 associated genes within 500 kb (FDR ≤ 0.1) (Table 4). Among the 42 cancer risk loci that function as cis-eQTLs, 8 were breast cancer risk loci, 31 were prostate cancer risk loci and another 1 each from lung adenocarcinoma and kidney cancer. The analyses also identified one colon cancer risk locus (20q13.33) associated with the expression of a miRNA (Table 4, Supplementary Material, Table S3).
Table 4.
Summary of the cis-eQTL associations of the GWAS risk loci of five cancer types
| Cancer type | Number of association test | Risk loci | Chromosome | Cytoband | Most significant associated transcripts | P-values | FDR |
|---|---|---|---|---|---|---|---|
| Breast invasive carcinoma | 394 | rs889312 | 5 | q11.2 | C5orf35 | 9.59 × 10−7 | 0.000188 |
| 394 | rs720475 | 7 | q35 | OR2A7 | 5.78 × 10−8 | 2.27 × 10−5 | |
| rs3817198 | 11 | p15.5 | TH | 0.00213 | 0.0559 | ||
| rs3903072 | 11 | q13.1 | DKFZp761E198 | 0.000734 | 0.0289 | ||
| rs3803662 | 16 | q12.1 | TOX3 | 0.000147 | 0.00966 | ||
| rs13329835 | 16 | q23.2 | DYNLRB2 | 0.000701 | 0.0289 | ||
| rs4808801 | 19 | p13.11 | LRRC25 | 0.00317 | 0.0731 | ||
| rs3760982 | 19 | q13.31 | ZNF155 | 0.00342 | 0.0731 | ||
| Colon adenocarcinomaa | 139 | rs4925386 | 20 | q13.33 | hsa-mir-1-1 | 0.00697 | 0.0901 |
| Kidney renal clear cell carcinoma | 18 | rs718314 | 12 | p11.23 | BHLHE41 | 0.000764 | 0.00817 |
| Lung adenocarcinoma | 43 | rs7216064 | 17 | q24.2 | C17orf58 | 0.000536 | 0.00764 |
| Prostate adenocarcinoma | 216 | rs1218582 | 1 | q21.3 | GBA | 0.00303 | 0.018 |
| rs4245739 | 1 | q32.1 | PIK3C2B | 0.0445 | 0.0469 | ||
| rs10187424 | 2 | p11.2 | GNLY | 0.00294 | 0.0178 | ||
| rs1465618 | 2 | p21 | LOC100129726 | 0.0149 | 0.0389 | ||
| rs12621278 | 2 | q31.1 | ITGA6 | 0.0182 | 0.0393 | ||
| rs7584330 | 2 | q37.3 | MLPH | 0.000659 | 0.00526 | ||
| rs3771570 | 2 | q37.3 | HDLBP | 0.0249 | 0.0393 | ||
| rs17181170 | 3 | p11.2 | CHMP2B | 0.0498 | 0.0498 | ||
| rs7611694 | 3 | q13.2 | GRAMD1C | 0.0192 | 0.0393 | ||
| rs10934853 | 3 | q21.3 | RUVBL1 | 0.0448 | 0.0469 | ||
| rs6763931 | 3 | q23 | ACPL2 | 0.0101 | 0.032 | ||
| rs12500426 | 4 | q22.3 | BMPR1B | 0.000394 | 0.00333 | ||
| rs12653946 | 5 | p15.33 | IRX4 | 5.11 × 10−14 | 7.13 × 10−12 | ||
| rs2273669 | 6 | q21 | SESN1 | 1.41 × 10−7 | 1.17 × 10−5 | ||
| rs1933488 | 6 | q25.2 | RGS17 | 1.75 × 10−6 | 5.69 × 10−5 | ||
| rs651164 | 6 | q25.3 | PNLDC1 | 0.0384 | 0.0437 | ||
| rs6465657 | 7 | q21.3 | LMTK2 | 0.0463 | 0.0476 | ||
| rs11135910 | 8 | p21.2 | EBF2 | 9.58 × 10−5 | 0.00157 | ||
| rs1512268 | 8 | p21.2 | LOXL2 | 0.00119 | 0.00833 | ||
| rs4242384 | 8 | q24.21 | MYC | 0.0323 | 0.0411 | ||
| rs817826 | 9 | q31.2 | RAD23B | 0.0196 | 0.0393 | ||
| rs3123078 | 10 | q11.23 | NCOA4 | 0.0101 | 0.032 | ||
| rs3850699 | 10 | q24.32 | AS3MT | 0.00016 | 0.00223 | ||
| rs7130881 | 11 | q13.2 | FGF19 | 0.0298 | 0.0401 | ||
| rs10875943 | 12 | q13.12 | C1QL4 | 0.00251 | 0.0159 | ||
| rs902774 | 12 | q13.13 | KRT5 | 0.00439 | 0.024 | ||
| rs8008270 | 14 | q22.2 | PSMC6 | 0.0379 | 0.0435 | ||
| rs684232 | 17 | p13.3 | VPS53 | 1.40 × 10−6 | 5.59 × 10−5 | ||
| rs11650494 | 17 | q21.33 | ZNF652 | 0.000209 | 0.00237 | ||
| rs11672691 | 19 | q13.2 | CEACAM21 | 0.0262 | 0.0393 |
aThe 20q13.33 risk locus of COAD is associated with miRNA expression.
Fine mapping cancer risk loci that are acting as eQTLs
The identification of causal alleles presents a challenge because of the intrinsic property of linkage disequilibrium (LD)—according to the 1000 genomes data, on average, a given risk locus is correlated with 56 other variants (14). eQTLs provide rational additional information for candidate causal allele detection. eQTL–gene associations represent a biological unit comprised of a regulatory element and its target gene. This relationship is important for two reasons: it provides (i) the appropriate cell type in which to perform functional work; and (ii) a clear-cut readout for causal allele identification—the causal variant would be expected to influence expression levels whereas a correlated variant would not.
We performed a fine-mapping analysis for the 42 significant cancer risk loci identified above. The goal of the analysis is to identify candidate causal alleles that could be prioritized for downstream functional evaluation. We specifically hypothesized that the target gene is under the control of a single variant. Because DHS sites are an accepted way of annotating regulatory elements, we used this feature to prioritize variants residing in potential regulatory elements. We used the ENCODE DNaseI track that is a compilation of genome-wide DNaseI HS profiles from125 cell types (15). Our rationale was to minimize false negatives by not having to rely upon a profile from one cell line (derived from one individual), which may not adequately capture the diversity of DHS. For each variant, we derived a posterior probability to summarize the eQTL level of significance (the association data) and the regulatory potential (DHS) (Materials and Methods).
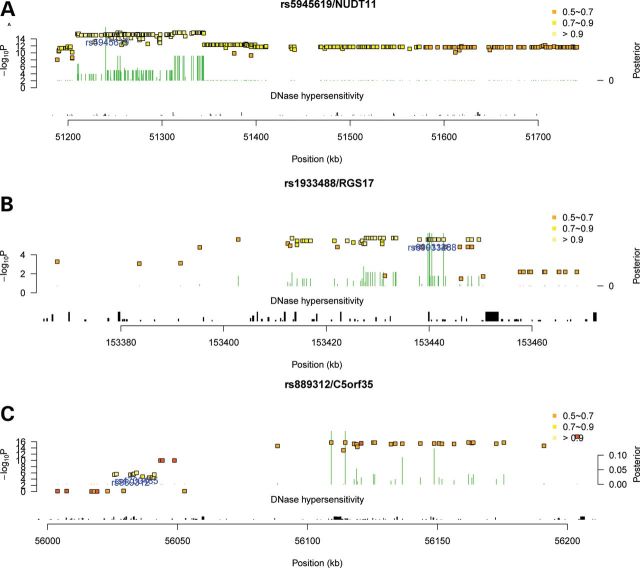
For the 42 risk loci where we identified significantly associated gene(s), we imputed the genotypes of all correlated polymorphisms with an r2 larger than 0.5 (Materials and Methods) and located within 1 Mb of the risk locus (N = 2181) (Materials and Methods). We calculated the posterior probability for each of the 42 eQTL target–gene associations. The posterior values showed a strong bimodal distribution (Supplementary Material, Fig. S3). To select the variants with the highest likelihood of being causal, we applied an unsupervised partition algorithm (Materials and Methods). Thus, we identified 408 candidate causal alleles for the 35 cancer risk loci with each locus having 1 to 81 candidates (Table 5, Supplementary Material, Table S4). While the rank order list of the posterior should serve as a guidepost for downstream functional follow-up, we note that 65 variants had a posterior value of at least one order of magnitude higher than the posterior of reported risk loci. Figure 2 illustrates various scenarios. The NUDT11 and RGS17 plots show loci with a large number of highly correlated variants (Fig. 2). Incorporating DHS data greatly reduces the number of candidate causal variants for functional testing. The C5orf35 locus demonstrates that the strongest candidate variants are moderately correlated with the index variant and are 41.5 kb (rs11960484) ∼ 81.1 kb (rs252920) away (Fig. 2).
Table 5.
Summary of fine mapping of causal variants for each risk loci as cis-eQTLs
| Cancer type | Risk loci | Transcripts associated in cis | Number of variants tested | Number of candidates | Posterior risk loci | Posterior candidates |
|---|---|---|---|---|---|---|
| Breast invasive carcinoma | rs889312 | C5orf35 | 38 | 5 | 1.37 × 10−11 | 0.0442–0.131 |
| rs720475 | OR2A1 | 8 | 1 | 0.0167 | 0.0167–0.0167 | |
| rs720475 | OR2A7 | 8 | 1 | 1.12 × 10−6 | 1.12 × 10−6–1.12 × 10−6 | |
| rs720475 | OR2A9P | 8 | 1 | 4.67 × 10−6 | 4.67 × 10−6–4.67 × 10−6 | |
| rs3903072 | DKFZp761E198 | 40 | 7 | 0.00710 | 0.0071–0.0168 | |
| rs3903072 | OVOL1 | 40 | 17 | 0.00548 | 0.00548–0.0243 | |
| rs3903072 | SNX32 | 40 | 6 | 3.53 × 10−6 | 0.00202–0.00553 | |
| rs3817198 | TH | 5 | 1 | 0.00371 | 0.00371–0.00371 | |
| rs13329835 | CDYL2 | 48 | 14 | 9.74 × 10−5 | 0.00064–0.00239 | |
| rs13329835 | DYNLRB2 | 48 | 5 | 0.0163 | 0.0422–0.0668 | |
| rs3803662 | TOX3 | 21 | 5 | 0.0148 | 0.0446–0.071 | |
| rs4808801 | CCDC124 | 72 | 28 | 0.00754 | 0.00754–0.0246 | |
| rs4808801 | LRRC25 | 72 | 7 | 0.00387 | 0.00387–0.0231 | |
| rs3760982 | ZNF155 | 45 | 3 | 6.51 × 10−9 | 1.81e-07–5.07 × 10−7 | |
| Kidney renal clear cell carcinoma | rs718314 | BHLHE41 | 30 | 2 | 0.00487 | 0.0757–0.107 |
| Lung adenocarcinoma | rs7216064 | C17orf58 | 145 | 14 | 0.0206 | 0.0206–0.0402 |
| Prostate adenocarcinoma | rs4245739 | PIK3C2B | 106 | 52 | 0.000108 | 0.000578–0.00422 |
| rs10934853 | RUVBL1 | 102 | 25 | 0.00232 | 0.00232–0.0151 | |
| rs17021918 | BMPR1B | 55 | 9 | 0.00304 | 0.00304–0.006 | |
| rs17021918 | HPGDS | 55 | 1 | 0.000134 | 0.00255–0.00255 | |
| rs12653946 | IRX4 | 12 | 1 | 0.00293 | 0.958–0.958 | |
| rs1933488 | RGS17 | 69 | 5 | 0.0145 | 0.0779–0.0788 | |
| rs2273669 | SESN1 | 204 | 33 | 0.00123 | 0.0103–0.0705 | |
| rs6465657 | LMTK2 | 57 | 6 | 0.00472 | 0.00472–0.00472 | |
| rs11135910 | EBF2 | 35 | 3 | 0.05146 | 0.154–0.154 | |
| rs3850699 | AS3MT | 50 | 16 | 0.000241 | 0.000565–0.00184 | |
| rs3850699 | C10orf32 | 50 | 5 | 8.18 × 10−5 | 0.00246–0.0046 | |
| rs3850699 | SUFU | 50 | 5 | 0.000321 | 0.000801–0.00108 | |
| rs3850699 | TMEM180 | 50 | 2 | 1.62 × 10−9 | 1.62 × 10−9–3.32 × 10−9 | |
| rs10875943 | C1QL4 | 5 | 3 | 5.52 × 10−9 | 5.52 × 10−9–6.98 × 10−9 | |
| rs10875943 | FKBP11 | 5 | 3 | 0.00561 | 0.00561–0.00772 | |
| rs684232 | FAM57A | 64 | 1 | 0.0416 | 0.302–0.302 | |
| rs684232 | GEMIN4 | 64 | 3 | 0.0184 | 0.0184–0.0254 | |
| rs684232 | VPS53 | 64 | 3 | 0.101 | 0.101–0.324 | |
| rs5945619 | NUDT11 | 334 | 113 | 0.00862 | 0.00862–0.0464 |
Figure 2.
Illustration of the fine-mapping candidates of three cancer risk loci based on an integrated posterior probability combining association and epigenetic data. Each point represents a correlated germline variant of the initially reported risk locus (labeled by blue text); the height of the points corresponds to the eQTL level of significance (−log10 P values); the DNaseI HS scores are shown beneath the posterior; the green bars show the posterior probabilities. (A) Xp11.22/rs4945619 with NUDT11 in prostate cancer; (B) 6q25.2/rs1933488 withRGS17 in prostate cancer and (C) 5q11.2/rs889312 with C5orf35in ER-positive breast cancer. NUDT11 and RGS17 demonstrate how incorporating DNaseI HS data can prioritize variants for functional testing in areas with extensive LD. For C5orf35, the strongest candidates are a considerable distance away from and are moderately correlated with the initial risk SNP.
The fine mapping results also suggested that variants not correlated with the risk alleles are associated with the same target gene as the risk variant. Specifically, out of 35 target genes associated with cancer risk loci, 16 have minimally correlated (r2 < 0.3) variants that possess at least one order of magnitude higher significance than the index variant (Supplementary Material, Table S5).
DISCUSSION
eQTLs can serve as an organizing principle. They provide a logical framework for identifying both candidate causal genes and candidate causal loci. They help to identify the particular cell type where the trait-associated allele is acting providing a rationale for selecting the appropriate cell line for follow-up experiments. In addition, eQTLs provide a clear experimental readout (transcript levels) for fine mapping and causal allele identification. In this study, we generated a list of candidate causal genes and causal loci that should be considered as strong candidates for functional evaluation in appropriate assays.
We leveraged the strength of TCGA data. TCGA is notable for amassing multiple data elements on tumors in a quality-controlled fashion. Because expression measurements are performed in tumors, it is important to adjust for somatically acquired alterations as we have previously shown (8). In this study, we used expression as measured by RNA-sequencing as opposed to microarray data in our prior study. Importantly, two of the three cis-eQTLs discovered in our prior study were significant in this study. Interestingly, MYC levels are associated with one of the prostate cancer 8q24 risk loci, rs4242384. Most studies evaluating 8q24 cancer risk loci (across multiple tumor types) and MYC levels are negative (16,17). Despite negative eQTL data, other data such as long-range physical interactions, as well as mouse data for the colon cancer 8q24 risk locus, implicate MYC involvement in 8q24 driven tumors (16,18–20).
The current eQTL analyses are restricted to common variants due to sample size limitations. However, rare variants may also play critical roles to determine the phenotype and should be included in future studies. Another important question is the comparison of eQTLs from the tumor and the normal tissue, which is hindered by lack of matched normal samples in TCGA. Therefore, comparison of eQTL target–genes in both normal and tumor tissue of the same cell type will be interesting. Efforts such as the Genotype-Tissue Expression project will be informative in this regard (4).
Multiple prostate eQTL target–gene pairs that have been previously identified in other studies are observed in this data set, including rs12653946/IRX4 and rs5945619/NUDT11 (21,22). Other previously reported eQTL target–genes, however, did not replicate in the prostate data set such as HNF1B and MSMB (21). Discordant results between studies may be due to power (the current TCGA prostate sample size is smaller than some of the prior studies) and/or because the association is only observed in normal tissue. For prostate cancer associations in particular, some of the discrepancies may be explained by variants that are associated with PSA levels, but not with prostate cancer as have been shown for a handful of polymorphisms (23). Additionally, different analytic and statistical techniques can influence the comparisons between studies. Lastly, it is conceivable that some of the prior associations are false positive results.
Certain genes deserve mention as plausible candidates mediating cancer risk. DYNLRB2 is a gene that has been implicated in the progression from in situ to invasive breast cancer (24). BMPRIB is a serine/threonine kinase that has been shown to inhibit growth and proliferation of prostate cell lines (25). SESN1 is part of a family of genes induced by the p53 protein and has been shown to be repressed by the androgen receptor (26). RGS17, a member of the regulator of G-protein signaling family, shRNA knockdown in prostate cells inhibited proliferation (27). FGF19, a member of the fibroblast growth factor family, was recently shown to function as an autocrine growth factor. Exogenous FGF19 promoted growth, invasion, adhesion and colony formation, whereas decreasing FGF19 decreased invasion and proliferation in vitro and tumor growth in vivo (28). Only one risk locus was associated with a microRNA—the colon cancer risk locus, rs4925386, with miR-1-1. Interestingly, miR-1-1 is frequently methylated in colorectal cancers and it has been suggested to act as a tumor suppressor (29).
A notable observation is that the number of significant eQTL target–gene associations differs substantially among the five tumor types. Despite having similar numbers of risk loci as breast cancer (and fewer samples to analyze), the prostate samples had the greatest number of associations. Trait-associated variants may exert their effects in a cell automonous or non-cell autonomous fashion. Perhaps prostate cancer risk variants are more likely to act in a cell autonomous manner. Further studies will be necessary to definitively address this issue.
While eQTLs are being used to annotate trait-associated loci, they are potentially equally powerful for prioritizing candidate causal loci for downstream functional testing. As stated above, due to LD, a purely genetic approach is unlikely to definitively identify the causal variant. Because an eQTL target–gene association represents a regulatory element and its target gene, a straightforward experimental test with a quantitative readout (i.e. gene expression levels) can be designed. Using genome editing tools such as transcriptional activator like effector nucleases (TALENs) or Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technologies, it is possible to directly engineer single nucleotide changes with pinpoint precision into the nuclear genome without selectable markers (30,31). The expectation is that the causal allele will alter transcript levels whereas a correlated variant will not.
We devised a posterior probability that incorporates both the strength of an association between a variant and transcript levels as well as whether or not the variant was located in a DHS. This test is similar to another recently published test that also accounts for the association signal and epigenetic features (10). Over the past several years, many epigenetic marks have been shown to correlate with various types of regulatory activity (e.g. promoters, enhancers, silencers and insulators). Because we did not want to be biased toward a particular type of regulatory element, we chose to use DHS, a generic mark of open chromatin. In order to increase the sensitivity of the fine mapping analysis, we used the union DHS from 125 cell types to define the prior. This selection is probably made at the cost of specificity. On the other hand, an investigator may select tissue-specific DHS to increase specificity, at the possible cost of sensitivity. Importantly, the final proof of the success or failure of any algorithm that proposes to identify causal alleles will be determined by comparing the top variants from the eQTL analysis with the top variants from case–control fine mapping as well as functional validation. Until more causal alleles are definitively identified and we gain a deeper understanding of how various epigenetic features track with actual causal alleles, individual investigators will be required to make an educated guess as to which feature they believe to be the most important. Our goal was to provide a model that is flexible enough to accommodate investigators' choices for various features.
The data generated in this study provide a list of genes and variants that can be immediately prioritized for functional evaluation. However, some caveats exist. For approximately half of the associations, we have identified for the risk loci, the significance of the risk loci is less (at least one magnitude) than independent variants from the same region. This observation is consistent with two plausible scenarios: first, it is possible that the target gene has nothing to do with the particular phenotype under study. That is, the risk locus can be an eQTL for the target gene and this eQTL target–gene association is completely independent from risk; second, the independent variant is also a risk locus that acts on the same target gene. As is now being observed by many fine mapping studies, genomic risk loci often harbor multiple independent risk alleles (12,32,33). Fine mapping in large-scale case–control studies will distinguish these two scenarios.
Lastly, many risk loci are not significantly associated with a target gene. Possible reasons include evaluation of the wrong cell type (e.g. either a different cell type or subset of morphologically similar cells such as stem cells), transcripts that are outside the tested interval, other mechanisms driving risk (e.g. influencing long non-coding RNA levels), evaluating the incorrect state (tumor instead of normal), measuring expression at the wrong time point, or a false negative due to sample size and/or assay sensitivity.
MATERIALS AND METHODS
Data sets
Five tumor types were included in the eQTL analyses (Table 1). The data were downloaded from ‘The Cancer Genome Atlas’ (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp). A total of 906 600 common variants were genotyped from matched normal DNA samples using Affymetrix SNP 6.0. The expression profiles of mRNA and miRNA in matched tumor samples were obtained from pre-processed RNA sequencing data; the somatic copy number and methylation are also measured and inferred from matched tumor samples.
eQTL analysis
For each cohort of tumors, we determined the ancestry based on the germline genotype data using EIGENSTRAT software with 415 HapMap genotype profiles as control set. Only populations of Northern and Western European ancestries were included in the analyses.
We first performed cis-eQTL analyses for the five tumor types using a method we described previously, in which the association between 906 600 germline genotypes and the expression levels of mRNA or miRNA (located within 500 kb on either side of the variant) were evaluated using linear regression model with the effects of somatic copy number and CpG methylation being deducted. (For miRNA expression, the effect of CpG methylation is not adjusted for since the data are not available.) To adjust for multiple tests, we adjusted the test P-values using the Benjamini–Hochberg method based on the total number of cis-associations that has been tested. A significant association is defined by a FDR of <0.1. For each eQTL analyses, we generated the Q–Q plot based on the distribution of test P-values and the distribution of expected P-values. The inflation factor (λ statistics) is used to indicate possible confounding effects in the analyses.
Following the cis-eQTL analyses, we collected a set of risk loci of the five tumor types which were derived from previous GWAS with P-values below 10−7 (Table 1) (13). For each tumor type, we retrieved the correlated variants (r2 > = 0.7) of the corresponding risk loci that are represented in the germline genotype data as a test set. Again, we adjusted the test P-values using the Benjamini–Hochberg method based on the number of cis-associations tested for the risk loci only. The significant associations are defined by a FDR of <0.1.
Imputation
For each risk locus to which a gene has been found associated in cis, we retrieved the genotype of all SNPs on the Affymetrix 6.0 array within 2 Mb of either side of the risk locus. Using these genotypes and the impute2 March 2012, 1000 Genomes Phase I integrated variant cosmopolitan reference panel of 1092 individuals (haplotypes were phased via SHAPEIT), we imputed the genotypes of SNPs in the 1000 Genomes Project in the target regions for TCGA samples (34–37). For each risk locus to which a gene had been found associated in cis, we retrieved the imputed variants within 500 kb of either side of the risk locus. In order to control for the accuracy, variants of low imputation quality (<0.7) are excluded from the following fine-mapping analysis. We tested for association between imputed SNPs and gene expression using the linear regression algorithm described above, where each imputed SNP was coded as an expected allele count (38).
Fine mapping
For each SNP locus i, we considered two factors: the eQTL level of significance, which is represented by the test P-values from the eQTL analysis; and the potential regulatory activity in the locus (prior), which is represented by the DNaseI hypersensitivity.
We select the candidate causal alleles by a posterior probability which takes into account both aforementioned factors. Let pi denote the P-value resulting from the test between the genotype of a SNP locus i and the target gene, and let χi2 = zi2, where zi = Φ−1(1 − pi/2), and Φ−1 is the inverse of the normal cumulative distribution function. Under the assumption that there is exactly one causal variant in the set of SNPs tested, the posterior probability that SNP i is the causal allele can be approximated by the following expression:
where the sum is over all SNPs in the tested set, δi = 1 if a DNaseI HS site is located within 50 bp of SNP i and δi = 0 otherwise, and πi is a relative prior weight for SNPs in or near DNaseI HS sites (39). DNaseI HS sites were identified from 125 cell types by the ENCODE project (15). To determine πi, we sampled one million random variants from 1000 Genome Project (CEU population) that are uniformly represented in known DNaseI HS sites; and another one million variants in non-DNaseI HS sites. We evaluated the distribution of annotated eQTL activity (B. Stranger, personal communication) in either set. As a result, 11.3% of the variants in or near DNaseI HS sites (δi = 1) turned out to be eQTLs compared with 2.3% of the variants outside any DNaseI HS sites (δi = 0). Thus, we set πi = 0.113/0.023 = 4.91. The higher the posterior, the more likely the variants are the causal.
To identify candidate causal variants, we retrieved all correlated variants (r2 ≥ 0.5) of a known risk locus from the 1000 genomes phase I data and minor allele frequency >0.05 (based on the CEU population). We then calculated the posterior probability for each of the correlated variant i as described.
For each risk locus, we stratified the posterior probabilities of all the correlated variants using a medoid-based partition method (40). Then, we selected the subset of variants with higher posterior as the fine mapping of the original reported risk locus.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Confilict of Interest statement. None declared.
FUNDING
This work was supported by the Elucidating Loci Involved in Prostate Cancer Susceptibility ELLIPSE (U19 CA148537) and Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) project (U19 CA148065). M.L.F., P.K. and C.A.H. on behalf of the GAME-ON Network; the National Natural Science Foundation of China (31371289) (Q.L.) the Fundamental Research Funds for the Central Universities (ZK10121) (Q.L.); AAPC WG-seq grant (C.A.H. and M.L.F.); the Mayer Foundation (M.L.F.), the H.L. Snyder Medical Foundation (M.L.F.), the Kohlberg Foundation, and the A. David Mazzone Awards Program (M.L.F.). M.L.F. was a Howard Hughes Medical Institute Physician-Scientist Early Career Awardee (www.hhmi.org) and a recipient of a 2009 Claudia Adams Barr award (www.dana-farber.org).
Supplementary Material
REFERENCES
- 1.Stranger B.E., Forrest M.S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S., et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G., et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 3.Cheung V.G., Conlin L.K., Weber T.M., Arcaro M., Jen K.Y., Morley M., Spielman R.S. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 4.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nica A.C., Parts L., Glass D., Nisbet J., Barrett A., Sekowska M., Travers M., Potter S., Grundberg E., Small K., et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A., et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q., Seo J.H., Stranger B., McKenna A., Pe'er I., Laframboise T., Brown M., Tyekucheva S., Freedman M.L. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross D.S., Garrard W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 10.Gaffney D.J., Veyrieras J.B., Degner J.F., Pique-Regi R., Pai A.A., Crawford G.E., Stephens M., Gilad Y., Pritchard J.K. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eeles R.A., Olama A.A., Benlloch S., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Ghoussaini M., Luccarini C., Dennis J., Jugurnauth-Little S., et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet. 2013;45:385–391. doi: 10.1038/ng.2560. 391e381-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361. doi: 10.1038/ng.2563. 361e351-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hindorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA (E.B.I.) (European Bioinformatics Institute) 2013. A catalog of published genome-wide association studies, Vol. 2013.
- 14.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomerantz M.M., Ahmadiyeh N., Jia L., Herman P., Verzi M.P., Doddapaneni H., Beckwith C.A., Chan J.A., Hills A., Davis M., et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomerantz M.M., Beckwith C.A., Regan M.M., Wyman S.K., Petrovics G., Chen Y., Hawksworth D.J., Schumacher F.R., Mucci L., Penney K.L., et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009;69:5568–5574. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserman N.F., Aneas I., Nobrega M.A. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright J.B., Brown S.J., Cole M.D. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol. Cell. Biol. 2010;30:1411–1420. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sur I.K., Hallikas O., Vaharautio A., Yan J., Turunen M., Enge M., Taipale M., Karhu A., Aaltonen L.A., Taipale J. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 21.Grisanzio C., Werner L., Takeda D., Awoyemi B.C., Pomerantz M.M., Yamada H., Sooriakumaran P., Robinson B.D., Leung R., Schinzel A.C., et al. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proc. Natl Acad. Sci. USA. 2012;109:11252–11257. doi: 10.1073/pnas.1200853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen H.H., Takata R., Akamatsu S., Shigemizu D., Tsunoda T., Furihata M., Takahashi A., Kubo M., Kamatani N., Ogawa O., et al. IRX4 at 5p15 suppresses prostate cancer growth through the interaction with vitamin D receptor, conferring prostate cancer susceptibility. Hum. Mol. Genet. 2012;21:2076–2085. doi: 10.1093/hmg/dds025. [DOI] [PubMed] [Google Scholar]
- 23.Gudmundsson J., Besenbacher S., Sulem P., Gudbjartsson D.F., Olafsson I., Arinbjarnarson S., Agnarsson B.A., Benediktsdottir K.R., Isaksson H.J., Kostic J.P., et al. Genetic correction of PSA values using sequence variants associated with PSA levels. Sci. Transl. Med. 2010;2:62ra92. doi: 10.1126/scitranslmed.3001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao S., Desouki M.M., Gaile D.P., Shepherd L., Nowak N.J., Conroy J., Barry W.T., Geradts J. Differential copy number aberrations in novel candidate genes associated with progression from in situ to invasive ductal carcinoma of the breast. Genes Chromosomes Cancer. 2012;51:1067–1078. doi: 10.1002/gcc.21991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazaki H., Watabe T., Kitamura T., Miyazono K. BMP signals inhibit proliferation and in vivo tumor growth of androgen-insensitive prostate carcinoma cells. Oncogene. 2004;23:9326–9335. doi: 10.1038/sj.onc.1208127. [DOI] [PubMed] [Google Scholar]
- 26.Wang G., Jones S.J., Marra M.A., Sadar M.D. Identification of genes targeted by the androgen and PKA signaling pathways in prostate cancer cells. Oncogene. 2006;25:7311–7323. doi: 10.1038/sj.onc.1209715. [DOI] [PubMed] [Google Scholar]
- 27.James M.A., Lu Y., Liu Y., Vikis H.G., You M. RGS17, an overexpressed gene in human lung and prostate cancer, induces tumor cell proliferation through the cyclic AMP-PKA-CREB pathway. Cancer Res. 2009;69:2108–2116. doi: 10.1158/0008-5472.CAN-08-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng S., Dakhova O., Creighton C.J., Ittmann M. Endocrine fibroblast growth factor FGF19 promotes prostate cancer progression. Cancer Res. 2013;73:2551–2562. doi: 10.1158/0008-5472.CAN-12-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki H., Takatsuka S., Akashi H., Yamamoto E., Nojima M., Maruyama R., Kai M., Yamano H.O., Sasaki Y., Tokino T., et al. Genome-wide profiling of chromatin signatures reveals epigenetic regulation of MicroRNA genes in colorectal cancer. Cancer Res. 2011;71:5646–5658. doi: 10.1158/0008-5472.CAN-11-1076. [DOI] [PubMed] [Google Scholar]
- 30.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joung J.K., Sander J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bojesen S.E., Pooley K.A., Johnatty S.E., Beesley J., Michailidou K., Tyrer J.P., Edwards S.L., Pickett H.A., Shen H.C., Smart C.E., et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371–384. doi: 10.1038/ng.2566. 384e371–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaneau O., Marchini J., Zagury J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 36.Delaneau O., Zagury J.F., Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 37.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchini J., Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 39.Burnham K, A.D. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. New York: Springer-Verlag; 2002. [Google Scholar]
- 40.Reynolds A.P., Richards G., de la Iglesia B., Rayward-Smith V.J. Clustering rules: a comparison of partitioning and hierarchical clustering algorithms. J. Math. Model. Algorithm. 2006;5:475–504. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.