Abstract
Odorant receptor (OR) gene choice is a paradigmatic example of stochastic regulation in which olfactory neurons choose one OR from > 1,000 possibilities. Recent biochemical, mathematical, and in vivo findings have revealed key players, introduced new axes of control, and brought the core mechanisms of the process into sharper focus.
Keywords: chromatin and transcription, Eukaryotic transcription
Introduction
The ability to perceive the chemical world is a universal feature of life. In mammals, the olfactory system detects and discriminates between vast numbers of distinct volatile molecules (recently estimated at an astonishing > 1012 in humans1) providing critical information that allows an organism to respond appropriately to changes in its environment. The universe of volatile odorants is accommodated by a large gene family of greater than 1,000 odorant receptors (ORs), employed by the olfactory sensory neurons (OSNs) to commence a cascade that ends in the perception of smell.2 The ORs are seven-transmembrane G-protein coupled receptors encoded by genes found in large and small arrays that are scattered throughout the genome.3,4 ORs are expressed in the olfactory epithelium in an intriguing manner: each OSN chooses a single OR family member and transcribes it from only one allele.5,6 This monogenic and monoallelic mode of gene regulation has captivated the interest of neuroscientists and biologists across disciplines.
Stochastic Choice
The pattern of receptor expression in the nose, in which any given OR is expressed sparsely and irregularly across a limited sub-region spanning the olfactory epithelium, suggested early in the study of the gene family that a stochastic mechanism may underlie selection.7,8 Further supporting evidence came from the demonstration that OR expression is monoallelic, with unbiased transcription from either allele.6 Finally, several labs have derived mouse lines bearing OR transgenes that were shown to recapitulate the monogenic and monoallelic regulation, with exogenous and endogenous OR alleles expressed exclusively from each other in a manner reminiscent of the antigen receptor genes.9,10,11,12 Together these findings strengthened the proposition that while deterministic regulation constrains receptor choice to a particular zone in the epithelium, a stochastic mechanism lies at the core of its monogenic and monoallelic selection.
Initiation and Maintenance of Expression
The initial choice of a receptor is suggested to involve a random process that limits expression of OR to just one allele, and once chosen, a maintenance mechanism ensures its faithful expression for the life of the cell.13 Genetically modified lineage-marking alleles that can reveal the history of expression of an OR in an OSN similarly demonstrated sparse expression, suggesting that regulation does not involve an initial pervasive activation followed by a winnowing process.14 These lineage-marking alleles further demonstrated that the choice of non-functional, pseudogenized OR was not maintained in the epithelium but was instead shut down, with the OSN continuing to choose until it expressed a functional receptor.14 This process, termed OR gene “switching” suggested that only functional receptor may maintain its expression, and together with similar analyses of OR pseudogenes15,16 established the idea that feedback control is critical for maintenance of OR expression.17 Intriguingly, functional OR alleles were observed to undergo low-frequency switching by the sensitive lineage-marking strategy, with the MOR28 receptor switching in 10% of the OSNs that initially selected it.14 This “wild type switching” was attributed to an inherently dynamic system, tuned to efficiently remove non-functional OR alleles from the final expressed OR repertoire. Thus OR regulation is considered to result from a random initial choice followed by a feedback mechanism that halts the process after the expression of a functional OR allele.
Initiation of OR Choice
Two types of mechanisms have been suggested to yield random, singular selection: one using kinetic constraints to limit OR expression and an alternate spatial model proposing the existence of a single selection machinery able to interact with one allele at a time.13 In the kinetic model, initial OR expression is proposed to be inefficient such that only one OR allele is likely to be expressed within a given window of time. In a biochemical tour de force, Magklara et al. revealed the dynamic epigenetic marks of OR chromatin in OSNs. OR chromatin in olfactory stem cells and non-olfactory tissue is enriched in histone H3K9me2, which gives way to more repressive H3K9me3 and H4K20me3 marks in newborn, OR negative OSNs, that then persist on non-selected OR genes throughout the life of the neuron.18 The chromatin of expressed ORs loses these heterochromatic marks and instead is found to be enriched in H3K4me3. These epigenetic findings are consistent with a kinetic model of OR regulation and suggest a delicate tuning between activation and repression could underlie initiation. What could mediate the inflection point between permissive and non-permissive chromatin in the OSN? A logical candidate would be a histone demethylase and recent elegant experiments by Lyons et al. identified the transient expression of the lysine-specific demethylase 1 (Lsd-1) in the immature OSN population as a key event.19 At the same time as OR chromatin in immature OSNs is becoming highly repressed through H3K9 and H4K20 trimethylation, the lysine demethylase Lsd-1 is transiently expressed in this population.19 The critical event in this model of activation is the slow conversion of repressed OR chromatin to a permissive state, which is believed to involve the demethylation of histones H3K9me2 and H4K20me2, prior to their conversion to the more fully repressed trimethylated state. Lsd-1 can remove methyl groups from H3K9me2 and H4K20me2 and thus could act as a key transcriptional coactivator.
We have recently used a genetic approach to functionally “interrogate” an OR allele in vivo, asking whether the OR chromatin was permissive for activation by an exogenous, non-OR promoter inserted into the locus.20 After placing the tetracycline-dependent transactivator responsive promoter (teto) at the start site of the P2 receptor gene by homologous recombination to generate the tet-P2 mouse line, we observed that the OR genomic milieu imposed constraints on teto-mediated activation of the locus by the tetracycline-dependent transactivator (tTa) including zonal restriction and sparse initial activation. Thus the tet-P2 allele obeyed the rules of the OR locus, recapitulating the phenomenology of OR expression, despite the ability for ubiquitous activation across the epithelium by the pervasive presence of tTa. These data are consistent with the existence of a kinetic constraint placed upon the OR locus to limit initial activation, which may be mediated by the unique epigenetic state of OR chromatin. Together these in vivo and in vitro findings are leading to a consensus in the field in which kinetic limitations on OR transcription, through restrictive OR chromatin, potentially coupled with transient expression of Lsd-1, may generate the sparse and random activation required to set in motion the initiation of monogenic, monoallelic OR expression.
Zonal Control
The zonal regulation of OR choice, in which a given OR is expressed only in a limited sub-region of the olfactory epithelium, suggests an added level of restriction not yet explained by epigenetic analyses, but whose mode was revealed by our in vivo analyses. Using the tet-P2/tTa set up, we observed a graded frequency of expression of tet-P2, with the highest likelihood of activation coming from within the wild type P2 zone. Outside of this zone, the frequency of tet-P2 expression diminishes despite the uniform high-level presence of tTa, suggesting that zonal regulation may be accomplished by increasing repression of the OR locus outside of its home zone.20 It is not likely that Lsd-1 expression alone could account for zonal regulation as the enzyme is expressed uniformly across the neuroepithelium. Additional zonal epigenetic marks could explain these data, and the implications of our single-cell resolution in vivo analyses await biochemical confirmation. Nonetheless, the simplest explanation for zonal regulation is a gradient of repression extending outside of the zone.
Location, Location, Location
The first olfactory enhancer element identified, a 2.1kb element termed H, is conserved between humans and mice and regulates an OR cluster which includes the MOR28 receptor gene.21 Remarkably, the H element which resides on chromosome 14 in mice, was shown to associate at high frequency in trans with expressed OR genes from disparate loci from across the genome.22 This finding propelled thinking about “spatial” models of singular OR activation in which one allele of H might act as a master OR trans-enhancer. Subsequent genetic ablation studies revealed a more limited, cis-acting role for H23 but recent FISH studies have resurrected a function for inter-chromosomal interactions and nuclear architecture in OR regulation. Using a pan-OR DNA FISH probe Clowney et al. have shown that OR loci across the genome congregate in ~5 discrete foci that decorate the chromocenters in mature OSN.24 The expressed OR allele is predominantly found excluded from these clusters, which are enriched in repressive epigenetic marks. Further, the lamin b receptor (LBR), observed to be expressed in immature OSNs, was implicated in OR choice as ectopic expression of LBR in mature OSNs disrupts the tertiary inter-chromosomal structure and leads to a generalized decrease in OR expression along with violations in the one-receptor-per-neuron phenomenology of OR regulation.24
Feedback Control
Stochastic mechanisms may have an inherent requirement for feedback to halt the random process after a desired outcome is achieved. In the generation of antigen receptor in the immune system, that outcome is expression of a single, in-frame rearrangement whereas in odorant receptor gene choice it is expression of a single, functional OR. In the absence of feedback in antigen receptor choice, allelic exclusion may be breached as the Rag recombinase continues to rearrange genomic sequences.25 In the case of OR regulation, additional OR alleles would eventually be expressed in the kinetic model of choice while a single selection machinery may continue to randomly hop between different OR alleles and activate them in the sptial model; in either of these unconstrained situations the monogenic and monoallelic regulation of OR would be abrogated. A simple feedback circuit was recently revealed by Lyons et al.19 in which expression of functional OR triggers activation of adenylyl cyclase III expression which in turn downregulated Lsd-1. Remarkably, it was revealed that this circuit is mediated by the induction of the unfolded protein response (UPR) by functional OR protein and upon its relief, leads to activation of adenyly cyclase III and shutdown of Lsd-1.26 This has led to a conception of feedback control in which non-selected OR alleles are repressed prior to selection and the functional outcome of feedback, as mediated through the UPR, is the downregulation of Lsd-1 and thus the suppression of further de-repression and subsequent expression of additional OR alleles.19,26,27
We took advantage of the conditional expression capabilities of our tet-P2 mouse line to detect changes in OR chromatin as OSNs mature. Using staged doxycycline feeding, a developmental change in the permissiveness of the OR chromatin was observed, corresponding to repression of the locus at later stages of OSN maturation. OR chromatin in younger OSNs showed higher levels of permissiveness for tet-P2 activation.20 These findings differ somewhat from Lyons et al. and suggest that only at the stage when the OR repertoire is stably expressed is the chromatin of non-selected OR alleles more heavily repressed. The epigenetic analyses to date reveal H3K9me3 and H4K20me3 marks are laid down on OR chromatin in newborn, immature OSN and persists on non-selected alleles for the life of the cell.18 Our functional dissection of OR permissiveness during OSN development suggests that OR chromatin remains permissive until shortly after OMP expression, after which complete repression is observed. Thus there are likely additional dynamic features of OR chromatin to be revealed through biochemical analyses. Finally, it is possible that feedback commits non-selected OR loci to further descent into the unique, repressive subnuclear compartments revealed by DNA FISH analyses.24
In a revealing mathematical exploration of OR regulation Tan et al. assessed the parameters required for establishing singular receptor expression.27 This theoretical approach provided an opportunity to assess how the timing of individual events (initiation, epigenetic remodeling, feedback) influence singular OR gene choice. Dividing OR gene choice into a three-state process (off, intermediate, and on), the authors demonstrated that to achieve singularity of expression the OR had to spend most of its time in an ‘off’ state, a brief time in the ‘intermediate’ state, and once in the ‘on’ state a relatively quick feedback had to be achieved. This modeling highlighted the importance of tightly regulated relative timings of the individual events involved in OR gene choice and confirmed the feasibility of the kinetic model as deduced by in vitro and in vivo work. These analyses also zeroed in on the critical rate-limiting event in the initiation of OR choice, proposing an alternate linchpin step. Rather than the activation of an OR gene by demethylation of its dimethylated histone H3K9 and H4K20 by Lsd-1, these authors proposed that an alternate, slow conversion of H3K9me3 and H4K20me3 to H3K9me2 and H4K20me2 would more efficiently generate rare initial activation, albeit through a yet unidentified histone demethylase.27
When the Kinetics Slip Up
The feedback model of OR regulation intuitively predicts that timing is critical for success: once a single OR is chosen, feedback must be swiftly accomplished prior to the activation of any additional receptor alleles. Mathematical modeling has confirmed this logic and has demonstrated the boundary values for the timing of activation and feedback necessary for a high probability of singular expression. What happens however when these critical kinetic parameters fail to yield activation of just a single OR in the defined window of time? In this case the neuron would have an indeterminate identity, with two or more ORs activated at once. This presents a predicament for the OSN which would likely be blind to the biallelic expression, not having the ability to distinguish that the feedback signal, which leads to the activation of adenylyl cyclase III expression and Lsd-1 downregulation, was coming from more than one OR. An additional axis of regulation would need to be in place to refine OSN cell fate. Such a refinement process would likely be an autonomous mechanism of the expressed OR alleles that would winnow down expression to just one, during a meta-stable period of expression.20 This refinement could employ an allelic competition for a limiting factor or compartment or a regulatory non-coding RNA that would act to effect shutdown of all but one OR.
There is evidence for such a scenario. It can be argued that the puzzling observation of “wild type” switching represents just such an event. In wild type switching, lineage marking analyses revealed a history of two functional ORs having been active in the same OSN but with only one OR having persisted. This phenomenon was originally interpreted to have resulted from the serial expression of functional OR alleles, one shut off before the next was selected, through the same feedback process that prevented pseudogene expression (Fig. 1A and14). It is equally feasible that receptor expression could have occurred in parallel, with two ORs activated at once and the subsequent shutdown of one or the other. Further evidence may be seen when we attempted to force biallelic expression of the tet-P2 allele through the pervasive expression of the tTa. Despite the genetic potential for 100% coexpression of the the two tet-modified P2 alleles in each cell in the homozygous animal, biallelic expression was observed only 3% of the time, and in a predominantly younger subpopulation of OSNs.20 These findings suggest a developmental window exists after feedback and prior to full OSN maturation in which a refinement or failsafe mechanism may ensure singular OR choice. In this conception, wild type switching is not a consequence of the negative feedback program which, removes OR pseudogenes from the final expressed repertoire of the epithelium, but rather a result of a refinement process, possible resulting from an allelic competition, when more than one OR is expressed at the same time (Fig. 1B).
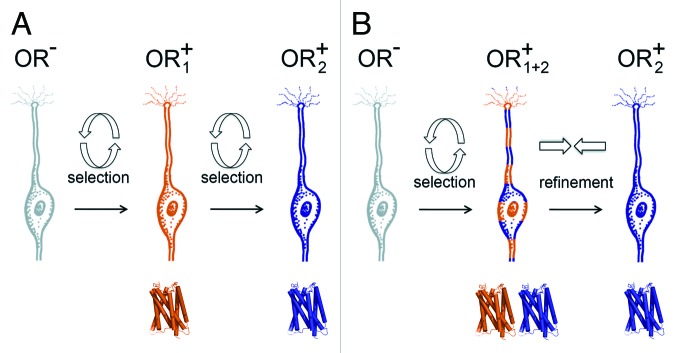
Figure 1. Switching vs. Refinement. (A) Switching between two functional ORs was considered to represent the serial expression of OR alleles in which the neuron conducts a second random initiation of gene activation (circle of arrows) after the shutdown of the OR chosen by a prior selection. The neuron is initially OR1+ but then switches to become OR2+, maintaining the one-receptor-per-cell rule. (B) Refinement is proposed to maintain monogenic selection after a failure of the initiation process to generate the choice of a single OR allele. The neuron is temporarily double positive, OR1+OR2+, prior to a competitive process (opposing arrows) through which only one OR emerges.
Future of the field
Cells may attain distinct identities through cascades of transcription factors that generate different fates. But such regulatory programs may fail to accommodate extreme biological pressure for diversity. In the face of greater requirements for diversification, stochastic mechanisms have evolved to allow for the maximal exploration of critical biochemical, genetic, or cellular space. Recent findings have lent support to a kinetic model of OR choice and revealed that the olfactory epithelium is a developmental palimpsest, where repressive epigenetic marks are written and rewritten over OR chromatin as OSNs develop from stem cell populations to mature OR+ sensory neurons. We look forward to exciting new answers to outstanding questions in the study of OR selection including: What roles do noncoding RNAs, which are implicated in every well-studied example of monoallelic gene regulation 28 play in OR expression? A role in maintenance of singular expression could easily be imagined for repressive ribonucleoprotein complexes. Do yet uncharacterized epigenetic states of OR chromatin explain zonal repression? Is there a unique and identifiable limiting event that generates activation of single OR alleles in a given window of time? And finally, what does the OSN do in the face of a breakdown in the normally tight singular regulatory process when more than one functional allele is expressed?
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Bushdid C, Magnasco MO, Vosshall LB, Keller A. Humans Can Discriminate More than 1 Trillion Olfactory Stimuli. Science (80-) [Internet] 2014 [cited 2014 Mar 20]; 343:1370–2. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24653035 [DOI] [PMC free article] [PubMed]
- 2.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–87. doi: 10.1016/0092-8674(91)90418-X. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1840504 [DOI] [PubMed] [Google Scholar]
- 3.Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB. The chromosomal distribution of mouse odorant receptor genes. Proc Natl Acad Sci U S A. 1996;93:884–8. doi: 10.1073/pnas.93.2.884. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8570653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–33. doi: 10.1038/nn800. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11802173 [DOI] [PubMed] [Google Scholar]
- 5.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–23. doi: 10.1016/S0092-8674(00)80581-4. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10089886 [DOI] [PubMed] [Google Scholar]
- 6.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–34. doi: 10.1016/S0092-8674(94)90562-2. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8087849 [DOI] [PubMed] [Google Scholar]
- 7.Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–18. doi: 10.1016/0092-8674(93)90422-M. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8343958 [DOI] [PubMed] [Google Scholar]
- 8.Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-G. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7683976 [DOI] [PubMed] [Google Scholar]
- 9.Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron. 2002;35:681–96. doi: 10.1016/S0896-6273(02)00793-6. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12194868 [DOI] [PubMed] [Google Scholar]
- 10.Qasba P, Reed RR. Tissue and zonal-specific expression of an olfactory receptor transgene. J Neurosci. 1998;18:227–36. doi: 10.1523/JNEUROSCI.18-01-00227.1998. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9412503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, et al. Mutually exclusive expression of odorant receptor transgenes. Nat Neurosci. 2000;3:687–93. doi: 10.1038/76641. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10862701 [DOI] [PubMed] [Google Scholar]
- 12.Shykind BM, Axel R. Unpublished Data. 1999
- 13.Shykind BM. Regulation of odorant receptors: one allele at a time. Hum Mol Genet. 2005;14:R33–9. doi: 10.1093/hmg/ddi105. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15809271 [DOI] [PubMed] [Google Scholar]
- 14.Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–15. doi: 10.1016/j.cell.2004.05.015. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15186780 [DOI] [PubMed] [Google Scholar]
- 15.Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci U S A. 2004;101:1069–74. doi: 10.1073/pnas.0307986100. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14732684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science (80-) [Internet] 2003; 302:2088–94. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14593185 [DOI] [PubMed]
- 17.Fleischmann A, Shykind BM, Sosulski DL, Franks KM, Glinka ME, Mei DF, Sun Y, Kirkland J, Mendelsohn M, Albers MW, et al. Mice with a “monoclonal nose”: perturbations in an olfactory map impair odor discrimination. Neuron. 2008;60:1068–81. doi: 10.1016/j.neuron.2008.10.046. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19109912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–70. doi: 10.1016/j.cell.2011.03.040. http://www.ncbi.nlm.nih.gov/pubmed/21529909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–36. doi: 10.1016/j.cell.2013.06.039. http://www.ncbi.nlm.nih.gov/pubmed/23870122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleischmann A, Abdus-Saboor I, Sayed A, Shykind B. Functional interrogation of an odorant receptor locus reveals multiple axes of transcriptional regulation. PLoS Biol [Internet] 2013 [cited 2014 Jan 14]; 11:e1001568. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3660300&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed]
- 21.Nagawa F, Yoshihara S, Tsuboi A, Serizawa S, Itoh K, Sakano H. Genomic analysis of the murine odorant receptor MOR28 cluster: a possible role of gene conversion in maintaining the olfactory map. Gene. 2002;292:73–80. doi: 10.1016/S0378-1119(02)00670-4. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12119101 [DOI] [PubMed] [Google Scholar]
- 22.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16873069 [DOI] [PubMed] [Google Scholar]
- 23.Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–84. doi: 10.1016/j.cell.2007.06.023. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17662950 [DOI] [PubMed] [Google Scholar]
- 24.Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–37. doi: 10.1016/j.cell.2012.09.043. http://www.ncbi.nlm.nih.gov/pubmed/23141535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nemazee D. Receptor selection in B and T lymphocytes. Annu Rev Immunol. 2000;18:19–51. doi: 10.1146/annurev.immunol.18.1.19. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10837051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–32. doi: 10.1016/j.cell.2013.09.033. http://www.ncbi.nlm.nih.gov/pubmed/24120133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan L, Zong C, Xie XS. Rare event of histone demethylation can initiate singular gene expression of olfactory receptors. Proc Natl Acad Sci U S A [Internet] 2013 [cited 2014 Mar 23]; 110:21148–52. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24344257 [DOI] [PMC free article] [PubMed]
- 28.Yang PK, Kuroda MI. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell 2007; 128:777–86. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17320513 [DOI] [PubMed]


