Graphical abstract
Abbreviations: IDP, intrinsically disordered protein; NMR, nuclear magnetic resonance; RBD, RNA-binding domain; SLBP, stem-loop binding protein
Keywords: Stem-loop binding protein (SLBP), RNA-binding domain (RBD), Intrinsically disordered protein (IDP), Phosphorylation, Histone mRNA, Phosphorus-31 NMR
Highlights
-
•
SLBP is an intrinsically disordered protein (IDP) in the absence of RNA.
-
•
A phosphothreonine in the SLBP RNA-binding domain stabilizes the SLBP–histone mRNA complex.
-
•
This phosphate exhibits torsional strain as revealed by its 31P NMR chemical shift.
-
•
Phosphates can play structural roles in stabilizing tertiary structure in IDPs.
-
•
31P NMR can be a good spectroscopic probe for folding of phosphorylated IDPs.
Abstract
Phosphorus-31 (31P) NMR can be used to characterize the structure and dynamics of phosphorylated proteins. Here, I use 31P NMR to report on the chemical nature of a phosphothreonine that lies in the RNA binding domain of SLBP (stem-loop binding protein). SLBP is an intrinsically disordered protein and phosphorylation at this threonine promotes the assembly of the SLBP–RNA complex. The data show that the 31P chemical shift can be a good spectroscopic probe for phosphate-coupled folding and binding processes in intrinsically disordered proteins, particularly where the phosphate exhibits torsional strain and is involved in a network of hydrogen-bonding interactions.
1. Introduction
Intrinsically disordered proteins (IDPs) are stably unfolded under physiological conditions and are highly prevalent in the eukaryotic genome [1,2]. Disordered regions in proteins can be sites of protein–protein or protein–nucleic acid interaction [3,4]. IDPs are known to undergo either disorder-to-order transitions [2,5] upon binding their partner, or they may form “fuzzy” complexes that remain dynamic in the complex [6–8]. Post-translational modifications or PTMs such as phosphorylation, acetylation, methylation, and ubiquitination also frequently occur in intrinsically disordered regions in proteins [9]. These PTMs can modulate the affinity of the IDP with its binding partner by either conformational selection [10] or induced fit mechanisms, or both [11]. Understanding how PTMs can modulate protein–protein or protein nucleic acid interactions in IDPs is important as it provides mechanistic understanding into how IDPs function in biological pathways.
Stem-loop binding protein (SLBP) is a histone mRNA specific RNA processing factor that forms a stable and specific complex with a 16 nucleotide stem-loop in the 3′ untranslated region of histone mRNA [12]. Human and Drosophila SLBPs are IDPs in the absence of RNA [13–16]. A unique feature of SLBP proteins is that they are phosphorylated at Thr171 (human SLBP numbering) in their RNA-binding domains (RBD) [17] and phosphorylation at this site is important for the kinetics of association with the RNA [17,15]. Dephosphorylation at Thr171 results in a ∼10-fold faster on rate and a ∼100-fold faster off-rate for the histone mRNA stem-loop [15]. The effect on the overall dissociation constant (Kd) is ∼7–11-fold [15,17] lower affinity for the histone mRNA stem-loop, although there is a larger effect on the microscopic association (kon) and dissociation (koff) constants, and hence the kinetics of RNA binding [15]. NMR studies in solution also provide evidence for cis–trans isomerization about the Thr171–Pro172 sequence [15], and heteronuclear NMR studies show that mutation of Pro172 to glycine results in a single major conformation in solution [15]. Therefore the intrinsic disorder observed in solution is at least in part due to proline isomerization. Mutation of Pro172 also results in loss of RNA binding and embryonic lethality in Caenorhabditis elegans [15,18]. Consistent with this, the Pro172Gly mutant does not efficiently bind histone mRNA stem-loop in an EMSA assay [15]. In the crystal structures [19], the Pro172 ring is in the trans configuration, and shows van der Waals interactions with Trp183, Ile187, and Trp190. Mutation of Pro172 or isomerization to the cis-conformer would likely disrupt hydrophobic packing in this region and hence destabilize the SLBP RBD–RNA complex, as previously reported [15,20].
Phosphorylation at Thr171 is important for subcellular localization of SLBP in the nucleus [20,21], for efficient histone mRNA processing [21] and mRNA decay [20], and SLBP protein stability in vivo [20] by regulating the stability of the SLBP–histone mRNA complex [22]. The structural environment around the phosphothreonine, as observed in the crystal structure of the human SLBP RBD complexed to histone mRNA stem-loop (Fig. 1), provides some insight into the role of threonine phosphorylation in RNA binding. When expressed in baculovirus, the 98-residue human SLBP RBD is phosphorylated at only one site (Thr171), as was previously confirmed by mass spectrometry [15,17]. In the crystal structure of the dephosphorylated SLBP–histone mRNA complex [19], no electron density is observed for 37 out of 99 residues in the SLBP RBD, particularly those surrounding the site of phosphorylation and the C-terminus. In addition, 57 out of 99 residues have high B-factors that may be attributed to the flexibility of the SLBP RBD. Phosphorylation of Thr171 stabilizes the structure of the RBD (PDB code 4QOZ), particularly around the phosphate. The phosphate is involved in a network of hydrogen-bonding interactions with residues in the three α-helices of the SLBP RBD in the presence of the RNA ligand, which helps stabilize the protein–RNA complex. The crystal structure is consistent with previous biophysical and NMR studies that showed that both phosphorylation and RNA binding of human [15] and Drosophila SLBP [13] proteins is important for stable recognition of the histone mRNA stem-loop.
Fig. 1.
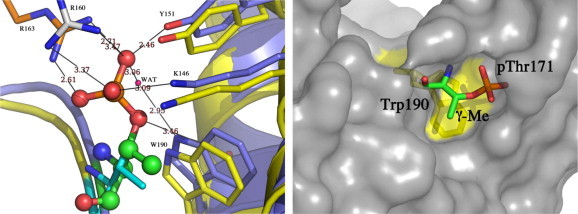
(Left) Electrostatic and aromatic environment around the phosphothreonine as observed in the crystal structure of the phosphorylated SLBP RBD–histone mRNA stem-loop-3′hExo ternary complex (PDB code 4QOZ) is shown in purple. The structure of the unphosphorylated SLBP RBD–histone mRNA stem-loop-3′hExo ternary complex (PDB code 4L8R) is superimposed in yellow. Hydrogen bonding interactions and distances to the phosphoryl oxygens (shown in ball and stick) are indicated. The γ-methyl group of the threonine is shown in green ball and stick. (Right) The surface of the SLBP RBD is shown. The phosphothreonine lies in a pocket where the phosphate group is solvent exposed while the γ-methyl group lies directly above the indole ring within van der Waals contact distance. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
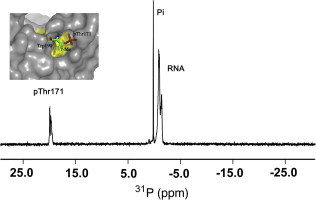
Here I report that the 31P resonance for the phosphothreonine resonates 20 p.p.m downfield of H3PO4 in the SLBP–RNA complex. We previously reported this chemical shift in the 31P NMR spectrum of the baculovirus expressed Drosophila SLBP (dSLBP) RBD–histone mRNA stem-loop complex [13]. However, it was not possible to unambiguously attribute the shift to a single phosphate at the time since baculovirus-expressed dSLBP RBD is phosphorylated at the analogous threonine (T230 in dSLBP) [17] as well as four serines in the extreme C-terminus [23]. Here, 31P NMR has been used to describe the chemical nature of the phosphate corresponding to phosphorylated Thr171 and monitor the response of this phosphate in the human SLBP RBD to the presence of RNA. The 31P NMR data indicate that the orthophosphate that is covalently bonded to the threonine exhibits torsional strain in solution. The results have important implications for the role of phosphorylation in other IDPs. I propose that since many IDPs are phosphorylated, phosphates may play an important structural role in stabilizing the tertiary fold of such proteins, particularly in the presence of their ligands, and may also exhibit anomalous 31P chemical shifts as is observed for SLBP.
2. Materials and methods
2.1. Protein expression, purification, and NMR sample generation
A 128-residue pseudo-wild-type hSLBP RBD construct was designed to increase expression and ensure stoichiometric phosphorylation at Thr171. The hSLBP RBD (residues E118–E219) were cloned into the Nco1 and Xho1 restriction sites of the vector pFastBac™HTA (Invitrogen) and was expressed in Sf9 cells using the Bac-to-Bac expression system (Invitrogen) as previously described [15]. The protein was expressed in sf9 cells and purified using standard protocols used for Ni2+ affinity chromatography followed by gel filtration. Phosphorylation of the protein was confirmed by Electrospray Ionization Mass Spectrometry (ESI-MS) which gave a measured monoisotopic mass of 15255.40 Da corresponding to removal of the N-terminal Met (−131), acetylation of the new Ser (+42) N-terminus and phosphorylation of Thr171 (+80) as expected from previous studies [15,17]. Samples were concentrated and buffer exchanged using a G25 column into the NMR buffer (see below).
2.2. NMR spectroscopy
One-dimensional 31P NMR experiments were performed on a Varian Inova 500 MHz spectrometer using a broadband probe operating at a phosphorus frequency of 202 MHz. Unless otherwise noted, all measurements were made at 25 °C. For each experiment between 1000–20,000 transients were collected with a 65° excitation pulse, a recycle delay of 3 s, and a sweep width of 98.7 p.p.m with proton decoupling, unless otherwise noted. All 31P chemical shifts were referenced to 85% phosphoric acid. Experiments were recorded on 1–3 mM protein/peptide samples dissolved in 20 mM deuterated Tris, 50 mM NaCl, 0.1% sodium azide and 100% D2O. The hSLBP RBD–RNA complex samples contained a sixfold molar excess of RNA relative to protein.
3. Results and discussion
In the absence of RNA, two 31P NMR resonances are observed for a single phosphate in the hSLBP RBD (Fig. 2) at basic pH (pH > 8.5) at 3.00 p.p.m and 4.19 p.p.m, both of which lie within the range of that expected for o-phosphothreonine (3–5 p.p.m) [15]. The linewidths for these resonances are broad, consistent with the hypothesis that this domain undergoes conformational exchange between multiple states as previously reported for both Drosophila [13] and human [15] SLBP RBDs. In the absence of RNA, unphosphorylated and phosphorylated hSLBP and dSLBP RBDs are intrinsically disordered [13,15] with a large hydrodynamic radius that is characteristic of a pre-molten globule state [13]. At pH 8.1, the upfield-shifted resonance has an apparent linewidth of 28.2 Hz while the downfield resonance has an apparent linewidth of 89.5 Hz. This broad resonance comprises a number of conformational sub-states that are only slightly different in chemical shift at pH 8.1, but become more apparent at pH 7.0 (Fig. 2). When RNA is titrated into the free protein, the phosphothreonine resonance undergoes a remarkable downfield-shift (Fig. 2) to a resonance position (∼20 p.p.m downfield of 85% H3PO4) that may be attributed to a deviation of the O–P–O σ-bond angle from that of a perfect tetrahedron (109°28′) to that observed in a five-membered cyclic phosphate ester (I) [24] (Fig. 3) or an increase in chemical-shift anisotropy due to the electronegativity of the phosphate due to next-nearest-neighbor ligands as seen in cation–phosphate interactions [25]. It is not the chemical shift expected for a free dianionic orthophosphate (II) (Fig. 3). At least two conformations are observed in the RNA bound complex as well (at 20.02 and 20.3 p.p.m) suggesting that it remains dynamic when bound to RNA in solution.
Fig. 2.
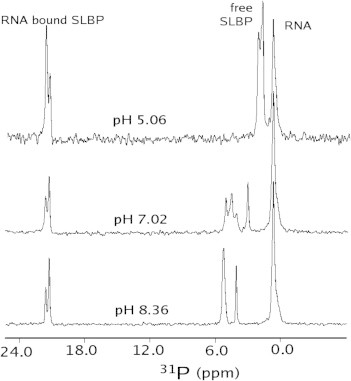
31P NMR spectra of the SLBP RBD–RNA complex collected at 25 °C at a spectrometer frequency of 202 MHz and at different pH values as indicated. A detailed pH titration is shown in Supplementary materials (Fig. S2).
Fig. 3.
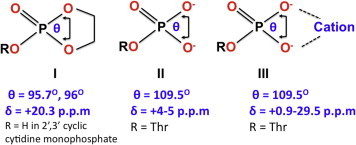
Expected 31P chemical shifts and dihedral angles due to the effect of structure and cations on an orthophosphate. The shifts are taken from references [25–28].
To determine the ionization states of the phosphoryl groups, the 31P chemical shifts were followed as a function of pH in a pH range of 4–10 (Fig. 2, Supplementary Fig. S2). At neutral pH, the phosphoryl groups corresponding to free SLBP exist in multiple conformations in solution (Fig. 2) that are in slow exchange on the NMR timescale. Similar to inorganic phosphate (pKa 7.0 ± 0.01) and the o-phosphothreonine standard (pKa 6.3 ± 0.02), these 31P resonances go through a single ionization event corresponding to an apparent pKa of 6.9. The pH profile at pH values below 4.3 were not determined due to limited protein–RNA complex solubility under these conditions. Therefore, in the absence of RNA, and at neutral pH, the phosphothreonine in free SLBP favors a dianionic form that is dominant in all the observed conformational states. In contrast, the pH response of the two 31P resonances in the RNA bound form of the protein is flat and the chemical shift does not titrate with pH as would be expected for a dianionic phosphate inferred from the crystal structure (Fig. 2, Supplementary Fig. S2). The introduction of electrostatic interactions between a phosphoserine and a tyrosine and lysine in the KID–KIX complex has been reported to decrease the pKa of the phosphate to 4.3 ± 0.2 [26]. Therefore, it is not surprising that the network of electrostatic interactions mediated by the phosphate in the hSLBP RBD–RNA complex decreases the pKa of the phosphate to <4. Although the 31P resonances in the bound complexes do not exchange with pH, the relative populations of the two forms does change such that the downfield shifted conformer is preferred at acidic pH, whereas the upfield shifted conformer is preferred at basic pH.
The unusual 31P shift observed in the SLBP–RNA complex is intriguing since such a downfield shift has never been previously reported for an orthophosphate. The only other protein for which an unusual 31P chemical shift has been reported is alkaline phosphatase where the catalytic phosphoserine was reported to resonate 8.5 p.p.m down field of H3PO4 and this effect was attributed to torsional strain [27,28]. No resonance is observed between +5 and +30 p.p.m in the spectrum of the free RNA or the NMR buffer (Supplementary Fig. S3). Temperature titration of the SLBP RBD in the presence of RNA (Supplementary Fig. S1) shows that the phosphate resonances of the free SLBP protein in solution exchange with those of the bound complex. No downfield shift is observed for a complex of non-phosphorylated SLBP and RNA (not shown). The addition of a denaturant such as urea to the hSLBP RBD–RNA complex results in a change in peak intensities for the resonances at 20 p.p.m and the free protein (Supplementary Fig. S4), as would be expected if the resonance came from the protein. 31P chemical shifts in RNA usually cluster between −3 and −1 p.p.m (upfield of orthophosphoric acid), however it is conceivable that unusual backbone torsional angles can shift a phosphate resonance originating from the RNA. The tetraloop structure of the RNA hairpin is significantly distorted and unfolded in the crystal structures of both phosphorylated and non-phosphorylated SLBP–RNA complexes [19,29]. To determine whether the shift can be attributed to the protein or the RNA, the SLBP RBD–RNA complex was purified over a gel filtration column to remove excess free SLBP RBD from the bound complex. A 31P NMR spectrum of the purified SLBP RBD complex shows only the downfield shifted resonance at 20.3 p.p.m and resonances between −3 and −1 p.p.m for the RNA (Fig. 4). No phosphate resonances are observed between 2 and 4 p.p.m for the phosphothreonine, as would be expected for a phosphothreonine in the free SLBP RBD. Taken together, the data shows that the anomalous 31P chemical shift in the SLBP–RNA complex can be directly attributed to the environment surrounding the site of phosphorylation and cannot be attributed to either the RNA or to a contamination in the protein/RNA complex.
Fig. 4.
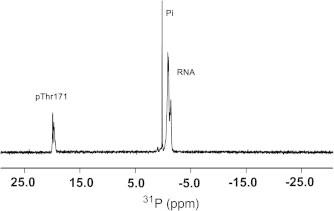
1D 31P NMR spectrum of the phosphorylated SLBP RBD–RNA complex collected after removal of free SLBP by gel filtration.
Based upon quantum-mechanical calculations, Lechter and Van Wazer proposed 40 years ago that 31P chemical shift differences Δδ arise from the sum of three contributing effects: the difference in the electronegativity of the P–X bond (Δχx) due to nearest-neighbor ligands, the change in the π-bonding orbitals about the phosphorus atom (Δnπ), and Δθ or the change in the O–P–O σ-bond angle according to Eq. (1), where C, k, and A are constants,
| (1) |
The depressed pKa of the phosphothreonine in the SLBP RBD–RNA complex suggests that these phosphates may exist in a strong electrostatic interaction with neighboring residues. In the crystal structure (PDB code 4QOZ), the phosphoryl oxygens are involved in a network of hydrogen bonds with Arg163, Tyr151, and Lys146 (Fig. 1). A weak hydrogen bonding interaction may also be present with Arg160 (the guanidinium ηN–O bond distances are 3.47 Å and 3.37 Å). A phosphoryl oxygen also makes a hydrogen bond to a water molecule, which is also coordinated to the Trp190 indole nitrogen. All known SLBPs have threonine conserved at position 171 (but never a serine) and tryptophan is also conserved at position 190 and is important for RNA binding [30]. The observed γ-methyl–π interaction likely contributes favorably to the SLBP–RNA complex stability, and also increasing the chemical-shift anisotropy on the phosphate, likely contributing to the downfield shift of the phosphorus-31 resonance.
No metal ion is observed in the vicinity of the phosphate in the crystal structure (PDB code 4QOZ). To test whether coordination of a metal ion to the phosphate in solution could explain this chemical shift behavior, up to 50 mM EDTA was titrated into the SLBP–RNA complex. Addition of EDTA had no appreciable change on the spectrum (data not shown). Addition of Mg2+ ions also did not have any appreciable effect on the downfield shifted peaks. Therefore perturbation of the π-electron cloud via metal-ion coordination does not explain the observed anomaly.
The contribution of electronegative effects to the 31P chemical shift is generally considered to be small [31]. A ∼2 p.p.m downfield shift (to 6 p.p.m) is reported for the KID–KIX complex where the phosphate participates in two hydrogen bonds [26]. Although the multitude of interactions mediated by the phosphoryl oxygens observed in the hSLBP RBD–RNA complex i.e. the change in the π-electron overlap (Δnπ) due to the hydrogen bonding interactions between the phosphoryl oxygens as well as the favorable γ-methyl interaction with the indole ring of Trp190 likely contributes to the large change in chemical shift observed for the phosphate resonance, it is unlikely to be the dominant or sole contributing factor.
The anomalous chemical shift observed in solution is most likely attributed to steric strain imposed by salt-bridging interactions as previously reported for alkaline phosphatase. Previous studies have shown that there is an empirical correlation between the O–P–O bond angle and the 31P chemical shift such that a decrease in the O–P–O bond angle by ∼3° is correlated with a downfield shift of the 31P resonance by ∼4 p.p.m [24]. The chemical shifts I report for the phosphate in SLBP are very close to those reported for five-membered cyclic phosphate esters in tetra co-ordinated phosphate compounds where the chemical shifts range between +10 and 20 p.p.m (Fig. 3(I)). The O–P–O bond angle surprisingly deviates from the ideal value of 109.5° and ranges between 89.49° and 127.77° over 74 PDB structures that have phosphothreonine for which the bond angles were measured (Supplementary Table 1) with several crystal structures showing decreased O–P–O bond angles. However, the measured O–P–O angles in the SLBP–RNA complex crystal structure are close to tetrahedral geometry (107.64°, 109.95°, 110.64°, and 109.92°). Contrary to this, the NMR data indicates that the phosphate in the hSLBP RBD exhibits torsional strain in solution, suggesting that the stereochemistry around the phosphate in solution may differ from that observed in the crystal.
4. Conclusions
The database of reported 31P chemical shifts for phosphorylated Ser/Thr/Tyr residues in proteins is small. The studies reported here along with previous studies on alkaline phosphatase suggest that orthophosphates can show anomalous chemical shifts in proteins due to the propensity of the phosphoryl oxygen to engage in a network of electrostatic interactions via nearest-neighbor effects on the oxygens as well as the geometry of the O–P–O bond. The presence of torsional strain on the phosphate may be particularly relevant in the case of intrinsically disordered proteins such as SLBP, where the phosphate brings together elements of secondary structure, thereby stabilizing the overall tertiary fold.
Acknowledgements
This research was supported by the National Institutes of Health (NIH) Grant 1RO1-GM076660 (Phosphorylation-dependent recognition of a histone mRNA stem-loop by SLBP). RT conceived, designed, and performed all experiments, analyzed and interpreted the data and wrote the paper.
Appendix A. Supplementary data
This pdf file contains Supplementary Table S1 and Figs. S1–4.
References
- 1.Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem. Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 3.Hsu W.L., Oldfield C., Meng J., Huang F., Xue B. Intrinsic protein disorder and protein–protein interactions. Pac. Symp. Biocomput. 2012:116–127. [PubMed] [Google Scholar]
- 4.Minezaki Y., Homma K., Kinjo A.R., Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 5.Spolar R.S., Record M.T., Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 6.Mittag T., Kay L.E., Forman-Kay J.D. Protein dynamics and conformational disorder in molecular recognition. J. Mol. Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- 7.Fuxreiter M., Tompa P. Fuzzy complexes: a more stochastic view of protein function. Adv. Exp. Med. Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 8.Mittag T., Orlicky S., Choy W.Y., Tang X., Lin H. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pejaver V., Hsu W.L., Xin F., Dunker A.K., Uversky V.N. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23:1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bui J.M., Gsponer J. Phosphorylation of an intrinsically disordered segment in Ets1 shifts conformational sampling toward binding-competent substates. Structure. 2014;22:1196–1203. doi: 10.1016/j.str.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battle D.J., Doudna J.A. The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA. 2001;7:123–132. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thapar R., Marzluff W.F., Redinbo M.R. Electrostatic contribution of serine phosphorylation to the Drosophila SLBP–histone mRNA complex. Biochemistry. 2004;43:9401–9412. doi: 10.1021/bi036315j. [DOI] [PubMed] [Google Scholar]
- 14.Thapar R., Mueller G.A., Marzluff W.F. The N-terminal domain of the Drosophila histone mRNA binding protein, SLBP, is intrinsically disordered with nascent helical structure. Biochemistry. 2004;43:9390–9400. doi: 10.1021/bi036314r. [DOI] [PubMed] [Google Scholar]
- 15.Zhang M., Lam T.T., Tonelli M., Marzluff W.F., Thapar R. Interaction of the histone mRNA hairpin with stem-loop binding protein (SLBP) and regulation of the SLBP–RNA complex by phosphorylation and proline isomerization. Biochemistry. 2012;51:3215–3231. doi: 10.1021/bi2018255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bansal N., Zhang M., Bhaskar A., Itotia P., Lee E. Assembly of the SLIP1–SLBP complex on histone mRNA requires heterodimerization and sequential binding of SLBP followed by SLIP1. Biochemistry. 2013;52:520–536. doi: 10.1021/bi301074r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borchers C.H., Thapar R., Petrotchenko E.V., Torres M.P., Speir J.P. Combined top-down and bottom-up proteomics identifies a phosphorylation site in stem-loop-binding proteins that contributes to high-affinity RNA binding. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3094–3099. doi: 10.1073/pnas.0511289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kodama Y., Rothman J.H., Sugimoto A., Yamamoto M. The stem-loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans. Development. 2002;129:187–196. doi: 10.1242/dev.129.1.187. [DOI] [PubMed] [Google Scholar]
- 19.Tan D., Marzluff W.F., Dominski Z., Tong L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science. 2013;339:318–321. doi: 10.1126/science.1228705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan N., Titus M.A., Thapar R. The prolyl isomerase pin1 regulates mRNA levels of genes with short half-lives by targeting specific RNA binding proteins. PLoS One. 2014;9:e85427. doi: 10.1371/journal.pone.0085427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanzotti D.J., Kupsco J.M., Yang X.C., Dominski Z., Marzluff W.F. Drosophila stem-loop binding protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol. Biol. Cell. 2004;15:1112–1123. doi: 10.1091/mbc.E03-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnan N., Lam T.T., Fritz A., Rempinski D., O’Loughlin K. The prolyl isomerase pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP–histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol. Cell. Biol. 2012;32:4306–4322. doi: 10.1128/MCB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominski Z., Yang X.C., Raska C.S., Santiago C., Borchers C.H. 3′ End processing of Drosophila melanogaster histone pre-mRNAs: requirement for phosphorylated Drosophila stem-loop binding protein and coevolution of the histone pre-mRNA processing system. Mol. Cell. Biol. 2002;22:6648–6660. doi: 10.1128/MCB.22.18.6648-6660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blackburn G.M., Cohen J.S., Weatherall I. Phosphorus-31 nuclear magnetic resonance studies of cyclic derivatives of phosphorus oxy-acids. Tetrahedron. 1971;27:2903–2912. [Google Scholar]
- 25.Turner G.L., Smith K.A., Kirkpatrick R.J., Oldfield E. Structure and cation effects on phosphorus-31 NMR chemical shifts and chemical-shift anisotropies of orthophosphates. J. Magn. Reson. 1986;70:408–415. [Google Scholar]
- 26.Mestas S.P., Lumb K.J. Electrostatic contribution of phosphorylation to the stability of the CREB–CBP activator–coactivator complex. Nat. Struct. Biol. 1999;6:613–614. doi: 10.1038/10655. [DOI] [PubMed] [Google Scholar]
- 27.Bock J.L., Sheard B. 31P NMR of alkaline phosphatase. Biochem. Biophys. Res. Commun. 1975;66:24–30. doi: 10.1016/s0006-291x(75)80289-0. [DOI] [PubMed] [Google Scholar]
- 28.Chlebowski J.F., Armitage I.M., Tusa P.P., Coleman J.E. 31P NMR of phosphate and phosphonate complexes of metalloalkaline phosphatases. J. Biol. Chem. 1976;251:1207–1216. [PubMed] [Google Scholar]
- 29.Thapar R., Denmon A.P., Nikonowicz E.P. Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins. Wiley Interdiscip. Rev. RNA. 2014;5:49–67. doi: 10.1002/wrna.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominski Z., Erkmann J.A., Greenland J.A., Marzluff W.F. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and for histone pre-mRNA processing. Mol. Cell. Biol. 2001;21:2008–2017. doi: 10.1128/MCB.21.6.2008-2017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorenstein D. Academic Press Inc.; Orlando, Florida: 1984. Phosphorus-31 NMR Principles and Applications. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This pdf file contains Supplementary Table S1 and Figs. S1–4.


