Cleavage and polyadenylation specificity factor (CPSF) is the central component of the 3′ processing machinery for polyadenylated mRNAs in metazoans. Schönemann et al. determined that four polypeptides (CPSF160, CPSF30, hFip1, and WDR33) are necessary and sufficient to reconstitute a CPSF subcomplex active in AAUAAA-dependent polyadenylation. WDR33 is required for binding of reconstituted CPSF to AAUAAA-containing RNA and can be specifically UV cross-linked to such RNAs.
Keywords: RNA processing, 3′ end formation, polyadenylation, poly(A) site, poly(A) polymerase
Abstract
Cleavage and polyadenylation specificity factor (CPSF) is the central component of the 3′ processing machinery for polyadenylated mRNAs in metazoans: CPSF recognizes the polyadenylation signal AAUAAA, providing sequence specificity in both pre-mRNA cleavage and polyadenylation, and catalyzes pre-mRNA cleavage. Here we show that of the seven polypeptides that have been proposed to constitute CPSF, only four (CPSF160, CPSF30, hFip1, and WDR33) are necessary and sufficient to reconstitute a CPSF subcomplex active in AAUAAA-dependent polyadenylation, whereas CPSF100, CPSF73, and symplekin are dispensable. WDR33 is required for binding of reconstituted CPSF to AAUAAA-containing RNA and can be specifically UV cross-linked to such RNAs, as can CPSF30. Transcriptome-wide identification of WDR33 targets by photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) showed that WDR33 binds in and very close to the AAUAAA signal in vivo with high specificity. Thus, our data indicate that the large CPSF subunit participating in recognition of the polyadenylation signal is WDR33 and not CPSF160, as suggested by previous studies.
All eukaryotic mRNAs, with the exception of histone mRNAs, undergo a 3′ end maturation step consisting of a specific endonucleolytic cleavage of the precursor followed by polyadenylation of the upstream cleavage fragment; the downstream fragment is degraded (Wahle and Rüegsegger 1999; Zhao et al. 1999; Millevoi and Vagner 2009; Proudfoot 2011).
In mammalian cells, the pre-mRNA cleavage site is determined by at least four sequence elements (Tian and Graber 2012): The central and most highly conserved signal is AAUAAA or a close variant located ∼20 nucleotides (nt) upstream of the cleavage site. The preferred sequence at the cleavage site is CA. GU- or G-rich downstream elements are important, and sequences upstream of AAUAAA, such as UGUA, can also contribute. RNA sequencing (RNA-seq) experiments revealed that, in many organisms, the majority of protein-coding genes have multiple polyadenylation sites generating either different protein isoforms or mRNA isoforms differing in the lengths of their 3′ untranslated regions (UTRs) and consequently in their interaction with RNA-binding proteins or microRNAs. Thus, there is substantial interest in the mechanism of poly(A) site recognition and of alternative poly(A) site choice (Campigli Di Giammartino et al. 2011; Shi 2012; Elkon et al. 2013; Lianoglou et al. 2013; Tian and Manley 2013).
In mammalian cells, at least sixteen polypeptides are dedicated to the cleavage and polyadenylation (CP) reaction (Chan et al. 2011; Xiang et al. 2014). Among these, cleavage and polyadenylation specificity factor (CPSF) can be considered the central complex: It carries the catalytic activity for pre-mRNA cleavage, and its interaction with the AAUAAA sequence is essential for cleavage and the AAUAAA dependence of polyadenylation. The two-subunit cleavage factor I (CF I) recognizes the UGUA upstream element. CF II contains two subunits with poorly defined functions. Cleavage stimulation factor (CstF) has three different subunits and recognizes downstream elements. Symplekin is considered a scaffolding protein connecting CPSF and CstF. Poly(A) polymerase generates the poly(A) tail and can also contribute to cleavage. Although CPSF and poly(A) polymerase are sufficient for AAUAAA-dependent polyadenylation, the nuclear poly(A)-binding protein 1 (PABPN1) stimulates poly(A) tail extension and is essential for the synthesis of a poly(A) tail of the appropriate length. In Saccharomyces cerevisiae, a slightly larger but mostly overlapping set of proteins has been identified as being required for pre-mRNA 3′ processing. Genetic confirmation of the in vivo roles of these proteins in CP provides the most persuasive evidence for similar functions of their mammalian orthologs. A much larger set of ∼80 polypeptides has been identified by affinity purification of a mammalian 3′ processing complex and mass spectrometric analysis (Shi et al. 2009). Some of these polypeptides may contribute to the coupling of 3′ processing to transcription and other processes. How many polypeptides are essential for the reaction remains to be determined.
The subunit composition of CPSF has not been entirely clear. Purification of the factor, based on its activity in polyadenylation assays, initially revealed four subunits: CPSF160, CPSF100, CPSF73, and CPSF30 (Bienroth et al. 1991; Murthy and Manley 1992). A fifth putative subunit, hFip1, was discovered on the basis of its homology with the yeast 3′ processing factor Fip1p (Kaufmann et al. 2004). A sixth polypeptide, WDR33, was identified among the components of an affinity-purified 3′ processing complex (Shi et al. 2009) due to its similarity to the yeast 3′ processing factor Pfs2p (Ohnacker et al. 2000). The polyadenylation activity of nuclear extract was abolished by immunodepletion of WDR33 and restored by the addition of purified CPSF. Affinity purification of CPSF by means of Flag-tagged CPSF73 resulted in the copurification of CPSF160, CPSF100, CPSF30, hFip1, WDR33, and also symplekin (Shi et al. 2009). The plant ortholog of WDR33, the protein FY, has been genetically shown to be involved in 3′ processing (Simpson et al. 2003) and is associated with other CPSF subunits (Herr et al. 2006; Hunt et al. 2008; Manzano et al. 2009). In S. cerevisiae, the orthologs of CPSF160, CPSF100, CPSF73, CPSF30, WDR33, and hFip1, together with poly(A) polymerase and Mpe1p, form the polyadenylation factor I (PF I) (Preker et al. 1997), which is part of a larger assembly (holo-CPF) (Nedea et al. 2003). The symplekin ortholog Pta1p is not part of PF I but mediates its association with the rest of holo-CPF.
Biochemical assays established that CPSF binds the AAUAAA sequence (Bienroth et al. 1991; Keller et al. 1991; Murthy and Manley 1992). CPSF160 has been considered the subunit recognizing the AAUAAA signal based on two pieces of evidence: First, in UV cross-linking experiments, an AAUAAA-dependent signal was observed at a molecular weight of ∼160 kDa (Moore et al. 1988; Gilmartin and Nevins 1989; Keller et al. 1991). However, the identity of the cross-linked band was never confirmed. Moreover, mapping of the 160-kDa cross-link in one particular RNA revealed it to be within an upstream sequence element rather than AAUAAA (Gilmartin et al. 1995). Second, in pull-down assays, recombinant CPSF160 had a twofold preference for binding to an AAUAAA-containing RNA in comparison with a point mutant (Murthy and Manley 1995). CPSF30 and hFip1 are also RNA-binding proteins but prefer U-rich sequences (Barabino et al. 1997; Kaufmann et al. 2004). Surprisingly, a comprehensive analysis of the RNA interactions of 3′ processing factors by UV cross-linking and immunoprecipitation (CLIP) followed by deep sequencing revealed that none of the putative CPSF subunits tested (CPSF160, CPSF100, CPSF73, CPSF30, and hFip1) showed a clear specificity for the AAUAAA sequence. In contrast, the CLIP-derived preferences of other 3′ processing factors matched those determined biochemically (Martin et al. 2012). In addition to specific RNA binding, CPSF catalyzes pre-mRNA cleavage at the site of poly(A) addition; CPSF73 is considered the endonuclease (Mandel et al. 2006). CPSF100 has a related structure. It has mutations in its active site but is thought to contribute to the endonuclease activity of CPSF73 (Kolev et al. 2008; Yang and Doublié 2011). Finally, CPSF also recruits poly(A) polymerase to its substrates. Accordingly, both CPSF160 and hFip1 interact with poly(A) polymerase (Murthy and Manley 1995; Kaufmann et al. 2004).
Here, we expressed and purified combinations of putative CPSF subunits and determined that four of them—CPSF160, CPSF30, hFip1, and WDR33—are necessary and sufficient to reconstitute, together with recombinant poly(A) polymerase, AAUAAA-dependent polyadenylation. Both CPSF30 and WDR33 could be UV cross-linked to short AAUAAA-containing RNAs, and transcriptome-wide mapping by photoactivatable ribonucleoside-enhanced CLIP (PAR-CLIP) showed that WDR33 contributes to the recognition of the polyadenylation signal in vivo.
Results
Six subunits associate with and reconstitute CPSF active in polyadenylation
To define the subunit composition of CPSF and facilitate functional analyses, we sought to reconstitute the factor by overexpression in insect cells. For this purpose, the MultiBac system (Berger et al. 2004; Fitzgerald et al. 2006) was used, which allows the simultaneous expression of multiple polypeptides from a single virus. The success of the reconstitution attempts was gauged by specific polyadenylation assays in which a “precleaved” RNA fragment carrying an AAUAAA sequence and ending close to the natural cleavage site is polyadenylated by recombinant poly(A) polymerase; the reaction depends on CPSF (Christofori and Keller 1989). An RNA substrate with an AAgAAA point mutation served as a specificity control.
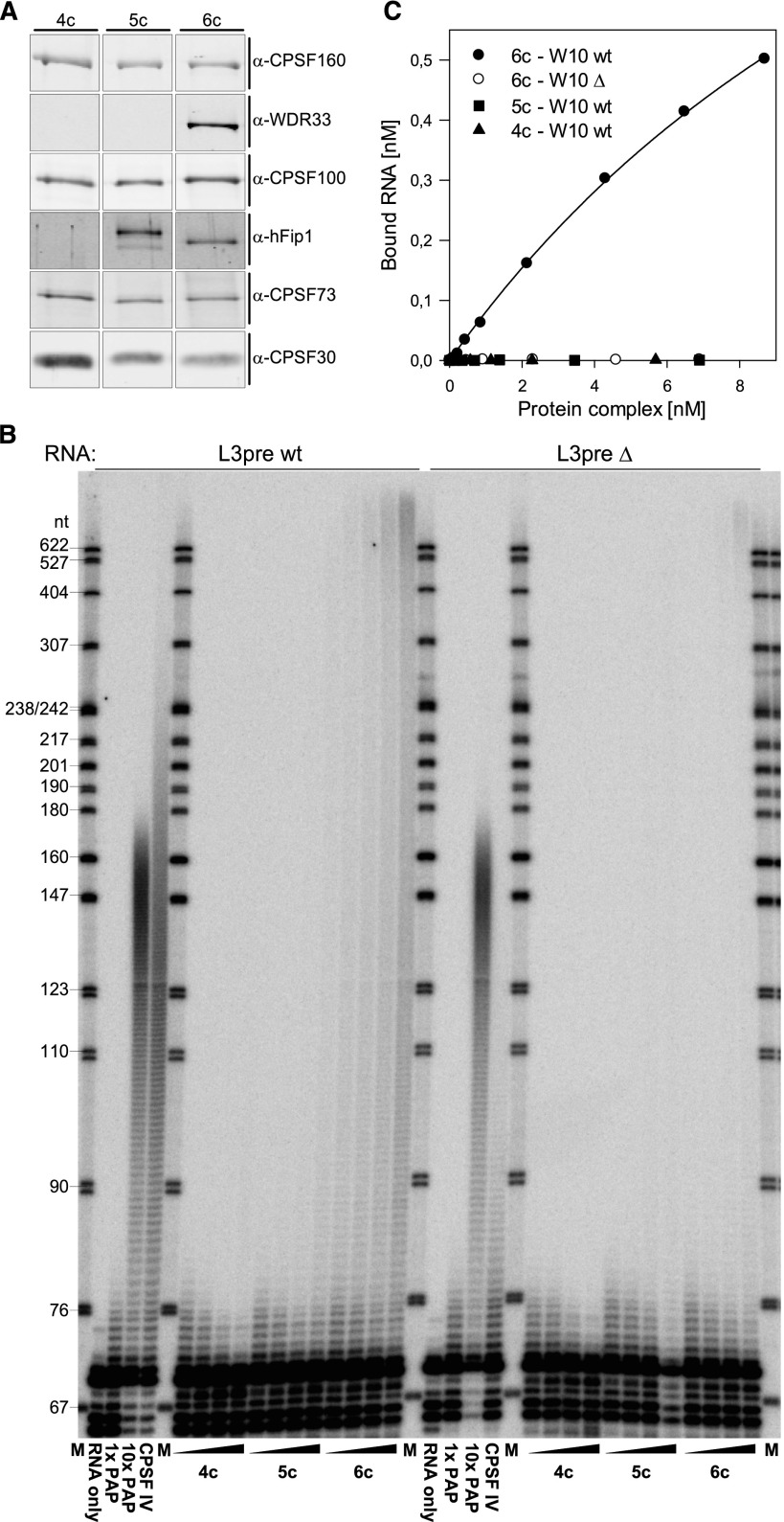
In initial experiments, three combinations of putative subunits were expressed: First, the four “classical” subunits CPSF160, CPSF100, CPSF73, and CPSF30; second, the four classical subunits plus hFip1; and third, these five polypeptides plus WDR33. In all three cases, affinity purification on the basis of Flag-CPSF160 resulted in the copurification of all subunits expressed in the respective experiment (Fig. 1A; Supplemental Fig. 1). The combinations of four and five subunits were inactive in polyadenylation assays, except that a weak AAUAAA-independent activity was observed in the preparation containing hFip1, in agreement with the activity of the isolated polypeptide (Kaufmann et al. 2004). In contrast, the preparation also containing WDR33 was much more active in polyadenylation and specific for the AAUAAA-containing substrate (Fig. 1B). This experiment establishes that six polypeptides are sufficient to reconstitute a CPSF that is active in polyadenylation. WDR33 is required. Symplekin is not essential for either polyadenylation or the association of the six CPSF subunits. The CPSF subunit requirements in pre-mRNA cleavage remain to be examined.
Figure 1.
Reconstitution of AAUAAA-dependent polyadenylation by coexpression of six polypeptides. Three different combinations of CPSF subunits, all including Flag-CPSF160, were expressed in a baculovirus system and purified by Flag affinity purification. (A) The main eluate fractions of the three preparations were examined by Western blot to verify the presence of the expected proteins. All signals are from the same blot that was stripped several times and probed separately with the antibodies indicated. All signals for the same subunit are from the same exposure. hFip1 migrated more slowly in the middle lane because the protein carried a Strep tag in this preparation but not in the six-subunit preparation. Silver-stained gels of the same fractions are displayed in Supplemental Figure 1. Viruses used for expression are listed in Supplemental Table 4A. (4c) Four-subunit complex (CPSF160, CPSF100, CPSF73, and CPSF30); (5c) 4c plus hFip1; (6c) 5c plus WDR33. (B) Polyadenylation assays were carried out with the same CPSF (sub)complexes shown in A and wild-type L3pre RNA (left) or a mutant control (right). Increasing amounts of the purified complexes indicated (0.5, 1, 2, and 4 µL of the respective fractions) were incubated with poly(A) polymerase and substrate RNAs as described in the Materials and Methods. Controls with either RNA included an “RNA-only” reaction, two with 1× or 10× poly(A) polymerase in the absence of CPSF, and one with 1× poly(A) polymerase plus CPSF purified from calf thymus (CPSF IV), as indicated. The very weak polyadenylation activity seen with the five-subunit assembly was not AAUAAA-specific; this is difficult to see in this experiment because RNA recovery was lower in the corresponding mutant sample. The difference in poly(A) tail length between the reactions containing calf thymus versus recombinant CPSF should be disregarded, as the tail length obtained in this type of assay depends on CPSF concentration and other variables. (C) The same three protein preparations as in A and B were used in nitrocellulose filter-binding assays with 1.5 nM labeled W10 (wild type [wt] or mutant) RNA as described in the Materials and Methods.
The ability of the three different subunit combinations to bind a synthetic AAUAAA-containing RNA (W10 RNA; AAUAAACCCA) (Wigley et al. 1990) was examined in nitrocellulose filter-binding experiments. The CPSF preparation containing all six subunits bound the wild-type RNA; the apparent KD was estimated as ∼20 nM. The affinity for the mutant control (AAgAAACCCA) was at least 20-fold lower (Fig. 1C). In contrast, the four-subunit and five-subunit complexes did not bind either RNA. We conclude that one reason for the inactivity of the CPSF subassemblies lacking WDR33 is their inability to bind the substrate RNA; WDR33, presumably in conjunction with other factors, appears to underlie the ability of CPSF to bind substrate RNA and trigger polyadenylation.
Although homology with the genetically confirmed 3′ processing factors Pfs2p and FY strongly suggests a role of WDR33 in pre-mRNA 3′ end formation, an RNAi experiment was performed as an additional test of the protein’s in vivo function. After treatment with a WDR33-specific pool of siRNAs, the abundance of uncleaved pre-mRNAs was tested by quantitative RT–PCR (qRT–PCR) across the cleavage/polyadenylation sites. A modest reduction of WDR33 levels led to an equally modest but reproducible accumulation of uncleaved precursor RNAs for three messages tested (Supplemental Fig. 2). These data support a role of WDR33 in 3′ end formation in vivo.
Four subunits are necessary and sufficient for the polyadenylation activity of CPSF
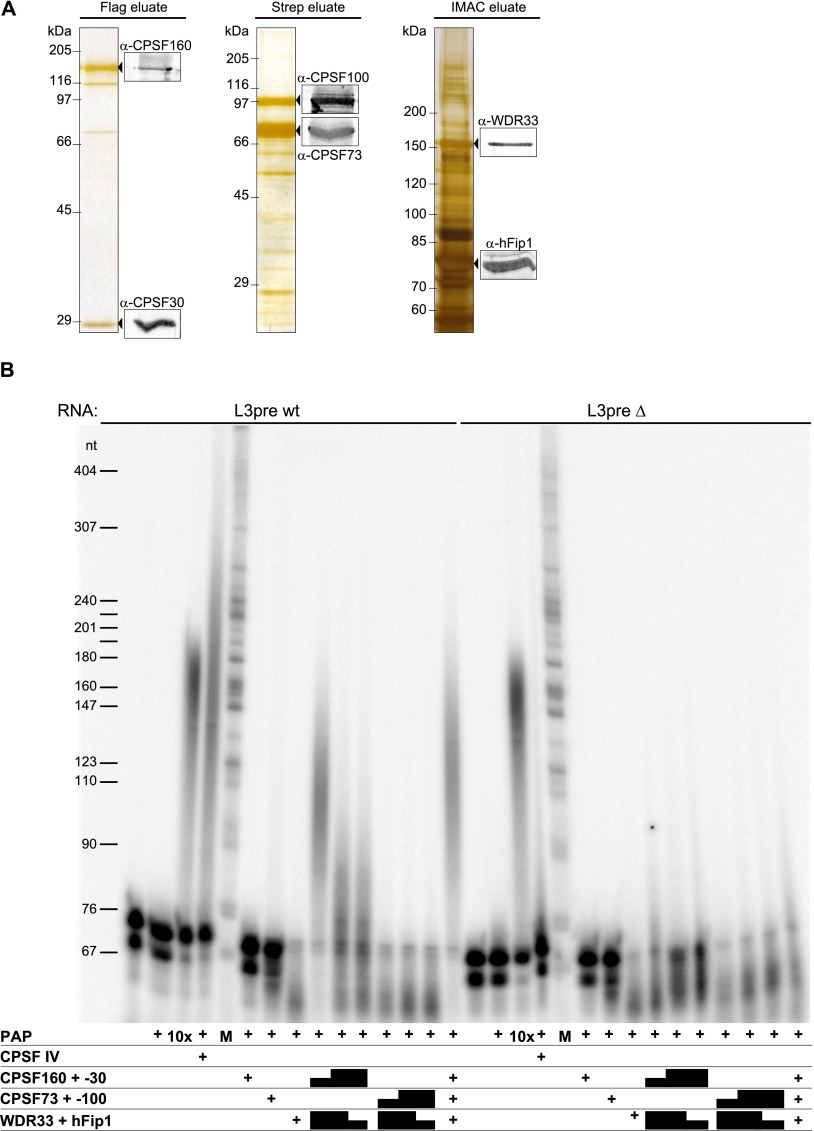
To further define the subunits necessary for polyadenylation, we expressed and purified three pairs of subunits separately: CPSF160 and CPSF30, CPSF100 and CPSF73, and hFip1 and WDR33. Each pair could be purified by means of an affinity tag carried on one subunit (Fig. 2A). In gel filtration, the CPSF100 and CPSF73 pair and the hFip1 and WDR33 pair behaved as soluble aggregates, eluting with the void volume (data not shown). In contrast, the CPSF160 and CPSF30 pair eluted at an apparent native molecular weight near 260 kDa, consistent with the association of one CPSF160 polypeptide with one or several CPSF30 subunits (Supplemental Fig. 3). The copurification results suggest nearest-neighbor relationships within CPSF but should be treated with caution, as the proteins tend to aggregate. In fact, even the CPSF160 and CPSF30 pair aggregated in another preparation (data not shown).
Figure 2.
Identification of mPSF, a CPSF subcomplex active in polyadenylation. Three pairs of CPSF subunits (Flag-CPSF160 and CPSF30; CPSF100 and Strep-CPSF73; and MycHis6-WDR33 and hFip1) were expressed and affinity-purified. (A) The polypeptide composition of the three preparations was analyzed in silver-stained SDS–polyacrylamide gels. The main eluate fractions are shown. Molecular weights of markers are indicated. Protein identities were verified by Western blotting. All Western blot signals were from the same membrane. Viruses used for expression are listed in Supplemental Table 4B. (B) The preparations shown in A were used for polyadenylation assays either separately or in combinations. For the pairwise combinations of subcomplexes, different ratios (1 μL:2 μL, 2:2, and 2:1) were used. RNA substrates were L3pre wild type (wt) or mutant as indicated. Controls (first four lanes for each RNA) were as in Figure 1B.
None of the three pairs of proteins was active in polyadenylation on its own. However, when all possible combinations were mixed in vitro and tested, the CPSF160 and CPSF30 pair together with the hFip1-WDR33 pair proved sufficient for the reconstitution of specific polyadenylation; addition of the CPSF100 and CPSF73 pair had no effect (Fig. 2B; data not shown). With the caveat that the WDR33–hFip1 pair was not very pure and might have been contaminated by host cell proteins contributing to the activity, the data suggest that CPSF100 and CPSF73 are dispensable for polyadenylation. Subsequently, a requirement for each of the four remaining subunits (CPSF160, CPSF30, hFip1, and WDR33) was tested by the coexpression of all possible combinations of three and, as a positive control, all four subunits. In repeated experiments, CPSF activity, assayed either directly in a crude extract of the infected cells or after affinity purification, was obtained only when all four subunits were coexpressed (Supplemental Fig. 4). Even the nonspecific activity expected for hFip1 was not consistently observed when any of the other subunits was missing. The four-subunit complex was also the only preparation able to bind AAUAAA-containing RNA in a gel shift experiment (data not shown). Thus, under the conditions used, CPSF160, CPSF30, hFip1, and WDR33 are necessary and sufficient to reconstitute CPSF active in RNA binding and AAUAAA-dependent polyadenylation. To facilitate discussion, we call this CPSF subcomplex “mammalian polyadenylation specificity factor” (mPSF).
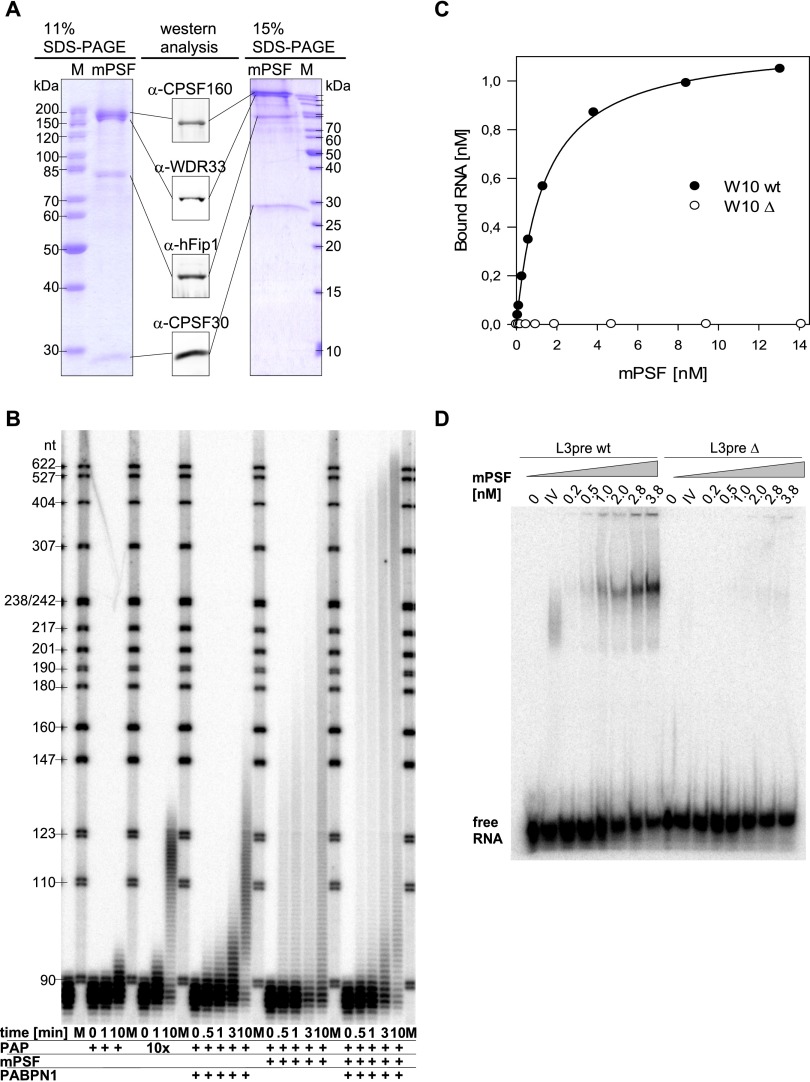
The four subunits of mPSF were then coexpressed from a single virus and purified by anion exchange chromatography followed by immobilized metal affinity chromatography (IMAC) based on an N-terminal His tag on WDR33 and Flag affinity chromatography based on tagged CPSF160. The eluate of the final column contained all four subunits with only minor contaminants and/or proteolysis products (Fig. 3A). As estimated from scans of the Coomassie-stained gels of two Flag columns, the subunits were present at roughly comparable amounts: Assuming equal amounts of CPSF160 and WDR33, which were barely resolved from each other, hFip1 was present at a 1.8-fold excess, and CPSF30 was present at a 2.5-fold and 3.1-fold excess. The preparations were active in AAUAAA-dependent polyadenylation. Addition of PABPN1 induced rapid and processive polyadenylation up to 250–300 nt and much slower elongation beyond this length (Fig. 3B; Kühn et al. 2009). Both preparations also bound AAUAAA-containing RNAs in nitrocellulose filter-binding and gel retardation assays with an apparent KD near 2 nM (Fig. 3C,D). The higher apparent affinity compared with the six-subunit assembly may reflect a higher proportion of active protein. In analytical gel filtration of the Flag eluate, all four subunits again eluted with the activity in AAUAAA-dependent polyadenylation and RNA binding (Supplemental Fig. 5). Much of the material eluted with the void volume, suggesting partial aggregation, but a peak was present at an apparent native molecular weight >600 kDa, which is larger than expected for a globular complex containing one copy of each subunit.
Figure 3.
Characterization of mPSF activities. CPSF160, CPSF30, WDR33, and hFip1 were expressed from a single virus (Supplemental Table 4C) and purified to near homogeneity (see the Materials and Methods). (A) The eluate of the final Flag affinity column was analyzed on two Coomassie-stained SDS–polyacrylamide gels. Molecular weights of markers (M) are indicated. The identities of protein bands were verified by Western blotting. All Western blot signals shown were obtained from a single gel lane. (B) The preparation shown in A was used for polyadenylation assays. For every 20-µL reaction, 80 fmol of L3preA15 wild-type (wt) RNA, 80 fmol of mPSF, 1200 fmol of PABPN1 and 8 or 80 fmol of poly(A) polymerase were used as indicated. After preincubation, reactions were started by ATP addition and stopped after the times indicated. In the first lane between the two marker lanes, only RNA was loaded. (C) Filter-binding assays were carried out with the Flag eluate and 1.5 nM W10 wild-type or mutant RNAs as described in the Materials and Methods. (D) Gel shift assays were carried out with the Flag eluate and 5 nM L3pre or L3preΔ RNA as described in the Materials and Methods. Purified calf thymus CPSF (CPSF IV) was used as a positive control.
Even though the mPSF preparation appeared quite pure in gel electrophoresis, there remained a concern that other, less abundant polypeptides might have been copurified from the eukaryotic expression system and might contribute to the activity. To address this question, we first estimated (from Coomassie-stained gels and comparison of the hFip1 band with a BSA standard curve) that the chemical concentration of mPSF in a particular Flag eluate fraction (E2) was 340 nM. The amount of active mPSF was estimated as follows: Polyadenylation time courses in which mPSF was preincubated with high concentrations of both RNA and poly(A) polymerase and the reaction started by ATP addition showed biphasic kinetics: A burst phase of 10 sec or less, obviously reflecting the number of active polyadenylation complexes assembled during the preincubation, was followed by a “steady-state” phase reflecting the turnover of RNA substrate. With a 10-sec reaction time to measure the burst phase, assays at increasing RNA concentrations suggested that fraction E2 contained between 290 and 410 nM active mPSF (Supplemental Fig. 6). This may be an underestimate, as we did not reach RNA saturation. Thus, Coomassie staining underestimated the concentration of mPSF, presumably due to poor dye binding of hFip1. Importantly, the experiments suggest that purified mPSF was fully active; thus, if any additional polypeptide had been essential for the activity, it should have been visible in amounts comparable with the four authentic subunits. The absence of such bands strongly suggests that CPSF160, CPSF30, WDR33, and hFip1 are indeed sufficient for the polyadenylation activity of CPSF and that other polypeptides, including CPSF100 and CPSF73, are dispensable.
The virus encoding all four subunits of mPSF was also used for coinfection of cells with a second virus encoding symplekin, and the complex was purified over the same sequence of three columns as the preparation lacking symplekin. Western blotting showed that IMAC largely separated symplekin in the flowthrough from mPSF in the bound fraction. On the final Flag affinity column, the remaining symplekin was again partially separated from mPSF, and the residual amounts left in purified mPSF were detectable by Western blotting but not by Coomassie staining (Supplemental Fig. 7; data not shown). Thus, symplekin does not associate stably with mPSF.
The AAUAAA signal is recognized by WDR33
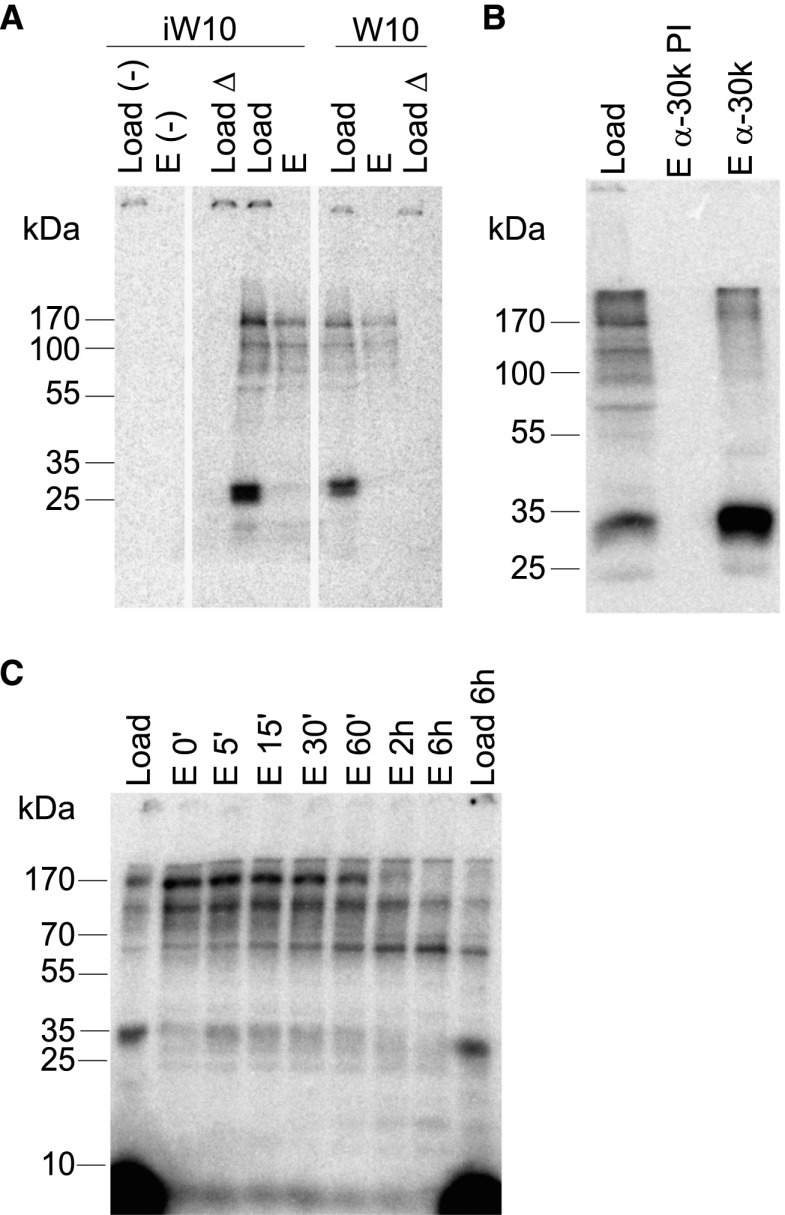
A W10 RNA derivative containing a 5-iodouridine (5-iodo) substitution (iW10; AA5iUAAACCCA) was used for UV cross-linking assays to identify RNA-binding polypeptides within reconstituted mPSF. A mutant 5-iodo-U-substituted RNA (iW10 Δ; Ac5iUcAACCCA) was used as a specificity control. Two major AAUAAA-dependent cross-linked bands were obtained at ∼30 kDa and 160 kDa. A similar cross-link pattern was observed with the W10 RNA lacking the 5-iodo substitution, but both signals were weaker, suggesting that both cross-links are mostly to the uridine in the AAUAAA signal (Fig. 4A). The WDR33 subunit in the mPSF preparation carried an N-terminal His tag, and the cross-linked 160-kDa band was bound under denaturing conditions by Ni-NTA beads, whereas the 30-kDa cross-link was not (Fig. 4A). Thus, the cross-link was formed by WDR33. Also, after denaturation, the 30-kDa cross-linked band was specifically enriched by an antibody directed against CPSF30 but not by preimmune serum (Fig. 4B). Thus, both CPSF30 and WDR33 may participate directly in RNA binding and contact the AAUAAA signal.
Figure 4.
WDR33 and CPSF30 can be cross-linked to AAUAAA-containing RNA. (A) Ten nanomolar mPSF containing MycHis6-WDR33 was incubated with 10 nM radiolabeled 5-iodo-modified iW10 RNA (wild type [wt] or mutant) or unmodified W10 RNA (wild type or mutant), UV cross-linked at 312 nm, and wild-type samples were used for a pull-down with Ni-NTA beads under denaturing conditions. Lanes labeled “load” contain 5% of the samples used for the pull-down. Lanes labeled “E” show 50% of the bead eluates. The first two lanes are no-protein controls (−). Lanes were cut from a single gel and rearranged. (B) Ten nanomolar mPSF was incubated with 10 nM radiolabeled iW10 RNA (wild type only), UV cross-linked at 312 nm, and used for a pull-down with antibody directed against CPSF30 or preimmune serum (see the Materials and Methods). “Load” contains 5% of the cross-link reaction, and “E” contains 50% of the eluate. (C) Ten nanomolar mPSF containing MycHis6-WDR33 was incubated with 10 nM radiolabeled iW10 RNA (wild type only), UV cross-linked at 312 nm, brought to 0.75 M urea, and digested with protease Lys-C as described in the Materials and Methods. Aliquots were taken at the time points indicated, and protein fragments were purified via Ni-NTA beads as in A. “Load” contains 10% of the cross-link reaction, and “E” contains 100% of the eluate.
In the Ni-NTA pull-down of the WDR33 cross-link, smaller radiolabeled bands were also precipitated, suggesting that the site of cross-linking was in the N-terminal portion of the protein. In fact, a radiolabeled, His-tagged protein fragment of nearly 60 kDa accumulated when, after cross-linking, mPSF was digested with protease Lys-C under mildly denaturing conditions (Fig. 4C). This maps the RNA-binding surface of WDR33 to the N-terminal region that includes seven or eight WD40 repeats.
The PAR-CLIP method (Hafner et al. 2010; Martin et al. 2012) was used for mapping the binding sites of WDR33 in vivo: HEK293 cells were incubated with 4-thiouridine, and RNA–protein cross-links were induced by exposure to 365-nm light. WDR33-containing complexes were immunoprecipitated, and cross-linked RNA fragments were extracted, ligated to adapters, and amplified by RT–PCR. After Illumina sequencing, reads were preprocessed and mapped to the human genome with CLIPZ (Khorshid et al. 2011). In total, >11 million reads could be assigned to a unique locus. The high rate of T-to-C transitions (>46%), which are introduced when the reverse transcriptase encounters a cross-linked 4-thiouridine, and the fact that the majority of reads originated from mRNAs with only a very minor fraction originating from ribosomal RNA (Supplemental Table 1) indicate a high data quality. On the basis of publicly available 3′ end sequencing data sets (Gruber et al. 2012; Martin et al. 2012), the 1000 most frequently used CP sites in HEK293 cells were determined. The density of PAR-CLIP reads along the regions centered on these sites showed a strong enrichment of WDR33 binding at the CP sites with a peak 16–18 nt upstream of the cleavage site, matching the position of the polyadenylation signal (Fig. 5A). To further investigate the sequence specificity of WDR33, we extracted 25-nt-long sequences that were centered on the 1000 genomic positions with the highest number of T-to-C transitions in the mapped reads. Compared with sequences with the same nucleotide composition, the sequences around the cross-linked positions were most enriched in the canonical polyadenylation signal AATAAA (Supplemental Table 2), with a number of other variants also enriched. To determine most specifically where WDR33 binds relative to the poly(A) signal, we obtained the 500 CP sites with the highest usage that additionally had a unique AATAAA or ATTAAA motif and no other variation of the poly(A) signal within 40 nt upstream of the CP site. The enrichment of T-to-C mutations relative to the frequency of T nucleotides at individual positions in these sequences indicated that WDR33 cross-links preferentially immediately downstream from and, to a smaller extent, within the poly(A) signal (Fig. 5B). Compared with other CPSF subunits that have been studied by PAR-CLIP before (Martin et al. 2012), WDR33 has a much higher specificity for the hexamer motif, although Fip1, CPSF100, and the Flag-tagged CPSF30 also cross-linked close to the hexamer (Martin et al. 2012).
Figure 5.
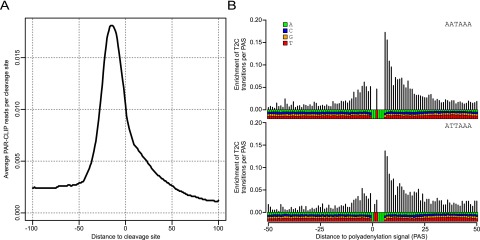
Transcriptome-wide binding of WDR33 to AAUAAA. (A) Average density of PAR-CLIP reads around the 1000 most abundantly used CP sites in HEK293 cells. WDR33 shows strong and specific positioning upstream of the CP sites, with a peak at nucleotides −16 to −18 upstream of the cleavage site. (B) Enrichment of T-to-C transition relative to the T nucleotide frequency as a function of distance with respect to the poly(A) signal. The analysis is based on the 500 most frequently used CP sites that have a single AATAAA (top panel) or ATTAAA (bottom panel) motif and no other variant poly(A) signal in the 40-nt region upstream of the cleavage site. These results indicate that WDR33 is most frequently cross-linked on U nucleotides that immediately follow the polyadenylation signal.
Discussion
We examined the composition and function of mammalian CPSF and carried out a complete reconstitution of the second step of pre-mRNA 3′ processing: polyadenylation. Our data justify two main conclusions: First, CPSF160, CPSF30, WDR33, and hFip1 are necessary and sufficient for the reconstitution of a CPSF subcomplex (mPSF) that is active in both specific binding to the polyadenylation signal AAUAAA and AAUAAA-dependent poly(A) addition. The functions of the CPSF100 and CPSF73 subunits remain to be clarified; presumably, they are restricted to pre-mRNA cleavage. Although known to associate with CPSF, symplekin is not required for polyadenylation either and does not bind mPSF in a stable manner. The second important conclusion is that the AAUAAA signal is recognized by WDR33, presumably in conjunction with CPSF30 and not by CPSF160, as was long considered. A model summarizing these data is shown in Figure 6.
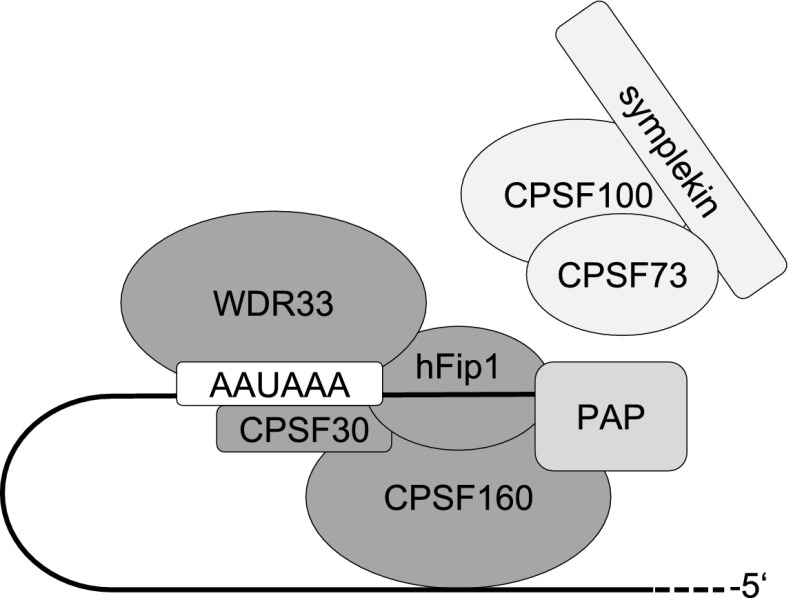
Figure 6.
Model depicting the polyadenylation complex. WDR33 and CPSF30 are shown binding to the AAUAAA signal. Poly(A) polymerase (PAP) is bound to the 3′ end. hFip1 is shown between AAUAAA and the polyadenylation site; experimentally, binding sites have been mapped both upstream of and downstream from AAUAAA (Kaufmann et al. 2004; Martin et al. 2012; Chan et al. 2014). CPSF160 is shown binding RNA in an upstream position for reasons discussed in the text. With the exception of WDR33 and CPSF30, which bind the same sequence element but are not known to interact, direct interactions have been reported for all other subunits shown touching each other. Interaction of hFip1 with CPSF30 and CPSF160 has been observed by Kaufmann et al. (2004). Evidence for all other interactions is discussed in the text. The CPSF100–CPSF73–symplekin complex is shown separately; while the complex is clearly part of CPSF, to the best of our knowledge, it is unknown how it associates with mPSF. During the transition from cleavage to polyadenylation, CPSF73, as the endonuclease, has to trade places with poly(A) polymerase.
Initially, six CPSF subunits (CPSF160, CPSF100, CPSF73, CPSF30, WDR33, and hFip1) were copurified by means of a single affinity tag on CPSF160. The preparation was active in RNA binding and polyadenylation but remains to be examined for cleavage activity. The reconstitution of this complex confirms the composition that was derived from Flag affinity purification of the endogenous complex from mammalian cell extracts, except for symplekin, which was copurified with the endogenous complex but not tested in our six-subunit reconstitution (Shi et al. 2009). Symplekin is dispensable for the integrity of the six-subunit complex and for polyadenylation. The reconstituted complex also matches the yeast PF I (yPF I) complex, although yPF I also contains Mpe1p and poly(A) polymerase as stable constituents (Preker et al. 1997; Nedea et al. 2003). Additional experiments showed that CPSF can be divided into two parts. CPSF160, CPSF30, WDR33, and hFip1 form mPSF, which can be considered a core complex within CPSF; mPSF is a relatively well-soluble assembly, carries the key function of AAUAAA binding, and is sufficient for recruiting poly(A) polymerase to AAUAAA-containing RNAs. CPSF100 and CPSF73 probably form a separable “cleavage module” within CPSF: The two polypeptides interact (Dominski et al. 2005; Hunt et al. 2008; Sullivan et al. 2009; Yang and Doublié 2011), and both are required for pre-mRNA cleavage (Mandel et al. 2006; Kolev et al. 2008) but not for polyadenylation. CPSF100 and CPSF73 also associate with symplekin (Kolev et al. 2008; Sullivan et al. 2009), and our results suggest that there are no stable CPSF–symplekin contacts outside these two subunits. Symplekin in turn mediates the interaction between CPSF and cleavage factors (Xiang et al. 2014). The possibility that a CPSF subcomplex lacking CPSF73 exists in vivo has been suggested (Dickson et al. 1999).
We were unable to generate functional complexes smaller than mPSF: Lysates of cells expressing all four subunits had clearly detectable polyadenylation activity, but no activity was seen when any individual subunit was omitted. Affinity-purified preparations from these expression experiments were also inactive in both RNA binding and polyadenylation unless they contained all four subunits. Individual subunits or combinations of them should at least display some RNA-binding activity (Murthy and Manley 1995; Barabino et al. 1997; Kaufmann et al. 2004), so the apparent inactivity may have been due to limited protein concentrations, exacerbated by solubility and/or folding problems. Whatever the specific reason, the data indicate that each of the four subunits makes an important contribution to the functionality of mPSF. With the caveat that the subunits have a propensity for potentially nonspecific aggregation, our purification data confirm the CPSF100 and CPSF73 interaction. They also indicate an association of CPSF160 with CPSF30, consistent with a similar interaction in Arabidopsis (Hunt et al. 2008), and of WDR33 with hFip1, consistent with an interaction between the yeast orthologs Pfs2p and Fip1p (Ohnacker et al. 2000). However, these pairs of polypeptides do not correspond to separable functions, as both CPSF160 and hFip1 contact poly(A) polymerase (Murthy and Manley 1995; Kaufmann et al. 2004), and all four subunits contribute to RNA binding. The native molecular weight of mPSF and its subunit stoichiometry could not yet be determined precisely because much of the purified material did not appear to be monodisperse.
That CPSF160 is the AAUAAA-binding subunit of CPSF was suggested mainly based on the detection of a 160-kDa polypeptide that could be specifically cross-linked to RNA containing an AAUAAA sequence (Moore et al. 1988; Gilmartin and Nevins 1989; Keller et al. 1991). When those experiments were carried out, it was not known that WDR33, which comigrates with CPSF160 in SDS–polyacrylamide gels, is a subunit of CPSF. To our knowledge, the identity of the cross-linked 160-kDa band was never examined directly. We found that CPSF subassemblies lacking WDR33 are unable to bind RNA. More specific evidence for a role of WDR33 in recognizing the polyadenylation signal is provided by cross-linking experiments. The short RNA oligonucleotides used contained only 4 nt outside the polyadenylation signal, and cross-linking to WDR33 was enhanced by a 5-iodo substitution in AAUAAA; thus, WDR33 makes direct contacts to this sequence. The same observation was made independently by Chan et al. (2014). Supporting a function of WDR33 in AAUAAA recognition, a PAR-CLIP experiment showed that WDR33 cross-links to polyadenylation signals and immediately downstream in vivo. This is in contrast to the other five subunits of CPSF that were examined previously: Only hFip1 had a weak positional preference (in relationship to the polyadenylation site) matching the position of the AAUAAA signal. CPSF100 and Flag-CPSF30 cross-linked close to the sequence, and the other subunits tended to cross-link further upstream (Martin et al. 2012). Also, hFip1 was the only subunit that showed some enrichment of AAUAAA sequences in its cross-linked sequences, but it was weaker than we now see for WDR33. Thus, the data overall support the idea that WDR33 recognizes the polyadenylation signal. In its N-terminal part, WDR33 contains seven or eight WD40 repeats. WD40 repeats mostly participate in protein–protein interactions, but interactions of WD40 domains with DNA and RNA have been described (Lau et al. 2009; Stirnimann et al. 2010). RNA binding of yeast Yhh1p/Cft1p, the ortholog of CPSF160, has also been mapped to its WD40 repeats (Dichtl et al. 2002). The WD40 repeats of WDR33, which are the most conserved part of the protein, may have a similar function. Consistently, partial proteolysis mapped WDR33–RNA crosslinks to the N-terminal part of the protein containing the repeats.
The other three subunits of mPSF are also RNA-binding proteins. Isolated CPSF30 prefers U-rich ligands (Barabino et al. 1997). The 30-kDa protein that was previously observed to cross-link to AAUAAA-containing RNAs (Moore et al. 1988; Gilmartin and Nevins 1989; Keller et al. 1991) has now been identified as CPSF30 by immunoprecipitation. As the RNA used for cross-linking here contained very few nucleotides outside AAUAAA and cross-linking of CPSF30 was also enhanced by the 5-iodo substitution, this polypeptide may cooperate with WDR33 in AAUAAA recognition. Although an earlier PAR-CLIP analysis did not provide strong evidence for AAUAAA specificity of CPSF30 binding (Martin et al. 2012), recent data clearly show that this polypeptide participates in AAUAAA recognition (Chan et al. 2014). Isolated hFip1 binds oligo(U) (Kaufmann et al. 2004). The protein tends to bind near AAUAAA (Kaufmann et al. 2004; Martin et al. 2012; Chan et al. 2014). Its contribution to poly(A) site selection remains to be analyzed. CPSF160 binds RNA in pull-down and cross-linking assays (Murthy and Manley 1995; Martin et al. 2012). CPSF purified from cells has been reported to recognize sequences outside AAUAAA (Bilger et al. 1994; Gilmartin et al. 1995), and cross-linking of CPSF160 can be relatively far upstream of AAUAAA (Gilmartin et al. 1995; Martin et al. 2012); the polypeptide might thus be responsible for upstream interactions of CPSF. The binding specificity of CPSF and the contribution of CPSF160 can now be re-examined with the help of the reconstituted, more rigorously purified factor.
Materials and methods
Baculovirus expression clones
MultiBac plasmids and methods for their use have been described (Fitzgerald et al. 2006). The cDNAs encoding CPSF subunits were of bovine or human origin and are listed in Supplemental Table 3. YFP, CFP, and mCherry were used as markers for infection. The resulting baculovirus clones and their use are listed in Supplemental Table 4, A–D. hFip1 was cloned into SalI/XbaI-opened pUCDM with the help of the In-Fusion Advantage PCR cloning kit (Clontech). The resulting vector was used for In-Fusion cloning of WDR33 into the XhoI/NheI-opened plasmid. For other clones, standard ligation-dependent cloning procedures were used with enzymes from New England Biolabs. Flanking restriction sites were introduced by PCR using Pwo DNA polymerase (Peqlab) and appropriate DNA primers (Invitrogen). In many cases, ORFs were first subcloned into additional vectors to introduce N-terminal tags for affinity purification (Supplemental Table 5) or additional restriction sites before transfer into the MultiBac system. Alternatively, phosphorylated oligonucleotides encoding tags were ligated directly into linearized MultiBac plasmids (Strep-CPSF73, MycHis-WDR33, and His-symplekin). After PCR, ORFs were verified by DNA sequencing. When more than two expression cassettes were integrated into plasmids, restrictions sites in the multiplication modules of the MultiBac plasmids and DNA ligase were used, or plasmids were fused by Cre recombinase (New England Biolabs). Detailed information about the cloning of individual ORFs as well as DNA oligonucleotides used will be provided on request.
Cre-loxP-mediated or Tn7-dependent integration of MultiBac plasmids carrying CPSF ORFs was performed as described (Fitzgerald et al. 2006). After selection of transformed Escherichia coli on agar plates, colonies were restreaked under selective conditions for isolation of single clones. Supplemental Table 4, A–D, lists the type of integration for each expression cassette and the plasmids used.
Bacmids were prepared by alkaline lysis from 4–5 mL of overnight cultures: E. coli cells were harvested (3000g for 15 min at 4°C); resuspended in 250 µL of 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 10 µg/mL RNase A; and mixed by inversion of the tubes with 250 µL of 200 mM NaOH and 1% SDS and then with 350 µL of 3 M potassium acetate (pH 5.0). Precipitates were pelleted (20,000 g for 20 min at room temperature), and supernatants were transferred to fresh tubes. Bacmid DNA was precipitated with 0.7 vol of isopropyl alcohol, and pellets were resuspended in 50 µL of 10 mM HEPES (pH 7.0).
Insect cell culture, virus propagation, and protein expression
Viruses were propagated in Sf21 cells (Invitrogen). Protein expression was carried out in either Sf21 or High Five cells (Invitrogen). Both cell lines were maintained as suspension cultures in ExCell 420 serum-free medium (Sigma-Aldrich) at densities between 0.8 × 106 and 4 × 106 (Sf21) or between 0.6 × 106 and 4 × 106 cells per milliliter (High Five). Viral titers were determined by plaque assays. Transfection of Sf21 cells with bacmid DNA was performed with the CellFectin II transfection reagent (Invitrogen) according to the manufacturer’s instructions. For virus propagation, cells were infected with a multiplicity of infection (MOI) <0.01. For expression cultures, each virus was used at a MOI between 0.1 and 1. Expression cultures were harvested between 72 and 96 h post-infection.
Protein purification
All procedures were carried out on ice or in a cold room. Column fractions were analyzed for CPSF subunits by Western blotting and polyadenylation assays. Procedures and buffer conditions were varied in initial purifications of CPSF (sub)complexes. The final purification of mPSF was carried out as follows: Baculovirus-infected cells (109) were harvested, resuspended in 100 mL of lysis buffer (50 mM Tris-HCl at pH 8.0, 10% sucrose, 3 mM MgCl2, 1 µg/mL pepstatin, 1 µg/mL leupeptin, 1 mM PMSF) containing 250 mM KCl, and lysed by sonication (up to 100 bursts at medium setting) (Branson Sonifier 250). Lysates were cleared by centrifugation (20,000g for 30 min) and passed over a 30-mL DEAE-Sepharose column (GE LifeSciences) equilibrated with the same buffer. The flowthrough was adjusted to 10 mM imidazole and incubated overnight with 3 mL of Ni-NTA agarose resin (Qiagen). The material was packed into a column and washed with lysis buffer containing 20 mM imidazole. Protein was eluted with the same buffer containing 250 mM imidazole. The mPSF-containing fractions were dialyzed against lysis buffer containing 200 mM KCl and no imidazole, and one half was loaded on 1 mL of anti-Flag agarose (Sigma-Aldrich). The column was washed with the same buffer and eluted with the same buffer containing 200 μg/mL Flag peptide. The same procedure was used for mPSF coexpressed with symplekin. Analytical size exclusion chromatography was carried out on a Superdex 200 HR 10/30 column (GE LifeSciences) in lysis buffer containing 200 mM KCl. The column was calibrated with proteins from the LMW and HMW gel filtration calibration kits (GE LifeSciences).
His-tagged full-length bovine poly(A) polymerase, CPSF from calf thymus (CPSF IV), and recombinant untagged PABPN1 were the preparations described (Fronz et al. 2008; Kühn et al. 2009).
Antibodies and Western blotting
Rabbit antibodies against human WDR33 (no. A301-152A) and symplekin (no. A301-465A) were from Bethyl Laboratories. Rabbit sera against recombinant bovine CPSF100 and CPSF30, purified under denaturing conditions, were made by Eurogentec. Rabbit sera against CPSF73, CPSF160, and hFip1 have been described (Jenny and Keller 1995; Jenny et al. 1996; Kaufmann et al. 2004). Secondary fluorescent antibodies (IRDye800CW donkey anti-rabbit) were from LI-COR Biosciences. Proteins were blotted to nitrocellulose membranes (Protran, Whatman) by the semidry procedure. Membranes were blocked with 2.5% milk powder in TN-Tween (20 mM Tris at pH 8.0, 150 mM NaCl, 0.05% Tween-20), incubated for at least 1 h at room temperature with primary antibodies diluted in TN-Tween with 0.5% milk powder, and washed five times with TN-Tween. They were incubated for 30 min in the dark at room temperature with the secondary antibody diluted 1:10,000 to 1:15,000 in TN-Tween, washed as above, and rinsed twice in the same buffer without Tween. Blots were scanned on an Odyssey infrared imaging system (LI-COR Biosciences), and fluorescence signals were analyzed by ImageQuant (GE Healthcare) software.
Polyadenylation and binding assays
Substrate RNAs L3pre, L3preΔ, L3preA15, and L3preA15Δ; their enzymatic synthesis; and conditions for polyadenylation assays have been described (Christofori and Keller 1989; Kerwitz et al. 2003; Kühn et al. 2009). Unless noted otherwise, 25-µL reactions contained 100 fmol of RNA, 20 fmol of poly(A) polymerase, and other proteins as indicated. Mixtures were preincubated for 5 min at 37°C, and reactions were started by the addition of 0.5 mM ATP. Standard reaction time was 30 min, and products were analyzed on 10% urea–polyacrylamide gels as described (Kühn et al. 2009). Sizes of DNA markers (in nucleotides) are indicated next to the gel images.
RNA oligonucleotides (W10, AAUAAACCCA; W10Δ, AAgAAACCCA; MWG Eurofins) were 5′-labeled with [γ-32P]-ATP and polynucleotide kinase and used for nitrocellulose filter-binding assays essentially as described (Kühn et al. 2003). For the determination of equilibrium dissociation constants, a fixed amount of RNA was titrated with increasing amounts of protein. Salt concentrations were adjusted by addition of appropriate amounts of saltless buffer. Data were fitted to a 1:1 association equilibrium with a single rectangular hyperbolic function (Sigma Plot version 12.5).
Gel shifts were performed as described (Kühn et al. 2009) with 5 nM internally labeled L3pre or L3preΔ RNA in a volume of 20 μL with amounts of mPSF reported in the figures.
5-iodo-modified, HPLC-purified RNA oligos (iW10, AA5iUAAACCCA; iW10Δ, Ac5iUcAACCCA; Eurogentec) were 5′-labeled as above and used in parallel with unmodified W10 and W10Δ for UV cross-linking assays. Cross-linking was carried out with 10 nM RNA and 10 nM mPSF in a volume of 100 μL under polyadenylation conditions but without ATP. The RNA–protein mix was preincubated for 10 min at 37°C and UV-irradiated at 312 nm for 5 min as described (Kühn et al. 2003). Cross-linked products were analyzed without RNase digestion via SDS–polyacrylamide gel electrophoresis and phophorimaging or used for affinity purification.
For antibody precipitation, 5 mg of protein A Sepharose CL-4B (GE Healthcare) per sample to be analyzed was washed in NT buffer (150 mM NaCl, 50 mM Tris/HCl at pH 7.4, 0.05% NP-40), mixed with 15 µL of polyclonal α-CPSF30 antibody serum or preimmune serum, and incubated for 1 h with end-over-end rotation. The beads were washed twice in NT buffer and once in NT buffer containing 2 M urea. One-hundred-microliter cross-linking reactions were mixed with 100 µL of NT buffer containing 8 M urea and brought to 8 M urea total by addition of 48 mg of solid urea. After 15 min, the mixtures were diluted to 2 M urea with NT buffer. Antibody-loaded protein A Sepharose was added and incubated for 1 h with end-over-end rotation. Beads were washed three times in NT buffer with 2 M urea and eluted with 2× SDS sample buffer for 5 min at 95°C. Cross-linked proteins were analyzed as above.
For pull-down of His-tagged WDR33, 100-µL cross-linking reactions were mixed with 1.1 mL of 6 M guanidinum-HCl, 100 mM NaH2PO4, 10 mM Tris/HCl (pH 8), and 0.05% NP-40 and incubated for 15 min. A twenty-microliter packed volume of Ni-NTA agarose (Qiagen) was added, and the mixture was incubated for 30 min with end-over-end rotation. The beads were washed once with 6 M guanidinium-HCl, 100 mM NaH2PO4, 10 mM Tris/HCl (pH 6.3), and 0.05% NP-40 and twice in the same buffer containing 8 M urea instead of guanidinium-HCl. Protein was eluted with 8 M urea, 100 mM EDTA, 100 mM NaH2PO4, 10 mM Tris/HCl (pH 5.9), and 0.05% NP-40 and analyzed as above.
For limited proteolysis, cross-linking reactions were carried out in a volume of 500 μL. The samples were brought to 0.75 M urea with NT buffer containing 8 M urea and incubated for 10 min at 37°C. Endoprotease Lys-C (Promega) was added at 1/50th the mass of WDR33. Digestion was carried out at 37°C, and aliquots equivalent to 50-µL cross-linking reactions were stopped at various time points by addition of 800 µL of 100 mM NaH2PO4, 10 mM Tris/HCl (pH 8), 0.05% NP-40, and 8 M urea. Pull-down of His-tagged protein fragments was completed as in the preceding section. Digestion products were separated via 10% Tricine–SDS-PAGE (Schägger and von Jagow 1987) and analyzed by phosphorimaging.
WDR33 knockdown and qRT–PCR analysis
HEK293 cells were grown in DMEM GlutaMax (Invitrogen) supplemented with 10% FCS (FCS Superior, Biochrome) at 37°C with 5% CO2. Cells were transfected with 5–25 pmol of siPoolRNAs (siTools Biotech) (Hannus et al. 2014) or 25 pmol of individual siRNAs using 5 µL of Lipofectamine RNAiMax (Invitrogen) in a six-well format. Total RNA was isolated with the Trizol method. Ten micrograms of total RNA was treated with RNase-free DNase I (Roche). Two micrograms of DNase-treated RNA was reverse-transcribed with random hexanucleotide primers and RNase H minus MMLV reverse transcriptase (Promega) in a 25-µL volume for 10 min at room temperature followed by an additional incubation for 55 min at 43°C. qPCRs were performed as triplicates in a 96-well format with the LightCycler 480 SYBR Green I Master Mix (Roche) in a LightCycler 480 II instrument (Roche) at an annealing temperature of 60°C for all primer pairs used. Cp values were calculated by the software provided by Roche. Relative quantification of mRNA/pre-mRNA levels was done by the ΔΔCp method (Livak and Schmittgen 2001). Additional details can be found in the Supplemental Material.
PAR-CLIP assays and data analysis
We obtained publicly available 3′ end sequencing data (GSM909242 and GSM986133) and used data and procedures from our in-house 3′ end sequencing data processing pipeline (Martin et al. 2012) to determine CP sites that are used in HEK293 cells. For each library, 3′ end sequencing reads were normalized to a library size of 1 million, and CP sites were then ranked by the sum of sequencing reads from the two libraries.
The PAR-CLIP assay was carried out as described (Martin et al. 2012) except that the KAPA HiFi HotStart ReadyMix PCR kit (KAPA Biosystems) was used for PCR. WDR33 was precipitated with 10 µg of antibody (Bethyl Laboratories, no. A301-152A). The epitope recognized by this antibody maps to a region between residues 1286 and 1336 of human WDR33 (representing the extreme C terminus according to NP_060853.3; GeneID 55339). The library was sequenced on an Illumina HiSeq 2000 platform. Preprocessing and mapping of reads to the human genome (hg19) was done with CLIPZ (Khorshid et al. 2011).
Genomic positions where cross-linking occurred were ranked according to the number of mapped reads with T-to-C transitions (alignment data obtained from the CLIPZ server). To obtain the 1000 most abundantly cross-linked genomic sites and extract each individual site once, we traversed this list from top to bottom, adding a site to our list of top sites only if it was at least 25 nt away from a site that was already in the set. Next, nucleotide sequences were extracted from the genome, and the occurrences of hexanucleotide motifs were counted. To estimate the background frequency, we shuffled each of these initial sequences 1000 times and calculated an average frequency of each hexameric motif in randomized sequence sets.
The Gene Expression Omnibus (GEO) accession number for the PAR-CLIP data is GSE61123.
Acknowledgments
We are grateful to Gudrun Scholz, Sarah Jurischka, and Andrea Ringel for help with cloning; Imre Berger and Tim Richmond for MultiBac materials; Sachio Ito, Marcel Köhn, and Christiane Rammelt for plasmids and other reagents; and Yongsheng Shi for sending us their manuscript before submission. The work was supported by grants from the Deutsche Forschungsgemeinschaft (grant no. WA 548/15-1) to E.W., and the Swiss National Science Foundation (grant no.31003A-143977) to W.K.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.250985.114.
References
- Barabino SML, Hübner W, Jenny A, Minvielle-Sebastia L, Keller W. 1997. The 30 kDa subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homologue are RNA binding zinc finger proteins. Genes Dev 11: 1703–1716 [DOI] [PubMed] [Google Scholar]
- Berger I, Fitzgerald DJ, Richmond TJ. 2004. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol 22: 1583–1587 [DOI] [PubMed] [Google Scholar]
- Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. 1991. Purification of the cleavage and polyadenylation specificity factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem 266: 19768–19776 [PubMed] [Google Scholar]
- Bilger A, Fox CA, Wahle E, Wickens M. 1994. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes Dev 8: 1106–1116 [DOI] [PubMed] [Google Scholar]
- Campigli Di Giammartino D, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S, Choi E-A, Shi Y. 2011. Pre-mRNA 3′-end processing complex assembly and function. WIREs RNA 2: 321–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR III, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev (this issue). doi: 10.1101/gad.250993.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G, Keller W. 1989. Poly(A) polymerase purified from HeLa cell nuclear extract is required for both cleavage and polyadenylation of pre-mRNA in vitro. Mol Cell Biol 9: 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Sadowski M, Hübner W, Weiser S, Keller W. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J 21: 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson KS, Bilger A, Ballantyne S, Wickens MP. 1999. The cleavage and polyadenylation specificity factor in Xenopus laevis is a cytoplasmic factor involved in regulated polyadenylation. Mol Cell Biol 19: 5707–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang X-C, Purdy M, Wagner EJ, Marzluff WF. 2005. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol 25: 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506 [DOI] [PubMed] [Google Scholar]
- Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I. 2006. Protein complex expression by using multigene baculoviral vectors. Nat Methods 3: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Fronz K, Otto S, Kölbel K, Kühn U, Friedrich H, Schierhorn A, Beck-Sickinger AG, Ostareck-Lederer A, Wahle E. 2008. Promiscuous modification of the nuclear poly(A) binding protein by multiple protein arginin methyl transferases does not affect the aggregation behavior. J Biol Chem 283: 20408–20420 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, Nevins JR. 1989. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev 3: 2180–2189 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, Fleming ES, Oetjen J, Graveley BR. 1995. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple contacts involved in poly(A) site definition. Genes Dev 9: 72–83 [DOI] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Keller W, Zavolan M. 2012. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol 9: 1405–1412 [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Korshid M, Hausser J, Berninger P, Rothballer A, Ascano MJ, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannus M, Beitzinger M, Engelmann JC, Weickert MT, Spang R, Hannus S, Meister G. 2014. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res 42: 8049–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Molnàr A, Jones A, Baulcombe DC. 2006. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci 103: 14994–15001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AG, Xu R, Addepalli B, Rao S, Forbes KP, Meeks LR, Xing D, Mo M, Zhao H, Bandyopadhyay A, et al. 2008. Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics 9: 220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Keller W. 1995. Cloning of cDNAs encoding the 160 kDa subunit of the bovine cleavage and polyadenylation specificity factor. Nucleic Acids Res 23: 2629–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Minvielle-Sebastia L, Preker PJ, Keller W. 1996. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274: 1514–1517 [DOI] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G. 1991. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J 10: 4241–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwitz Y, Kühn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. 2003. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J 22: 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorshid M, Rodak C, Zavolan M. 2011. CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res 39: D245–D252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Yario TA, Benson E, Steitz JA. 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end formation. EMBO Rep 9: 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U, Nemeth A, Meyer S, Wahle E. 2003. The RNA binding domains of the nuclear poly(A) binding protein. J Biol Chem 278: 16916–16925 [DOI] [PubMed] [Google Scholar]
- Kühn U, Gündel M, Knoth A, Kerwitz Y, Rüdel S, Wahle E. 2009. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem 284: 22803–22814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C-K, Bachorik JL, Dreyfuss G. 2009. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol 16: 486–491 [DOI] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27: 2380–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano D, Marquardt S, Jones AME, Bäurle I, Liu F, Dean C. 2009. Altered interactions within FY/AtCPSF complexes required for Arabidopsis FCA-mediated chromatin silencing. Proc Natl Acad Sci 106: 8772–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Reports 1: 753–763 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. 2009. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CL, Chen J, Whoriskey J. 1988. Two proteins crosslinked to RNA containing the adenovirus L3 poly(A) site require the AAUAAA sequence for binding. EMBO 7: 3159–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KGK, Manley JL. 1992. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem 267: 14804–14811 [PubMed] [Google Scholar]
- Murthy KGK, Manley JL. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev 9: 2672–2683 [DOI] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore C, Greenblatt J. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem 278: 33000–33010 [DOI] [PubMed] [Google Scholar]
- Ohnacker M, Barabino SML, Preker PJ, Keller W. 2000. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J 19: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W. 1997. A multisubunit 3′-end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J 16: 4727–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Shi Y. 2012. Alternative polyadenylation: new insights from global analyses. RNA 18: 2105–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. 2003. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787 [DOI] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Müller CW. 2010. WD40 proteins propel cellular networks. Trends Biochem Sci 35: 565–574 [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF. 2009. A core complex of CPSF73, CPSF100, and symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Graber JH. 2012. Signals for pre-mRNA cleavage and polyadenylation. WIREs RNA 3: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2013. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci 38: 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Rüegsegger U. 1999. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev 23: 277–295 [DOI] [PubMed] [Google Scholar]
- Wigley PL, Sheets MD, Zarkower DA, Whitmer ME, Wickens M. 1990. Polyadenylation of mRNA: minimal substrates and a requirement for the 2′ hydroxyl of the U in AAUAAA. Mol Cell Biol 10: 1705–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Tong L, Manley JL. 2014. Delineating the structural blueprint of the pre-mRNA 3′-end processing machinery. Mol Cell Biol 34: 1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Doublié S. 2011. Structural biology of poly(A) site definition. WIREs RNA 2: 732–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationship with other steps in mRNA synthesis. Microbiol Mol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]