Anand et al. examined break-induced replication (BIR) and template switching between highly diverged sequences in S. cerevisiae, induced during repair of a site-specific double-strand break (DSB). Template switches between highly divergent sequences appear to be mechanistically distinct from the initial strand invasions that establish BIR. BIR traversing repeated DNA sequences frequently results in complex translocations analogous to those seen in mammalian cells. These results suggest that template switching among repeated genes is a potent driver of genome instability and evolution.
Keywords: break-induced replication, chromosome rearrangements, chromothripsis, template switching
Abstract
Recent high-resolution genome analyses of cancer and other diseases have revealed the occurrence of microhomology-mediated chromosome rearrangements and copy number changes. Although some of these rearrangements appear to involve nonhomologous end-joining, many must have involved mechanisms requiring new DNA synthesis. Models such as microhomology-mediated break-induced replication (MM-BIR) have been invoked to explain these rearrangements. We examined BIR and template switching between highly diverged sequences in Saccharomyces cerevisiae, induced during repair of a site-specific double-strand break (DSB). Our data show that such template switches are robust mechanisms that give rise to complex rearrangements. Template switches between highly divergent sequences appear to be mechanistically distinct from the initial strand invasions that establish BIR. In particular, such jumps are less constrained by sequence divergence and exhibit a different pattern of microhomology junctions. BIR traversing repeated DNA sequences frequently results in complex translocations analogous to those seen in mammalian cells. These results suggest that template switching among repeated genes is a potent driver of genome instability and evolution.
Genome instability—commonly manifested as gain, loss, or translocation of chromosome segments and often involving entire chromosome arms—is a hallmark of cancer and other diseased cells (Lupski 2007; Negrini et al. 2010; Davoli et al. 2013). In some cases, there are dramatic chromosome rearrangements known as chromothripsis or chromosome shattering in which, within a single chromosome, there are dozens of inversions, deletions, and duplications of sequences characterized by very small numbers of shared nucleotides at their junctions. A question of utmost importance is what makes normal cells, which have a remarkably stable genome, undergo such drastic genetic changes. Recent studies have suggested that precancerous cells show evidence of replication stress in the form of an increased amount of ssDNA, terminated replication forks, and DNA double-strand breaks (DSBs), suggesting replication-generated DNA breaks as the initial trigger for genome instability. Regardless of the origins of the DSBs, it is clear that DNA ends are potent inducers of chromosome rearrangements that result in cancer and other diseases (Elliott and Jasin 2002; Aguilera and Gomez-Gonzalez 2008; Halazonetis et al. 2008).
Many chromosomal rearrangements in mammalian cells, including translocations and segmental duplications, exhibit junctions that share only very few base pairs (i.e., microhomology). Although some of these chromosome rearrangements can readily be explained by several nonhomologous end-joining mechanisms (Shaw and Lupski 2005; Lieber et al. 2006; Chiang et al. 2012; Forment et al. 2012), many others must have involved repair-mediated DNA replication to create genomes in which there are copy number increases (Lee et al. 2007; Payen et al. 2008; Hastings et al. 2009b). Hastings et al. (2009a) proposed that these alterations occur via microhomology-mediated break-induced replication (MM-BIR); a similar mechanism has been suggested to explain the origin of some segmental duplications in budding yeast (Payen et al. 2008). BIR occurs when one end of DSB is able to strand-invade a homologous or divergent (homeologous) sequence and to initiate recombination-dependent DNA replication, which can proceed ≥100 kb to a chromosome end, creating nonreciprocal translocations. In budding yeast, where BIR has been most extensively studied (Anand et al. 2013; Donnianni and Symington 2013; Saini et al. 2013; Wilson et al. 2013), most BIR events depend on the recombination protein Rad51 and are substantially dependent on the nonessential DNA polymerase δ (Pol δ) subunit Pol32 (Davis and Symington 2004; Malkova et al. 2005; Lydeard et al. 2007). Recently, we showed that the mammalian homolog of Pol32, POLD3, is essential for cell viability when they are subjected to replication stress, presumably using a BIR mechanism to repair and restart stalled and broken replication forks (Costantino et al. 2014). BIR can also occur via a less efficient Rad51-independent mechanism (Malkova et al. 1996; VanHulle et al. 2007). The repair replication fork established by BIR is not as accurate as a normal replication fork; consequently, BIR exhibits a highly elevated rate of mutation as well as evidence of template switching, in which a replicating strand jumps from copying one chromosome to begin copying a homologous chromosome (Smith et al. 2007; Ruiz et al. 2009; Deem et al. 2011). A similar level of template switching is found in another DSB repair mechanism, gene conversion, which is not Pol32-dependent (Hicks et al. 2010).
To examine template switching and recombination events that involve limited homology, we created systems in the budding yeast Saccharomyces cerevisiae in which a single DSB induced by HO endonuclease is repaired by a two-step process that involves an initial BIR event and a subsequent template switch, resulting in a complex nonreciprocal translocation that reconstitutes a functional and chimeric URA3 gene. We compared recombination involving homologous templates with those involving highly divergent (homeologous) templates. We coupled our genetic analyses with complementary physical, sequence, and array-based analyses. We show that template switching between highly diverse templates is a mechanism that gives rise to complex rearrangements.
Results
Frequent template switching during BIR
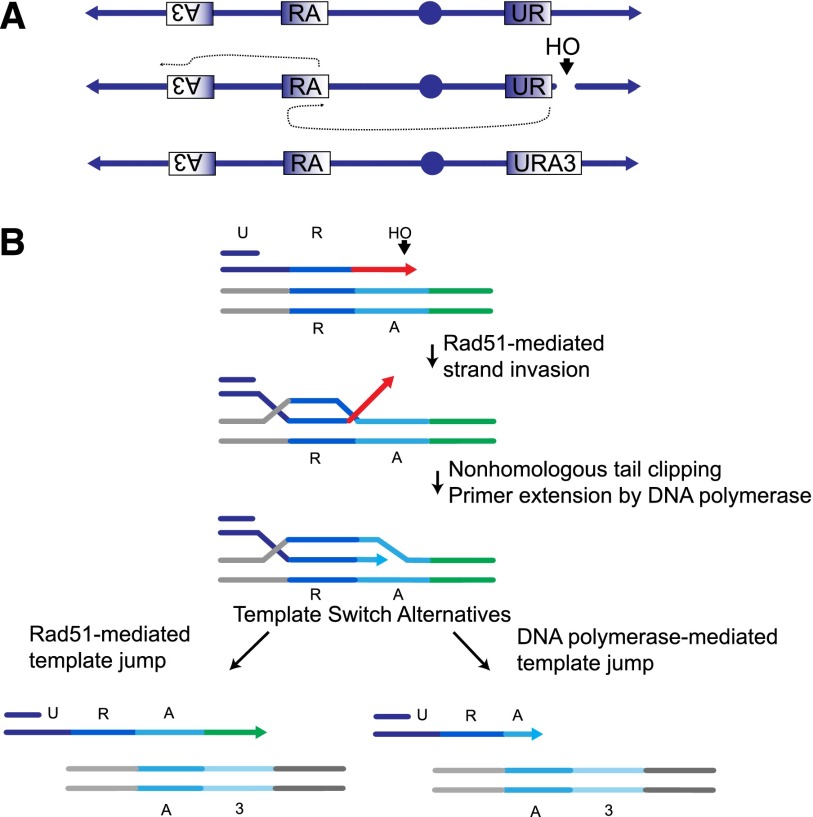
As a first step toward understanding the mechanism of template switching in more detail, we set up a genetic assay to measure the frequencies of template switching following a DSB (Fig. 1A,B). The assay system is based on the 804-base-pair (bp) URA3 gene from S. cerevisiae, which was split into three overlapping segments, each sharing ∼300-bp homology that we call “UR,” “RA,” and “A3” (Fig. 1; also see Supplemental Fig. 1). A DSB is induced by the galactose-inducible HO endonuclease adjacent to the UR locus, located at the CAN1 locus in a nonessential terminal region of chromosome 5 (Chr 5); this break can be repaired by a BIR mechanism using RA as the template sequence, located in the middle of the opposite chromosome arm. Template switching from RA to A3, located 50 kb more distally on the same chromosome arm, will result in the creation of a functional URA3 gene as part of a nonreciprocal translocation that can be screened by selection on media lacking uracil.
Figure 1.
Mechanisms of chromosome rearrangements via BIR and template switching. (A) Genetic assay to measure the frequencies of template switching following a DSB. The assay system is based on the 804-bp URA3 gene from S. cerevisiae, which was split into three overlapping segments, each sharing 300-bp homology, that we call “UR,” “RA,” and “A3” (see also Supplemental Fig. 1). A DSB is induced by the galactose-inducible HO endonuclease adjacent to the UR locus, located at the CAN1 locus in a nonessential terminal region of chromosome 5 (Chr 5); this break can be repaired by a BIR mechanism using, as the template, the RA sequence located in the middle of the opposite chromosome arm. Chr 5 is shown in reversed orientation from the conventional representation. Template switching from RA to A3, located 50 kb more distally on the same chromosome arm, will result in the creation of a functional URA3 gene as part of a nonreciprocal translocation that can be screened by selection on media lacking uracil. (B). Examining the mechanism of BIR and template switching. HO endonuclease creates a DSB next to the UR sequence. Following strand invasion into the RA template (dark-blue line followed by cyan line), clipping of the nonhomologous 3′ tail (red line) is a prerequisite for DNA synthesis. Template switching after the initial strand invasion can proceed by two alternative pathways. (Left) DNA synthesis proceeds past the RA template, and the subsequent template switch intermediate (cyan followed by green) carries a nonhomologous 3′ tail (green line). Subsequent DNA synthesis using the A3 sequence requires clipping of the nonhomologous 3′ tail (green line). (Right) Alternatively, a template switch into the A3 sequence (cyan followed by light-blue line) occurs during synthesis within the RA template. Subsequent DNA synthesis using the A3 sequence does not require tail clipping. With regards to tail clipping, the alternative shown on the left is similar to the first strand invasion event. In contrast, the alternative shown on the right is mechanistically different from the first strand invasion event, as it does not require tail clipping. The two alternatives are distinguished by examining the microhomology usage of the 3′ ends (see the text for details). Gray lines represent adjacent, nonhomologous, flanking sequences.
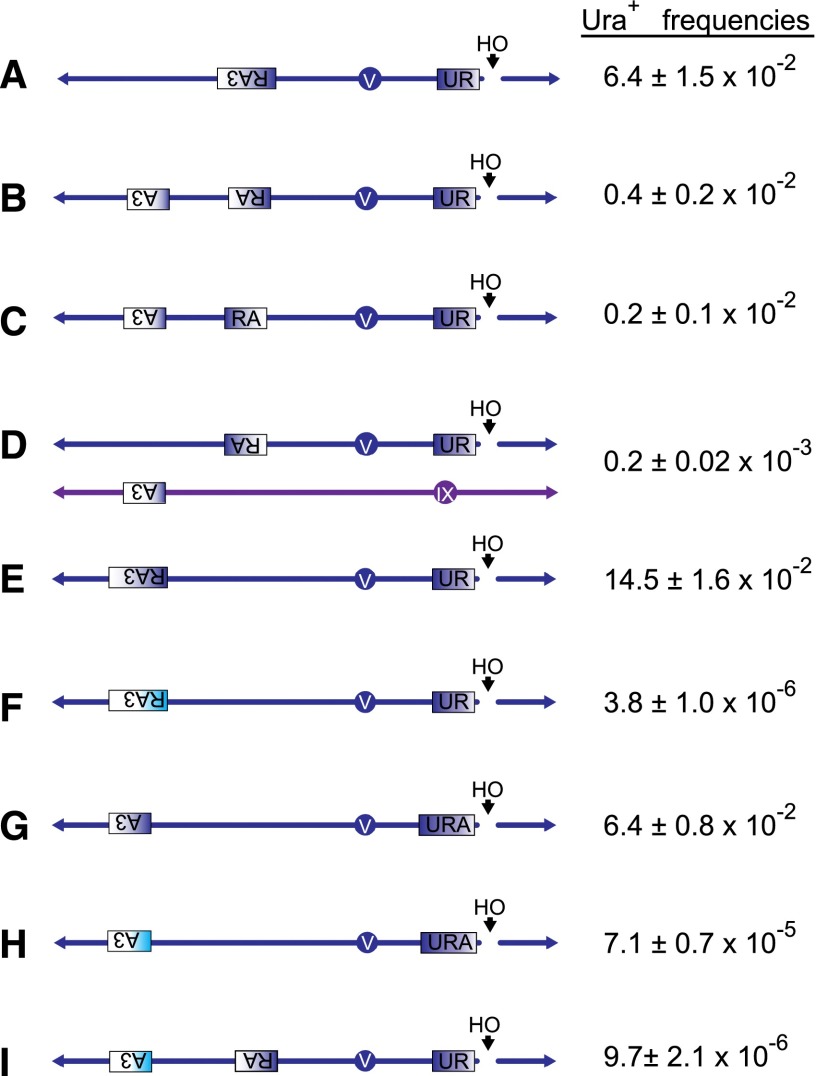
Based on this assay, we constructed a series of strains to examine template switch mechanisms (Fig. 2,; also see Supplemental Tables 1, 2). First, we compared a simple BIR event between UR and RA3 (Fig. 2A) on the opposite chromosome arm with the tripartite UR–RA–A3 system using templates that are 100% homologous, again with segments sharing 300 bp of homology. Repair efficiency by BIR (UR × RA3) was 6.4 × 10−2, consistent with a related BIR assay described previously (Lydeard et al. 2007). Nearly all viable cells were Ura+. Spontaneous recombination frequencies between the UR and RA3 are rare, ∼2 × 10−6 (Supplemental Fig. 2). In contrast, the frequency of BIR-provoked Ura+ outcomes in the UR × RA × A3 system was 0.4 × 10−2 (Fig. 2B). Moreover, in this case, most viable outcomes were Ura−, obtained at a rate of 4.7 × 10−2. The Ura− outcomes resulted from simple BIR events in which UR invaded the RA sequence to initiate BIR but then proceeded without the second template jump to replicate to the telomere (Fig. 2B). From these data, we conclude that when distant templates share 300 bp of complete homology, about one out of 12 BIR forks (0.4 × 10−2/4.7 × 10−2) undergoes template switching. This result is comparable with a 16% rate of template switching seen in a diploid system transformed with a chromosome fragment sharing ≥3 kb of homology with each homologous chromosome, repaired by BIR (Smith et al. 2007).
Figure 2.
Constructs to examine homologous BIR, homeologous BIR, and template switch mechanisms. (A) BIR-only construct (yRA107). An HO-induced DSB next to the UR sequence is repaired by BIR using the RA3 sequence located on the opposite arm of Chr 5 and ∼80 kb distal to the telomere. UR and RA3 shares 300-bp homology. Repair by BIR results in reconstitution of functional URA3 gene and loss of a chromosome fragment centromere-distal to the HO DSB. (B) Template switch construct (yRA53). DSB is repaired by BIR using the RA sequence located 80 kb distal to the telomere. Some of the BIR forks will switch templates to A3 located 50 kb away from RA and 30 kb distal to the telomere. UR shares 300-bp homology with RA, and RA shares 300-bp homology with A3. The template switch is assayed as a reconstitution of the functional URA3 gene. (C) Same as B except that the orientation of RA is flipped so that the invading BIR fork is directed toward the centromere (yRA55). (D) Interchromosomal template switch construct (yRA126) in which A3 is located on Chr IX 30 kb distal to the telomere. (E) BIR-only construct (yRA52). An HO-induced DSB next to the UR sequence is repaired by BIR using the RA3 sequence located on the opposite arm of Chr 5 and ∼30 kb distal to the telomere. UR and RA3 share 300-bp homology. (F) Homeologous BIR construct (yRA57). An HO-induced DSB next to the S. cerevisiae UR sequence is repaired by BIR using the RA3 sequence from Kluyveromyces lactis located on the opposite arm of Chr 5 and ∼30 kb distal to the telomere, reconstituting a chimeric URA3. Sc-UR and Kl-RA3 are 68.3% identical and share a maximum of 11-bp microhomology. (G) BIR-only construct (yRA213). DSB at URA is repaired by BIR using the A3 sequence located 30 kb distal to the telomere. URA shares 300-bp homology with A3. (H) Homeologous BIR-only construct (yRA192); similar to G except Kl-A3 is substituted for Sc-A3. Sc-URA and Kl-A3 are 76% identical and share a maximum of 17-bp microhomology. (I) Homeologous template switch construct (yRA58). HO-induced DSB next to UR is repaired by homologous BIR between Sc-UR and Sc-RA and homeologous template switching between Sc-RA and Kl-A3 reconstituting a chimeric URA3. The dark-blue and cyan rectangles represent the Sc-URA3 and Kl-URA3 sequences, respectively.
We predicted that the orientation of the RA template would have a minimal effect on the frequency of template switching, provided that the chromosomal environment of the bridging sequence remained the same. To test this prediction, we inverted the RA sequence so that DNA synthesis by the BIR fork proceeds toward the centromere. The rate of template switches was ∼0.2 × 10−2, about half of what was found in the other orientation; however, the two rates of template switching were statistically indistinguishable (t-test, P-value = 0.054) (Fig. 2C). As we show below, some of the BIR events initiated in this orientation may have undergone other rearrangements and thus would be removed from those yielding a Ura+ outcome.
To determine whether template switch frequencies are affected by an intrachromosomal versus interchromosomal location, we moved the location of A3 to the left arm of Chr 9, ∼30 kb distal to the telomere, so that the template switch must occur interchromosomally but will synthesize a length (∼30 kb) comparable with the intrachromosomal construct. The frequency of interchromosomal template switching was ∼18-fold lower (Fig. 2, cf. B and D). We conclude that (1) template switching is largely independent of the orientation of the bridging sequence and (2) intrachromosomal template switching is more efficient than interchromosomal switching. We note that one of the remarkable features of mammalian chromothripsis is that a single chromosome is subject to massive rearrangement; this may be explained in part by a strong bias to engage in intrachromosomal template switching. Previously, Sun et al. (2013) had reported a similar bias in mammalian rearrangements.
BIR between highly divergent templates
We sought to understand whether canonical strand invasion that establishes recombination-dependent replication is mechanistically different from template switching. Experimentally, this is a tricky question to answer, as strand invasion must precede template switching, and deletion of genes involved in canonical strand invasion will perforce preclude template switches. To tackle this question, we built a series of strains (Fig. 2; see also Supplemental Table 1) that require recombination between S. cerevisiae URA3 sequences and those of the 71% identical Kluyveromyces lactis URA3 gene, which encodes the same number of amino acids (Supplemental Fig. 3). We showed previously that all or nearly all Sc–Kl chimeric genes are functional (Hicks et al. 2010). Alignment of the sequences reveals many regions of microhomology, up to 17 nucleotides (nt) (Supplemental Fig. 3), so that breakpoint junctions stemming from template switches can be clearly assigned after sequencing the functional chimeric URA3 that is formed after the recombination event.
We first examined the microhomology usage in a simple “BIR-only” strain in which a DSB adjacent to Sc-UR can be repaired using either the homologous Sc-RA3 (Fig. 2E) sequence or a homeologous Kl-RA3 template (Fig. 2F). Here, BIR is accomplished with ∼30 kb of new DNA synthesis compared with ∼80 kb for the construct shown in Figure 2A. The rate of BIR decreases from 14.5 × 10−2 (Fig. 2E) to 6.4 × 10−2 when an additional 50 kb must be copied (Fig. 2A). When recombination occurs between the homeologous templates sharing 68% identity (Supplemental Fig. 4), the efficiency of repair by MM-BIR dropped nearly 38,000-fold, from 14.5 × 10−2 to 3.8 × 10−6 (Fig. 2F). This level of restraint is dependent on the degree of sequence divergence, as illustrated when we carried out a similar BIR assay but where Sc-URA recombines with Kl-A3. The A segments share 76% identity compared with the 68% identity between Sc and Kl R regions; within the A region there is a stretch of 86 bp with only single-base-pair differences (Supplemental Fig. 5). When recombination used the less-divergent A segment (76% identical) as opposed to the R segment (68% identical), homeologous BIR dropped only 900-fold compared with homologous BIR (Fig. 2, G vs. H). Our results are consistent with an earlier report by Datta et al. (1997), who reported a 4000-fold reduction in spontaneous recombination when the recombining sequences were 74% identical. We conclude that homeology severely inhibits canonical strand invasion and BIR, and the inhibition is dependent on the degree of sequence divergence.
Template switching is less sensitive to sequence divergence
We then asked whether homeology inhibits template switching. To address this question, we used the UR × RA × A3 strain but substituted Sc-A3 with Kl-A3. In this strain, the first step (strand invasion and initiation of BIR) occurs between the fully homologous sequences of Sc-UR and Sc-RA, whereas the second step (the template switch) is microhomology-dependent (Fig. 2I). We observed that the presence of highly divergent A3 sequences decreased the efficiency of template switching by ∼400-fold (Fig. 2, cf. B and I). This reduction, although highly significant, is ∼2.5-fold less than the 900-fold drop seen in Sc-URA × Kl-A3 homeologous BIR (Fig. 2, cf. G and H) and suggests that template switching is not as severely impaired as strand invasion by sequence divergence (t-test of comparison of the fold change between homeologous BIR and homeologous template switch, P = 0.014).
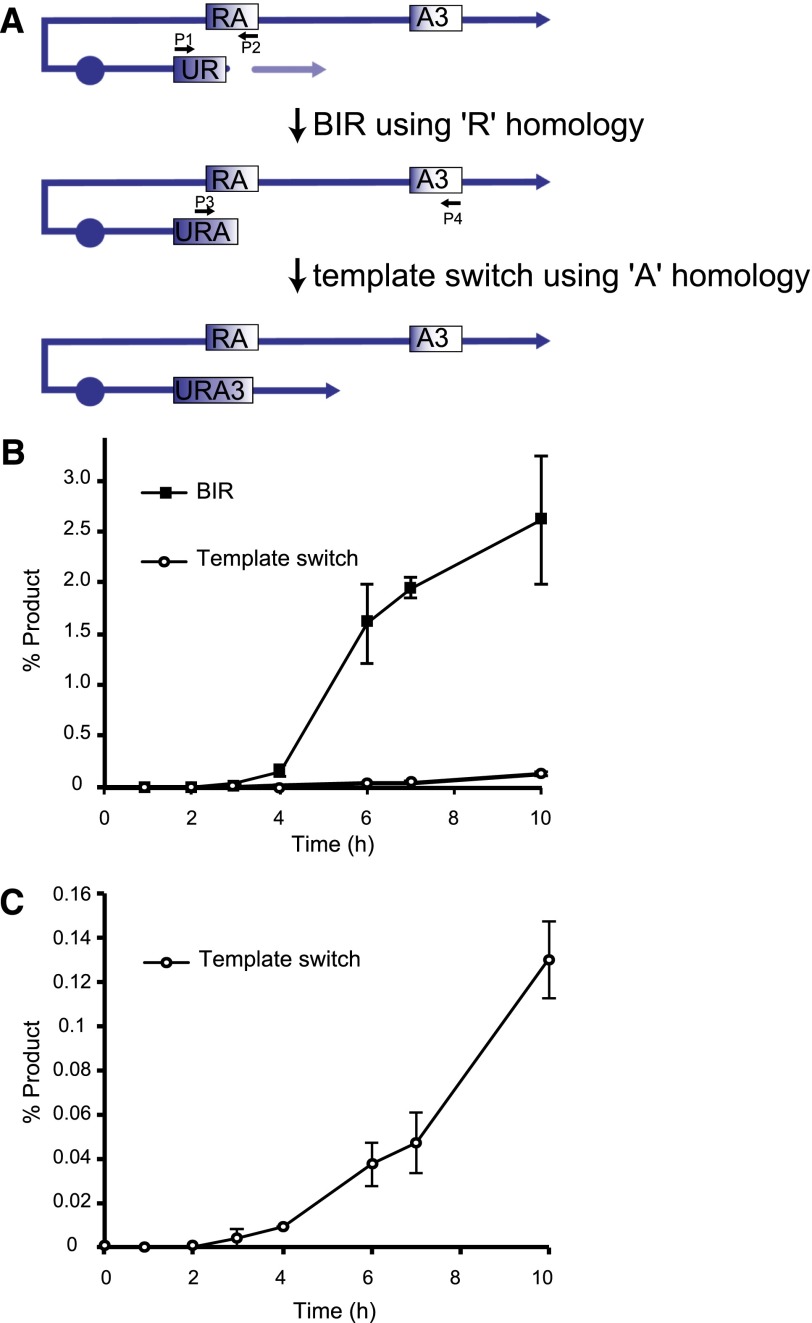
We also monitored the kinetics of BIR and template switching, as shown in Figure 3. Using a series of PCR primers, we could measure the time of appearance of both the simple BIR event (using primers p1 and p2) (Fig. 3) and that for the subsequent template switch (using primers p3 and p4) (Fig. 3). The level of BIR product was calculated to be ∼6%, relative to the PCR signal of URA3 from a survivor, consistent with the genetic result of ∼6% viability. Template switching proved to be ∼4% relative to the BIR outcome, somewhat lower than the 8% found by genetic assay. Importantly, there was no significant delay in the appearance of the signal for template switching compared with BIR (Fig. 3A,B). This result suggests that template jumping does not go through the same long delay as seen for the initial BIR event and lends support to the idea that the jump may not proceed through the same series of steps required to initiate BIR.
Figure 3.
Template switching followed the initial DNA synthesis of BIR without a significant delay. Kinetics of the initial DNA synthesis (BIR) and template switching were examined by primer extension assay with quantitative PCR. (A) Experimental scheme for quantitative PCR. The primer pairs employed are represented by arrows. (B) Kinetics of BIR and template switching are shown in the same graph. (C) Kinetics of template switching (zoomed in) are shown as a separate graph. The frequency of template switching was estimated to be ∼2%∼6% of the all-BIR invasion attempts, consistent with the results from the genetic assay. Error bars represent the range of two independent experiments.
Effect of homeology in gene conversion
We also note that the inhibitory effect of homeology is not unique to BIR but rather is a feature shared by mechanisms that involve canonical strand invasions. We constructed a related strain in which the DSB occurs between adjacent UR and A3 sequences and in which the DSB can be repaired by gene conversion from RA sequences located on the opposite chromosome arm (strains yRA97–yRA100) (Supplemental Table 1). When both DSB ends are homologous to the distant RA template, 64% of cells could repair the DSB and yield Ura+ cells (yRA97) (Supplemental Table 1; Supplemental Fig. 6). Similar to what was observed for BIR, homeology (when the Sc-RA template is changed to Kl-RA) severely reduced the efficiencies of gene conversion by ∼230,000-fold (yRA98) (Supplemental Table1; Supplemental Fig. 6). The decreased efficiency of gene conversion compared with BIR could be explained by the presence of homeology on both the ends compared with one-ended invasion in BIR.
Microhomology usage in strand invasion is influenced by sequence divergence
Recombination between homeologous sequences made it possible to determine the breakpoint junctions for both MM-BIR and template switch events. Many recent observations in various experimental systems and patient samples have revealed the presence of microhomologies at the breakpoint junctions (Lee et al. 2007; Payen et al. 2008; Hastings et al. 2009a; Liu et al. 2011; Stephens et al. 2011). It should be mentioned that, in most of the rearrangements examined in mammalian systems, the microhomologies are in unique sequences, whereas in our system, the microhomologies are embedded within diverged repeats.
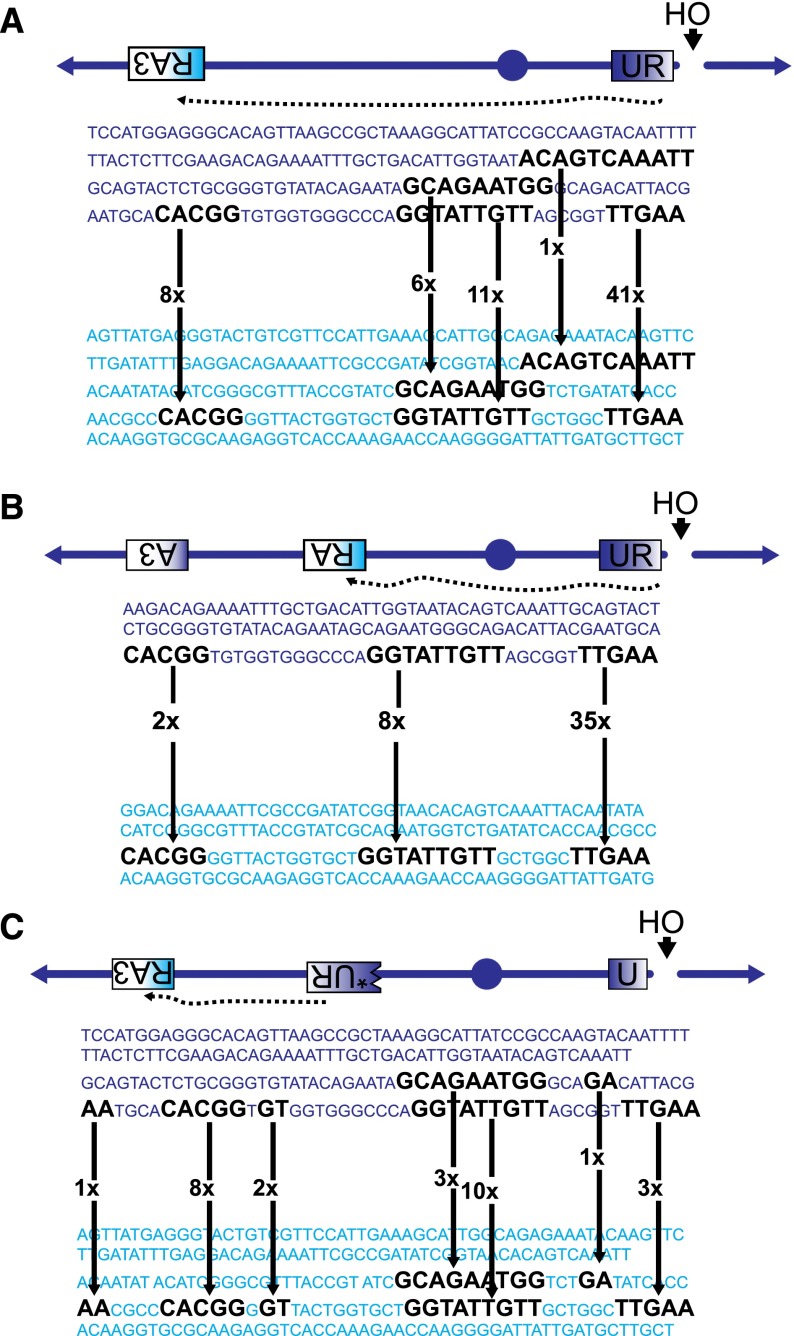
First, we gathered information regarding the sequence usage during BIR between Sc-UR and Kl-RA3. Among 67 Ura+ BIR events, there was an unexpected and clear preference for the use of microhomologies at the 3′ end of the DSB (Fig. 4A). In fact, 61% of Ura+ used the TTGAA sequence present at the very 3′ end of UR. This sequence is 68 bp away from the HO endonuclease cleavage site; these nonhomologous sequences between the DSB and the UR sequence must be clipped off during or after strand invasion so that the 3′ end can then prime new DNA synthesis along the template (Fig. 1B; Fishman-Lobell and Haber 1992; Paques and Haber 1997; Colaiacovo et al. 1999). Thus, it appears that strand invasion coupled to nonhomologous tail clipping forces the usage of the most 3′-terminal microhomology. This same bias was seen in a template switching strain in which Sc-UR initiates recombination with Kl-RA (Fig. 4B), with 78% of the events using the terminal TTGAA sequence.
Figure 4.
Sequence usage distinguishes canonical strand invasion from template switch. (A) Microhomology usage during strand invasion in a homeologous BIR strain. An HO DSB near the Sc-UR is repaired first by strand invasion into the homeologous Kl-RA3 sequence (yRA57). Arrows show the frequencies of microhomology usage during the invasion event. (B) Microhomology usage during strand invasion in homeologous BIR in a template switch strain (yRA54). An HO DSB near the Sc-UR is repaired first by strand invasion into the homeologous Kl-RA sequence. Arrows show the frequencies of microhomology usage during the invasion event. (C) Sequence usage during the template switch from Sc-UR to Kl-RA3 (yRA204). An HO DSB near the Sc-UR is repaired first by strand invasion into the homologous Sc-UR* sequence in which the promoter and 12 bp 5′ of the ORF are deleted (represented by the blue block with a jagged edge). The subsequent template switch between the Sc-UR* and the homeologous Kl-RA3 sequence reconstitutes a functional chimeric URA3. Arrows show the frequencies of microhomology usage during the template switch event.
Sequence preference in template switching differs from that seen for the initial BIR event
We then compared the sequence preference during strand invasion described above with that seen in template switching. Here we created a new strain in which BIR is initiated by recombination between Sc-U and a homologous, promoterless UR sequence on the opposite chromosome arm (Sc-U and Sc-*UR), after which there is a template switch to Kl-RA3 (Fig. 4C; Supplemental Table 1). In contrast to the strand invasion pattern, there was no preferential usage of any one particular sequence in template switching (Fig. 4C). This result suggests strongly that template jumps occur via a different mechanism from the initial strand invasion steps of BIR (see the Discussion).
The mismatch repair (MMR) gene MSH6 alters the spectrum of microhomology junctions
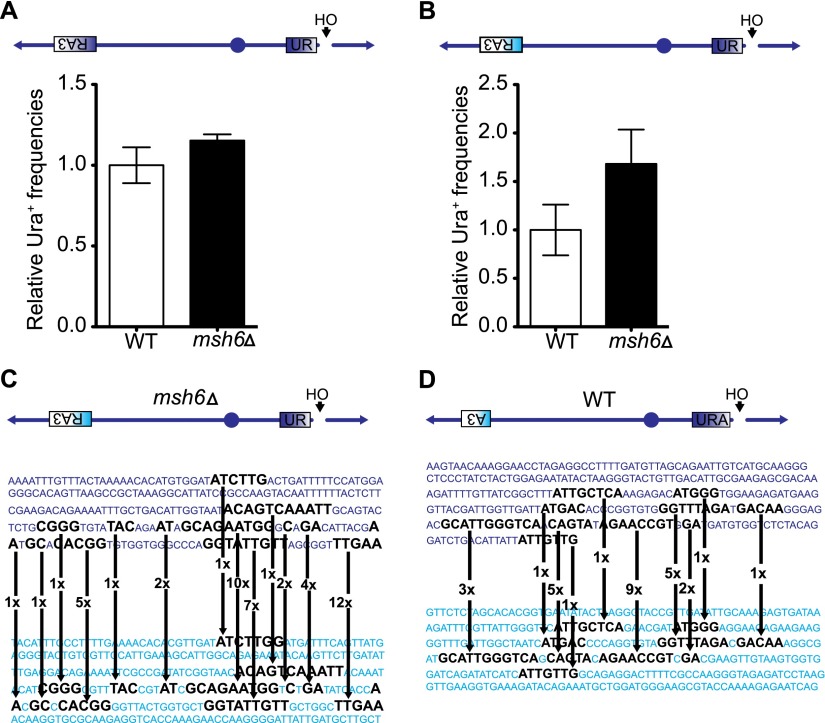
Mismatches in the recombining substrates can discourage recombination. Provided the degree of divergence is low (i.e., ∼10%), lack of MMR components has been shown to equalize the rates of spontaneous recombination between homeologous substrates to that of completely homologous substrates (Datta et al. 1997). To test whether MMR components could modulate the repair of the 68% divergent R–R substrate, we deleted MSH6 from both the homologous system and the homeologous system. The absence of Msh6 resulted in no change in the efficiencies of homologous BIR (Fig. 5A) but increased the efficiency in the homeologous BIR strain about twofold (Fig. 5B), consistent with an earlier report (Datta et al. 1997) showing improved rates of spontaneous recombination between highly mismatched (74%) substrates in the absence of the MMR components. Strikingly, compared with the wild-type strain, in the absence of Msh6, we observed a reduced 3′ end preference (cf. Figs 4A and 5C), showing a novel role for the MMR components in imposing a 3′ end preference during homeologous BIR between highly divergent sequences.
Figure 5.
The 3′ end preference during homeologous BIR between highly divergent sequences is reduced in the absence of Msh6. (A) Relative efficiencies of homologous BIR in wild type (WT) and msh6Δ. (B) Relative efficiencies of homeologous BIR in wild type and msh6Δ. (C) Microhomology usage in the absence of MSH6 during strand invasion in the homeologous BIR strain. Note the reduced 3′ end preference compared with the wild type (Fig. 4A). (D) Microhomology usage during strand invasion in a homeologous BIR strain with low sequence divergence (76%). Arrows show the frequencies of microhomology usage during the invasion event. Letters in dark blue and cyan represent the S. cerevisiae and K. lactis sequences, respectively.
Interestingly, we also found that the extent of mismatch between recombining segments also influenced the biased use of the 3′ end of the invading strand in BIR. When we examined BIR between the less divergent A–A segments (Sc-URA × Kl-A3) (Fig. 5D), we discovered that the location of the junctions between these 76% identical sequences did not display a marked use of the 3′-terminal microhomology (ATTGTTG), which is longer than the preferentially used TTGAA in the Sc-UR × Kl-RA3 case. BIR between the less divergent A–A sequences also showed an 18-fold improvement over the more divergent R–R sequences (Supplemental Fig. 7). These results suggest that the rules for establishing a stable strand invasion may be different as the degree of divergence increases.
Role of different recombination proteins in BIR and template switching
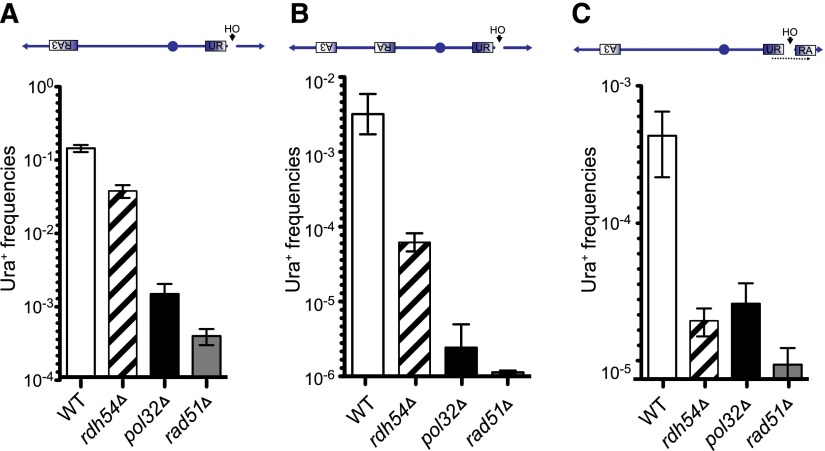
Our observations that strand invasion and template switch exhibit different features led us to test the role of Rad51 and Pol32 in template switching. The central recombination protein Rad51 is required for all gene conversion events and efficient BIR, although there is a Rad51-independent BIR pathway (Malkova et al. 1996; Davis and Symington 2004; Anand et al. 2013). The nonessential Pol32 subunit of the DNA Pol δ complex is not required for either normal DNA replication or gene conversion but is required for BIR (Davis and Symington 2004; Lydeard et al. 2007; Anand et al. 2013). We deleted Rad51 and Pol32 in our BIR-only strain (Sc-UR × Sc-RA3). Consistent with previous results (Davis and Symington 2004; Lydeard et al. 2007), deletion of Rad51 and Pol32 resulted in 360-fold and 97-fold reductions in Ura+ frequencies, respectively (Fig. 6A). We also tested the role of the recombination factor Rdh54 in BIR based on our recent finding that this homolog of Rad54 does not affect simple gene conversion but played a striking role in interchromosomal template switches between homeologous and homologous sequences that accompanied some gene conversion events (Tsaponina and Haber 2014). Deletion of Rdh54 reduced the efficiency of intrachromosomal BIR by about fourfold (Fig. 6A). We did not test the role of Rad54, since it has been shown to exhibit a phenotype similar to that of Rad51 in promoting gene conversion (Sugawara et al. 2003) and BIR (Davis and Symington 2004).
Figure 6.
BIR and template switch have different requirements for Rad51, Pol32, and Rdh54. (A) BIR frequencies in wild type (WT), rdh54Δ, rad51Δ, and pol32Δ. (B) Template switch frequencies in wild type, rdh54Δ, rad51Δ, and pol32Δ during BIR. (C) Template switch frequencies in wild type, rdh54Δ, rad51Δ, and pol32Δ during SSA.
We then examined the roles of Rad51, Pol32, and Rdh54 in BIR template switching. As mentioned previously, because canonical strand invasion precedes template switching and because deletion of the genes essential for canonical strand invasion will perforce preclude template switch events, we expected a dramatic decrease in the template switch outcomes in the strains lacking the above-mentioned activities. As mentioned before, our tripartite system (Figs. 1A, 2) to measure template switching between homologous sequences involves an initial recombination between the R and R segments (UR × RA) and a subsequent recombination between A and A segments (URA × A3). We first assessed the contribution of Rad51 in the initial step of recombination between R × R segments by deleting Rad51 from the yRA107 strain (Supplemental Table 1). Deletion of Rad51 in the yRA107 strain resulted in an ∼300-fold drop in BIR efficiency. We also assessed the contribution of Rad51 in recombination between A × A segments by deleting Rad51 from the yRA213 strain. Deletion of Rad51 resulted in ∼240-fold reduction in BIR efficiency. We reasoned that, in the rad51Δ strain, if the template switch step occurs with a degree of dependency similar to that of the strand invasion step of BIR, there would be a 300 × 240-fold (72,000-fold) reduction in the final Ura+ outcomes. Contrary to our prediction, deletion of Rad51 resulted in only a 3600-fold drop in the Ura+ outcomes (Fig. 6B). Thus, the template switch step (RA × A3) was only reduced by an additional factor of 12 over the 300-fold drop in initiating BIR. This result suggests that there is significantly less dependency on Rad51 for template switching than canonical strand invasion.
If the template switch step occurred with Pol32 dependence similar to that for simple BIR, we expected a 97 × 97 (9409)-fold reduction in the final Ura+ outcomes. Again, contrary to our expectations, deletion of Pol32 resulted in a 1265-fold reduction in the Ura+ outcomes. This result implies that pol32Δ reduced template switching ∼13 fold compared with 97-fold for simple BIR.
In contrast to rad51Δ or pol32Δ, deletion of RDH54 affected template switching considerably more than it affected BIR or gene conversion; deletion of RDH54 resulted in a 55-fold reduction in the rate of Ura+ recombinants compared with the expected 16-fold drop if template switching was affected as equally as strand invasion (Fig. 6A,B; Klein 1997; Signon et al. 2001). These data together with earlier findings (Tsaponina and Haber 2014) show the unique requirement of Rdh54 for template switching between fully homologous sequences.
It is possible that the absence of Rad51 or Pol32 significantly altered the BIR replication process so that we were measuring the frequencies of template switches stemming from an altered replication machine. We sought an alternative way to determine the Rad51 and Pol32 dependency of template switching by constructing a strain in which the first step is Rad51- or Pol32-independent. Single-strand annealing (SSA) occurs independently of Rad51 and Pol32 (Ivanov et al. 1996; data not shown). We therefore constructed new strains in which the step preceding template switching was Rad51- and Pol32-independent SSA (Fig. 6C,; Supplemental Fig. 8D). An HO-induced DSB was created between two direct repeats, UR and RA, which share 300-bp homology. The efficiency of repair by SSA is nearly 100% (data not shown). After strand annealing, SSA requires new DNA synthesis to fill in single-stranded regions beyond the annealed sequences, and we reasoned that filling in of the adjacent “A” region could result in a jump to an A3 segment on the opposite chromosome arm and would result in Ura3+ cells (Fig. 6C). We could also get URA3 by a secondary, spontaneous recombination event between SSA-generated URA and A3; however, we showed that such events were rare (mean recombination frequencies between URA and A3 are 2.6 × 10−6) (see Supplemental Fig. 2). In a wild-type background, template switching events that began with an SSA event and then resulted in Ura+ occurred at a frequency of ∼0.04 × 10−2, which is several orders of magnitude lower than the rate of template switching accompanying BIR. In the absence of Rad51 and Pol32, such events were reduced by ∼46-fold and ∼17-fold, respectively. Although we do not know whether the DNA polymerases engaged in filling in the ssDNA gaps after SSA are the same as required for template switching in BIR, the reductions in rad51Δ and pol32Δ are similar to those that we found for template switching (10-fold and 13-fold, respectively). Consistent with our previous data showing greater dependency on Rdh54 for template switching than for canonical strand invasions, deletion of Rdh54 resulted in a 20-fold decrease in SSA-generated URA3 (Fig. 6C) without affecting the efficiency of SSA per se (data not shown). Observed reduction was again similar to the 14-fold drop in the template switching step (Fig. 6B). These data support the idea that the process of template switching is distinctly different from DNA synthesis accompanying strand invasion that initiates BIR or gene conversion.
Frequent template switches between naturally repeated sequences
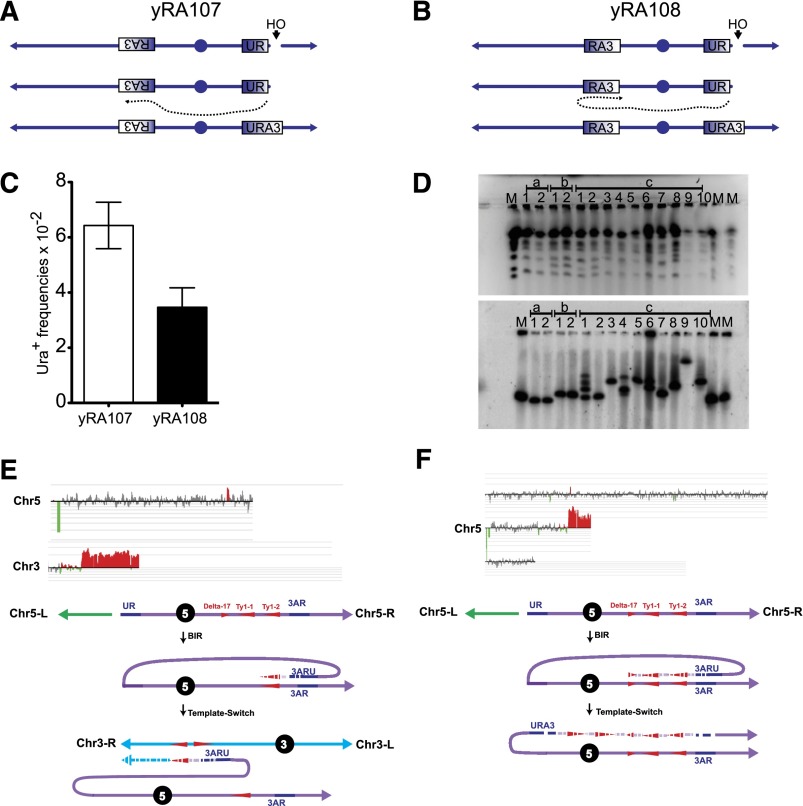
Previous studies of chromosomal rearrangements in budding yeast have shown that many rearrangements occur between dispersed copies of homologous retrotransposon sequences in yeast (Louis and Haber 1990; Fischer et al. 2000; Lemoine et al. 2005; VanHulle et al. 2007; Argueso et al. 2008; Chan and Kolodner 2011). We wished to determine how often such sequences would be engaged in template switching in BIR events. We created strain yRA108, in which Sc-UR × Sc-RA3 sequences would result in BIR proceeding toward the centromere of Chr 5 rather than toward the telomere, as in strain yRA107 (Supplemental Table 1). BIR to the opposite telomere would result in an unstable dicentric chromosome, assuming that replication could proceed past the centromere (Morrow et al. 1997). Thus, it should be impossible to recover Ura+ sequences without some later event to stabilize the BIR outcome (Fig. 7A,B). To our surprise, the two constructs, yRA107 and yRA108, had statistically indistinguishable Ura+ frequencies (6.4 × 10−2 vs. 3.5 × 10−2 respectively; P = 0.054) (Fig. 7C). We note that centromere-proximal to RA3 is a chromosomal region containing the repetitive and transcriptionally active Ty retrotransposon and a cluster of tRNA genes that might play a role in generating viable outcomes.
Figure 7.
Ty and δ element-mediated template switching. (A) yRA107 construct. DSB next to the UR sequence is repaired by BIR using the RA3 sequence, resulting in loss of the centromere-distal broken fragment and reconstitution of the URA3 gene. Note that DNA synthesis of the invading BIR fork is directed toward the telomere. (B) yRA108 construct. The DSB next to the UR sequence is repaired by BIR using the RA3 sequence, resulting in loss of the centromere-distal broken fragment and reconstitution of the URA3 gene. In contrast to A, the invading BIR fork is directed toward the centromere, potentially leading to dicentric chromosomes and subsequent chromosome instability. (C) BIR frequencies of the yRA107 and yRA108 constructs. (D) PFGE analyses of yRA107 and yRA108. (Top panel) (M) Chromosome size marker; (a1 and a2) uncut yRA107 and yRA108 controls; (b1 and b2) yRA107 BIR survivors; (c1 to c10) yRA108 BIR survivors. (Bottom panel) Southern blot analyses of the PFGE with URA3 ORF as the probe. Multiple bands seen in c1, c4, and c6 are putative unstable dicentric intermediates. (E,F) aCGH analyses of c3 and c5 from D. Green troughs represent deletion of the centromere-distal chromosome fragment. Red peaks represent duplications/triplications. Schematic interpretations of the respective aCGH analyses. Ty and δ elements that mediated the template switch events are shown as large and small red triangles, respectively. Hashed patterns represent new DNA synthesis (please see the text for details). The event shown in E results in the loss of the left arm of Chr 5 distal to the CAN1 locus and a concomitant duplication of a portion of the left of Chr 3. The event shown in F results in the loss of left arm of Chr 5 distal to the CAN1 locus and a concomitant triplication of a portion of right arm of Chr 5.
To understand how Ura+ survivors of strain yRA108 could appear, we subjected their DNA to pulse field gel electrophoresis (PFGE) analysis. Each of the 10 survivors that we tested had undergone one or more rearrangements of Chr 5 (Fig. 7D). Array comparative genome hybridization (aCGH) analyses of the same Ura+ survivors showed that these rearrangements were very likely mediated by recombination (presumably template switching) between the Ty retrotransposons adjacent to RA3 and Ty elements elsewhere in the genome (Fig. 7E,F). Figure 7E shows the following complex scenario: (1) BIR initiated by invasion of UR into RA3 with the concomitant loss of ∼30 kb to the left of the DSB; (2) the first BIR event results in duplication of an ∼10-kb sequence extending into Ty1-2; and (3) there is a subsequent template switch to a Ty element on Chr 3, resulting in a nonreciprocal Chr 5–3 translocation. In the event shown in Figure 7F, a BIR copies the sequences located centromere-proximal to RA3 until the second Ty element (Ty1-1) is reached. There is a template dissociation and a jump to a nearby oppositely oriented δ element (the flanking long terminal repeat [LTR] of a Ty element, likely YERWδ17). After this template switch, BIR must then continue to the end of the chromosome. The net result of these events is a triplication of ∼50 kb of Chr 5 sequences next to a duplication of ∼70 kb, consistent with the CGH results. About 70% (seven of 10) (Supplemental Fig. 9) of the survivors that we analyzed exhibited intrachromosomal template switching. Previously, Smith et al. (2007) had observed template switching between δ elements during BIR. These data show that Ty elements are hot spots for template switching, especially if they are located in the same chromosome.
Discussion
In this report, we describe model systems to investigate the mechanisms of BIR and template switch-mediated chromosome rearrangements. Our results show that when the sequences are 100% homologous, template switching can occur as often as one in every 12 BIR events. The high efficiency of BIR-associated template switching was also found when we analyzed survivors of BIR events that were forced to replicate toward the centromere, producing a variety of outcomes, all involving jumps between Ty elements dispersed in the genome.
Homeology poses a formidable barrier to BIR, reducing the rate of successful recombination from 6.4 × 10−2 between homologous sequences to 7.1 × 10−5, a 900-fold reduction. However, subsequent template jumps that began with recombination between identical substrates were reduced only 200-fold, suggesting that template switches are more tolerant to homeology than canonical strand invasion. The idea that template switches occur by a mechanism distinctly different from the initial strand invasion is supported by several additional observations.
First, with the most divergent substrates, the initial invasions exhibited a very strong preference to the very 3′ end, whereas no preferred sequences were found for template jumps. This result implies that, at least when sequences are 32% divergent, template switching uses different rules from strand invasion. Although previously the Msh2–Msh3 complex has been implicated in the process of clipping off a nonhomologous tail in several homologous recombination processes (Saparbaev et al. 1996; Kirkpatrick and Petes 1997; Sugawara et al. 1997; Studamire et al. 1999), here we show that the Msh6 and presumably Msh2 proteins also play a key role in determining the location of the microhomology junction. It is possible that the absence of Msh6 allows for a longer region of heteroduplex DNA to form between the invading strand and the diverged template (Tsaponina and Haber 2014), thus allowing different sites to be used as the 3′ terminus after tail clipping at different sites.
The preferential usage of the sequence at the very 3′ end for a highly divergent sequence clearly distinguishes canonical strand invasion from template switching, where no such preference was observed. The difference in strand invasion patterns can be explained in two ways. First template switch may be the result of DNA polymerase dissociating from the original template, directly entering the distant homeologous template, and reinitiating DNA synthesis (Fig. 1B). In this case, the template switch would occur before the DNA polymerase proceeded beyond the end of the region of shared homology, so there would be no need to clip off a nonhomologous single strand, as there is for the initial event. This scenario is consistent with our analyses of the kinetics of BIR and template switching (Fig. 3), where template switching was coincident with the initial BIR DNA synthesis. However, it does seem that the great majority of template switches requires Rad51 rather than being completely mediated by DNA polymerases, although the requirements for both Pol32 and Rad51 are less strong than for initiating BIR. Alternatively, if new DNA synthesis proceeded beyond the 300 nt copied from the homeologous region, then the end of the dissociated strand would again have a nonhomologous tail that would require end clipping. This would most likely be a Rad51-dependent event and might be expected to yield a similar 3′ end bias in the location of the homeologous junction. However, this was not the case when the same 68% divergent sequences were used as the target of the template switch in the U × *UR × RA3 construct (Fig. 4C). Taken together, we favor the idea that the template switch occurs prior to copying the full extent of the 300-bp region but still requires Rad51 and other repair factors nearly all the time.
In this regard, we found that deletion of Rdh54—a Swi2/Snf2 chromatin remodeler that has previously been shown to have no little or no effect on intrachromosomal gene conversion or DSB-induced gene conversion (Klein 1997; Signon et al. 2001; Tsaponina and Haber 2014)—plays an important role in BIR template switching, as it does in interchromosomal template switches during gene conversion (Tsaponina and Haber 2014). A recent description by Stafa et al. (2014) of a 3′–5′ helicase Mph1 in promoting template switching during BIR fits with the idea of template switching having different genetic requirements than canonical strand invasions.
Template switching can be a robust process mediating chromosome rearrangements, especially between dispersed repetitive elements in the genome. In one of our constructs, where the BIR fork has to travel a chromosomal region consisting of highly repeated Ty retrotransposon elements, all of the Ura+ survivors that we examined underwent at least one template switch; 0.6% of the cells that survived were Ura− and repaired the HO-induced break by nonhomologous end-joining. In certain cases (Fig. 7; additional examples in Supplemental Fig. 9), we could clearly assign multiple template switches between Ty elements. We think it is highly unlikely that the events that we recovered result from the formation, breakage, and subsequent repair of a dicentric chromosome that would occur if BIR proceeded through the centromere to the other end of the chromosome. There are several other Ty elements and δ sequences more centromere proximal to the cluster just adjacent to the RA3 sequence, but only the most adjacent ones were implicated in the rearrangements that we characterized. If a dicentric chromosome had formed and breakage occurred at random along the long arm of Chr 5, then we would have expected a variety of Ty × Ty events using the more centromere-proximal repeated sequences, but this was not the case.
Ty elements are known sources generating chromosome rearrangements under a variety of conditions: spontaneous and following DNA breaks. Malkova et al. (2001) showed that Rad51-independent BIR following a HO-induced DSB is mediated by Ty elements positioned in a head-to-head arrangement on Chr 3. Argueso et al. (2008) and Hoang et al. (2010) showed that repair of DNA breaks occurring within and without Ty elements could variedly proceed by nonallelic homologous recombination mechanisms, including BIR, between dispersed Ty elements, resulting in duplications, translocations, and deletions. Furthermore, under reduced Polα levels, Ty elements positioned in a head-to-head rearrangement on Chr 3 were shown to act as a fragile site that induces spontaneous deletions and nonallelic Ty-mediated recombination, resulting in duplications (Lemoine et al. 2005). Characterizing the genome makeup of the various Saccharomyces species, Fischer et al. (2000) showed that translocation breakpoints that resulted in chromosome rearrangements could be mapped to Ty elements/tRNA clusters in the genome. Our results clearly implicate Ty-mediated template switching during BIR as a prominent mechanism leading to chromosome rearrangements. In our study, it should be noted that the chromosomal regions that contained the Ty elements also contained tRNA clusters. Because tRNAs are known replication fork-blocking regions (Deshpande and Newlon 1996), we are currently in the process of determining whether tRNAs can trigger template switches independently of the Ty elements. Two likely reasons that Ty elements are an excellent substrate for template switching is that they are present in many copies and share long regions of sequence identity.
For the first time, we learned that homeology can differently affect canonical strand invasion and template switching. The presence of homeology restrained strand invasions at least 2.5-fold higher than template switches. We are currently investigating the reasons for such a severe restraint. Sgs1 and Msh2 were previously shown to discourage SSA between divergent sequences. It would therefore be interesting to test the role of the MMR machinery components in discouraging homeologous BIR and template switching. Sgs1, a 3′–5′ helicase shown previously to discourage SSA between divergent sequences (Sugawara et al. 2004) and spontaneous translocation between highly divergent sequences (Schmidt et al. 2006), did not improve the efficiency of homeologous BIR (data not shown). Interestingly, the absence of Msh6 reduced the 3′ end preference and modestly increased the efficiency of homeologous BIR. The MMR components Msh2/Msh3 acting with Rad1/Rad10 were previously shown to be involved in efficient nonhomologous tail clipping (Sugawara et al. 1997). One possibility is that during homeologous BIR, Msh6 could function at the level of tail clipping to impose the 3′ end preference. Another possibility is that mismatch recognition by Msh6 (following the strand invasion) could recruit other repair factors that in turn impose the 3′ end preference. Imposition of the 3′ end preference by Msh6 (and possibly by other MMR components) during strand invasion could serve to prevent the loss of sequence information at the 3′ end of the DSB and thus help in maintenance of genome stability.
Previous analyses of the breakpoint junctions of chromosome rearrangements in mammalian cells have revealed the presence of a few base pairs—typically 2–5 bp—of shared microhomologies between the rearranged segments. Previous reports have followed the convention of assigning the most terminal matching base pairs/microhomologies as the breakpoint junction. Following the previous conventions, the use of microhomologies ranging from 1 to 11 bp is evident during both the invasion and template switch steps (Fig. 4). However, on closer examination, it is apparent that additional islands of microhomologies flank the assigned breakpoint junctions (Supplemental Fig. 10). Therefore, it is possible that strand invasion is influenced by these additional microhomologies rather than the short sequence that marks the actual junction. In support of our above proposal, we were indeed able to detect patches of mismatches that went uncorrected during recombination, making it difficult to assign a single patch of microhomology as the breakpoint junction (Supplemental Fig. 11; Tsaponina and Haber 2014). Microhomologies of 2–5 bp are plentiful in the yeast genome of ∼12 million bp, and yet Hicks et al. (2010), using an unbiased assay system, were unable to detect template switches to unrelated sequences carrying 2- to 5-bp microhomology, suggesting that homeologous recombination proceeds mainly in the context of larger flanking microhomology patches that are present only in single copies in the genome. Together, our results lead us to believe that islands of multiple microhomologies, rather than an isolated singular patch of microhomology, guide homeologous recombination.
Materials and methods
All of the yeast strains described in the study have an identical background (CL11-7) that has been previously described by Lydeard et al. (2010) and are described in the Supplemental Material. Sc-URA3 and Kl-URA3 fragments were introduced into the CL11-7 background by a standard yeast transformation protocol and confirmed by PCR and sequencing (see below for the list of strains used in this study). Various URA3 fragments were constructed using a combination of yeast in vivo cloning techniques and plasmid rescue using Qiaprep (Qiagen) or GIBSON assembly (New England Biolabs). Data were graphed and analyzed using Graphpad software. Individual Ura+ cells were amplified using Phusion (New England Biolabs) and sequenced (Genewiz, Inc.). Sequence usage in MM-BIR and template switching were carried out by either the alignment feature contained in the Serial Cloner 2.6.1 (Franck Perez, Serialbasics) software or the online multiple sequence alignment tool. (http://www.genome.jp/tmp/mafft), with the Sc-URA3 and Kl-URA3 as the reference sequence and the survivor URA3 sequence as the query. Junction usage was manually curated for each of the individual sequences.
For the BIR efficiency calculation, cells were plated for individual colonies on YEPD + NAT to retain the HOcs (which is marked with the NATMX antibiotic marker). In strains yRA192 and yRA213, HOcs is marked with HPHMX marker; therefore, these strains were kept on YEPD + HYG plates. For the SSA strains only, cells were grown on YC-TRP (to retain the HOcs that is marked with TRP1). Cells were appropriately diluted and plated on YEPD plates to get the total cell count and on YEP-GAL plates for HO induction. Cells that grew on YEP-Gal plates (survivors) were counted and replica-plated to YC-URA plates to screen for the reconstitution of functional URA3. For each replicate, Ura+ frequencies were calculated as the total Ura+ cells over the total cells on YEPD plate.
PFGE was carried out using Bio-Rad CHEF gel kit (Bio-Rad) based on the manufacturer’s recommended protocol except that zymolyase (Zymolyase 20T, Sunrise Science Products) treatment was done outside of the agarose plugs prior to embedding the spheroplasts in 1% agarose. Running conditions were 0.25× TBE; 12°C; 200 V; initial and final switching times of 15 and 35 sec, respectively; and a 24-h run time. Subsequent to the run, the gel was autocross-linked using a Stratalinker UV cross-linker, wick-transferred in 0.4 M NaOH, and probed using radiolabelled URA3 ORF.
The analysis of gene dosage with microarrays was performed according to the procedures described in Lemoine et al. (2005). In brief, DNA isolated from the control Ura− strain yRA108 was labeled with Cy3-dUTP and mixed with DNA isolated from the experimental Ura+ yRA108 derivatives that had been labeled with Cy5-dUTP. The mixture was hybridized to an Agilent microarray (8x15K, AMID 028943). The slides were scanned with the GenePix 4000B scanner and analyzed using GenePix Pro 4.1 (Axon Instruments) and Gene Spring 5.1 (Silicon Genetics).
The details of the primer extension reaction are given below. For primer extension, 50 ng of genomic DNA prepared with the MasterPure yeast DNA purification kit (Epicentre, catalog no. MPY80200) was used in quantitative PCR reaction (Qiagen, Rotor-GeneQ). Signals were the averages of duplicates with 10 μL in each reaction. Genomic DNA of the Ura+ survivor was used to generate the standard curves. Signals were normalized to input, which was the signal amplified from the PGI1/TAF1 loci on Chr II. Primer pairs used in this analysis were as follows: RA_307 q_pcr jump 1 for (GCACGAAAAGCAAACAAACTTGT); RA_308 q_pcr jump 1 rev (CTTTGTTACTTCTTCTGCCGC); RA_309 q_pcr jump 2 for (GCCCAGGTATTGTTAGCGGT); CSL268-Jump2 rev (CATATATTACGATGCTGTCTATTAAATGCTTCC); Chr2, 625kb-10kb p1 (ACAAGGTGGTTTGATGAGGAGGCT); and Chr2, 625kb-10kb p2 (TTCCGTCTGGCTAATTTGGGACGA).
Acknowledgments
We acknowledge the members of the Haber laboratory for critical comments, and Yi Yin in the Petes laboratory for help with the depiction of the microarray data. We are grateful to the makers of the freely available Serial Cloner 2.6.1 (Franck Perez, SerialBasics). We thank Kirill Lobachev for tips on PFGE analyses. Work in the Haber laboratory is supported by National Institutes of Health (NIH)/National Institute of General Medical Sciences (NIGMS) grants 1F32GM096690, R01GM076020, and 5R37GM020056, and work in the Petes laboratory is supported by NIH/NIHMS grants R01GM24410 and RO1GM52319.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.250258.114.
References
- Aguilera A, Gomez-Gonzalez B. 2008. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Anand RP, Lovett ST, Haber JE. 2013. Break-induced DNA replication. Cold Spring Harb Perspect Biol 5: a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso JL, Westmoreland J, Mieczkowski PA, Gawel M, Petes TD, Resnick MA. 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci 105: 11845–11850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JE, Kolodner RD. 2011. A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet 7: e1002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger SRet al. 2012. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet 44: 390–397, S391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, Paques F, Haber JE. 1999. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics 151: 1409–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. 2014. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. 1997. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci 94: 9757–9762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Symington LS. 2004. RAD51-dependent break-induced replication in yeast. Mol Cell Biol 24: 2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. 2013. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155: 948–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. 2011. Break-induced replication is highly inaccurate. PLoS Biol 9: e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS. 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Donnianni RA, Symington LS. 2013. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci 110: 13475–13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B, Jasin M. 2002. Double-strand breaks and translocations in cancer. Cell Mol Life Sci 59: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. 2000. Chromosomal evolution in Saccharomyces. Nature 405: 451–454 [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J, Haber JE. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258: 480–484 [DOI] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. 2012. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer 12: 663–670 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. 2009a. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. 2009b. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE. 2010. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329: 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang ML, Tan FJ, Lai DC, Celniker SE, Hoskins RA, Dunham MJ, Zheng Y, Koshland D. 2010. Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet 6: e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DT, Petes TD. 1997. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature 387: 929–931 [DOI] [PubMed] [Google Scholar]
- Klein HL. 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598 [DOI] [PubMed] [Google Scholar]
- Lieber MR, Yu K, Raghavan SC. 2006. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 5: 1234–1245 [DOI] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ, Haber JE. 1990. Mitotic recombination among subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics 124: 547–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. 2007. Genomic rearrangements and sporadic disease. Nat Genet 39: S43–S47 [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, Haber JE. 2010. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev 24: 1133–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci 93: 7131–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Signon L, Schaefer CB, Naylor ML, Theis JF, Newlon CS, Haber JE. 2001. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev 15: 1055–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. 2005. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol 25: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow DM, Connelly C, Hieter P. 1997. ‘Break copy’ duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. 2010. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol 17: 6765–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JF, Gomez-Gonzalez B, Aguilera A. 2009. Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol Cell Biol 29: 5441–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. 2013. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502: 389–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saparbaev M, Prakash L, Prakash S. 1996. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1–RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics 142: 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KH, Wu J, Kolodner RD. 2006. Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom’s syndrome protein. Mol Cell Biol 26: 5406–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR. 2005. Non-recurrent 17p11.2 deletions are generated by homologous and non-homologous mechanisms. Hum Genet 116: 1–7 [DOI] [PubMed] [Google Scholar]
- Signon L, Malkova A, Naylor ML, Klein H, Haber JE. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol 21: 2048–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. 2007. Template switching during break-induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- Stafa A, Donnianni RA, Timashev LA, Lam AF, Symington LS. 2014. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics 196: 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire B, Price G, Sugawara N, Haber JE, Alani E. 1999. Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination. Mol Cell Biol 19: 7558–7567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Paques F, Colaiacovo M, Haber JE. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci 94: 9214–9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci 101: 9315–9320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Liu P, Jia X, Withers MA, Jin L, Lupski JR, Zhang F. 2013. Replicative mechanisms of CNV formation preferentially occur as intrachromosomal events: evidence from Potocki-Lupski duplication syndrome. Hum Mol Genet 22: 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaponina O, Haber JE. 2014. Frequent interchromosomal template switches during gene conversion in S. cerevisiae. Mol Cell 55: 615–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A. 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol 27: 2601–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. 2013. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 502: 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]